Abstract
Cats are the natural host for Bartonella henselae, an opportunistic human pathogen and the agent of cat scratch disease. Here, we have analyzed the natural variation in gene content and genome structure of 38 Bartonella henselae strains isolated from cats and humans by comparative genome hybridizations to microarrays and probe hybridizations to pulsed-field gel electrophoresis (PFGE) blots. The variation in gene content was modest and confined to the prophage and the genomic islands, whereas the PFGE analyses indicated extensive rearrangements across the terminus of replication with breakpoints in areas of the genomic islands. We observed no difference in gene content or structure between feline and human strains. Rather, the results suggest multiple sources of human infection from feline B. henselae strains of diverse genotypes. Additionally, the microarray hybridizations revealed DNA amplification in some strains in the so-called chromosome II-like region. The amplified segments were centered at a position corresponding to a putative phage replication initiation site and increased in size with the duration of cultivation. We hypothesize that the variable gene pool in the B. henselae population plays an important role in the establishment of long-term persistent infection in the natural host by promoting antigenic variation and escape from the host immune response.
The genomic heterogeneity within a bacterial species reflects lifestyle strategies, niche occupation, and the exposure to mobile elements, such as bacteriophages and plasmids, in the natural growth environment. A recent study suggested that the total genome content (the “pan-genome”) of all contemporary Streptococcus agalactiae strains is approaching infinity (64). In contrast, bacterial species adapted to restricted growth habitats, such as the aphid endosymbiont Buchnera aphidicola (63), have “closed” pan-genomes with few dispensable genes. Isolates of Bacillus anthracis also show little variation at the genomic level, consistent with a limited total gene count for the natural population as a whole (72). These examples demonstrate a necessary precondition for identifying the causes of genome plasticity in human pathogens; it is essential to consider the sequenced pathogen genomes in the context of the population-level dynamics of closely related environmental strains.
Species of the genus Bartonella are particularly attractive for studies of pathogen genome diversity, niche adaptation, and virulence because of their unique ecological characteristics: the approximately 20 known species have colonized a wide range of animal hosts and insect vectors (8, 11, 16, 68). More than 10 species and subspecies have been isolated from human clinical samples (14), but humans serve as the natural reservoir for only two of these: Bartonella quintana, the louse-borne agent of trench fever, and Bartonella bacilliformis, the agent of Carrion's disease complex. Bartonella henselae, which is closely related to B. quintana, naturally infects cats but may incidentally infect humans and cause a variety of symptoms including cat scratch disease (CSD) and bacillary angiomatosis. For both species, disease symptoms depend on the immune status of the host, and a variety of vasoproliferative disorders of the skin and the internal organs may develop following infections in immunocompromised individuals (37, 56).
A very low level of genotypic heterogeneity was found for a collection of B. quintana strains, in line with a recent emergence of this bacterium as a human pathogen (25). The B. henselae strains are divided into two 16S rRNA gene (rrs) genotypes (type I/Houston-1 and type II/Marseille) which correspond to two distinct human serotypes (19, 42). Both genotypes are present worldwide, but the Marseille genotype appears to be dominant in the European cat population, whereas the Houston-1 genotype is more common in Asia (7). However, the relative prevalence of the two genotypes in felines also shows regional variations (23, 31, 33).
Several studies have suggested that the Houston-1 genotype is overrepresented among human samples, as observed, for example, in Australia (17), Germany (59, 60), and The Netherlands (6). However, a study of isolates from CSD patients in France showed no overrepresentation of the Houston-1 strain (69). A more detailed characterization of Australian isolates by multilocus sequence typing (MLST) revealed at least seven distinct genotypes in the feline population, only a few of which were recovered among the human samples, suggesting that some sequence types may be predisposed to cause human infections (35). Hence, it was hypothesized that a few specific genotypes contribute disproportionately to the disease burden in humans (17, 35).
It has also been debated whether the Houston-1 genotype is more pathogenic for humans than the Marseille variant (59, 69). A recent comparison of individuals infected with the Marseille and Houston-1 isolates revealed no difference in clinical characteristics (69). However, another study of human immunodeficiency virus (HIV)-infected patients suggested that hepatosplenic vascular proliferative lesions are more common in patients with infections of the genotype I strains (12). Small sample sizes taken from geographically isolated regions may explain some of the discrepancies in the results. Another limitation is that the use of typing methods, such as MLST, provides no information about the variability of potential virulence traits in the B. henselae population.
The availability of genome sequence data for the human pathogen B. quintana and the human Houston-1 strain of B. henselae (2) now enables the genomic diversity for the species as a whole to be explored. As in other human pathogens, such as Rickettsia prowazekii (5, 53) and Bordetella pertussis (54), the emergence of B. quintana as a human pathogen has been associated with accelerated rates of sequence loss rather than with unique gene acquisitions (2). Thus, only remnants of a B. henselae prophage of 55 kb and genomic islands (GEIs) of 72, 34, and 9 kb could be identified in the B. quintana genome (2). The extent of sequence loss in the so-called chromosome II-like segment, which may have been integrated from an extra replicon and has a coding content of only 40 to 50%, was also more pronounced in B. quintana. A recent study of the gene content in Bartonella koehlerae, a feline-associated species with a phylogenetic placement near B. henselae, revealed a partial retention of GEIs, indicative of independent deletion events in divergent lineages (43).
The aim of this study was to examine the genomic variability of B. henselae strains and place it in the context of the population structure of the species. We describe gene content and genome structure variation for 38 B. henselae strains, including both feline and human isolates. We hypothesize that the observed genomic variation, which includes deletions and rearrangements in areas that encompass the GEIs, serves an important role in host colonization and immune evasion.
MATERIALS AND METHODS
Culture of strains and DNA isolation.
The B. henselae strains used in this analysis (Table 1) were kindly provided by Scott A. Handley (National Center for Infectious Diseases, Atlanta, Ga.), except the Houston-1 and Houston-2 strains, which were provided by Martin Holmberg (Uppsala University Hospital, Uppsala, Sweden), and the Marseille URLLY8 strain, which was supplied by Volkhard Kempf (Tuebingen, Germany). Included in the analysis were four different subcultures of the Houston-1 strain (Table 1). Bacteria were grown for 7 to 14 days on blood agar plates containing 5% horse blood, and DNA was extracted as reported previously (43).
TABLE 1.
Host and country of isolation of the 38 B. henselae strains analyzed
| Straina | Host | Origin | References |
|---|---|---|---|
| CA-1 | Human (HIV) | United States (California) | 15, 20, 58 |
| CA-8/DB.R | Human (CSD) | United States (California) | |
| Cheetah-1 | Cheetah | Africa | |
| FR96/BK3 | Cat | Europe (Germany) | 20, 60 |
| GA-1 | Human (CSD) | United States (Georgia) | 15, 58 |
| Goldie-1 | Cat of CSD patient | United States (Georgia) | 15, 58 |
| GreekCat-1 | Cat | Europe (Greece) | |
| GreekCat-9 | Cat | Europe (Greece) | |
| GreekCat-5 | Cat | Europe (Greece) | |
| GreekCat-23 | Cat | Europe (Greece) | |
| GreekCat-34 | Cat | Europe (Greece) | |
| Houston-1 | Human (HIV) | United States | 15, 17, 20, 35, 42, 58, 60, 69 |
| Subtype Houston-1980515 | Human (HIV) | United States, subcultivated in Sweden | |
| Subtype Houston-1Seq | Human (HIV) | United States, subcultivated in Sweden | |
| Subtype Houston-1Ref | Human (HIV) | United States, subcultivated in Sweden | |
| Subtype Houston-1ATCC | Human (HIV) | United States, subcultivated in Sweden | |
| Houston-2 | Human (HIV) | United States | 15, 20 |
| IndoCat-2 | Cat | Asia (Indonesia) | |
| IndoCat-5 | Cat | Asia (Indonesia) | |
| IndoCat-11 | Cat | Asia (Indonesia) | |
| Marseille, URLLY8 | Human (CSD) | Europe (France) | 15, 17, 20, 35, 42, 69 |
| MO-2 | Human (HIV) | United States (Missouri) | |
| SA-1 | Human (CSD) | United States (Texas) | |
| SA-3 | Human | United States (Texas) | |
| SD-2 | Human (HIV) | United States (San Diego, Calif.) | |
| Tiger-2 | Cat of CSD patient | United States (California) | 15, 20, 58 |
| Tx-4 | Human (CSD) | United States (Texas) | |
| UGA-3 | Cat | United States | |
| UGA-6 | Cat | United States | |
| UGA-7 | Cat | United States | |
| UGA-8 | Cat | United States (Ohio) | |
| UGA-9 | Cat | United States | |
| UGA-10 | Cat | United States (Virginia) | |
| UGA-11 | Cat | United States | |
| UGA-12 | Cat | United States | |
| UGA-13 | Cat | United States | |
| UGA-14 | Cat | United States (Montana) | |
| UGA-23 | Cat | United States | |
| UGA-24 | Cat | United States | |
| UGA-26 | Cat | United States | |
| UGA-28 | Cat | United States (Louisiana) | |
| ZimCat-25 | Cat | Africa (Zimbabwe) |
The Greek cat isolates (with the prefix GC) came from the Tessaloniki area in Greece and were collected by Aphrodite Tea. The UGA isolates came from a collection of cat blood samples from veterinary clinics in the United States; the samples were gathered by Craig Greene, University of Georgia, Athens, and were analyzed serologically by Perry Jamison.
MLST.
We have collected sequence data from these strains for nine selected loci previously used for genotyping (35). Internal fragments of 321 to 522 bp were amplified from each of the nine genes (rrs, batR, eno, ftsZ, gltA, groEL, nlpD, ribC, and rpoB) using primers for MLST of B. henselae as previously described (35). In addition, two spacer regions flanked by BH03830-BH3840 and BH15070-BH15080 were sequenced in the strains classified as sequence type 1 in the MLST analysis (Table 2) with primers sp_BH03830.f (GGATCGTCCCATAGACCATTG), sp_BH03830.r (CATTCTGTCCGAGTGTAAACG), sp_BH15070.f (TTTCCTGTGACATCAACATCATC), and sp_BH15070.r (TGTGTGATACGGCTGTGGC).
TABLE 2.
Multilocus sequence typing of the 38 B. henselae strains analyzed
| Isolate | Allelic varianta
|
STb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rrs | batR | eno | ftsZ | gltA | groEL | nlpD | ribC | RpoB | ||
| CA-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| CA-8 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| Cheetah | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 4 |
| FR96/BK3 | 2 | 4 | 1 | 3 | 2 | 1 | 2 | 2 | 1 | 7 |
| GA-1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| Goldie-1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| GreekCat-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| GreekCat-9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| GreekCat-23 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| GreekCat-25 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| GreekCat-34 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Houston-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Houston-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| IndoCat-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| IndoCat-5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| IndoCat-11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Marseille | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 3 | 8 |
| MO-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| SA-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| SA-3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| SD-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Tiger-2 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| Tx-4 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 |
| UGA-3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-10 | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 |
| UGA-11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| UGA-12 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 |
| UGA-13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-24 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| UGA-26 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| UGA-28 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 |
| ZimCat | 2 | 3 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 6 |
The numbers refer to the allelic variants by reference to the sequence of the corresponding gene in the Houston-1Seq genome (here called number 1). The allelic polymorphisms for each gene are detailed in Table 3, below.
ST inferred from the combination of allelic polymorphisms observed for each gene as described in reference 35.
The PCR mix contained 1× PCR buffer (Sigma), 1.5 μg MgCl2, 0.2 mM deoxynucleoside triphosphate, 0.33 μM of each primer, 4 ng of DNA template, and 2.5 U of Taq DNA polymerase (Sigma). The reaction conditions were 95°C for 10 min; 33 cycles of 95°C for 30 s, 50 to 55°C for 90 s, 72°C for 3 min; and a final extension step at 72°C for 10 min. Amplification products were purified by filtering through a MultiScreen PCR plate (Millipore Corporation). The sequencing reaction was performed with 3 μl of PCR products, 1 μl of one of the primers used for the PCR (above), and 4 μl of the DYEnamic ET-terminator sequencing premix containing labeled dideoxynucleotides and the enzyme (Amersham Bioscience, Germany). The reaction conditions were 30 cycles of 95°C for 30 s, 50°C for 15 s, and 60°C for 2 min. The products were purified by centrifugation (5 min, 2,400 rpm) through cleaned Sephadex G-50 Superfine columns placed in Multiscreen filter plates (Millipore Corporation). The Sephadex columns were cleaned by adding 150 μl of sterile water and centrifugation for 5 min at 2,400 rpm. The purified products were sequenced with the MegaBace1000 capillary sequenator.
Analysis of MLST data.
The sequences were assembled and edited with Phred, Phrap, and Consed (21, 22, 28) and compared to the known sequence types using BLAST (3). Alleles were named as described in reference 35, and polymorphic sites were numbered according to their position in the corresponding gene of the sequenced Houston-1 genome, since some of the nucleotide numberings in reference 35 did not agree with the GenBank entries to which they were supposed to refer. For each isolate, the combination of alleles at each of the loci examined was used to define the sequence type (ST), according to the methods in reference 35. For gltA, only one of the polymorphic sites (C1026T) reported in reference 35 for allele 2 was observed, the other (G648A) being located outside of the amplified region; the observed allele was nevertheless considered as allele 2. In addition, for nlpD, the polymorphism reported previously (35) (G1453A) was apparently mistyped, since 36 of the 37 strains had an A at the corresponding position (allele 1) while one isolate had a G (allele 2). For visualization of the MLST data, neighbor joining of the aligned concatenated allele sequences was performed using the PHYLIP programs dnadist, neighbor, seqboot, and consense (24) with 1,000 bootstrap replicates.
Microarray comparative genome hybridization (CGH).
Array probes (PCR products) were generated as reported previously (43) and spotted in six replicates (by Niclas Olsson at Uppsala University, Uppsala, Sweden) or three replicates (by Annelie Waldén at the Royal Institute of Technology, Stockholm, Sweden). Slides were UV cross-linked at 30 mJ/cm2 (slides scanned with ScanArray) or 250 mJ/cm2 (slides scanned with GenePix). DNA labeling was performed as previously reported (43), except that for the 5- and 10-day comparisons only 1 μg genomic DNA was used. Hybridization of labeled test strain and reference DNA was performed at 50°C overnight in hybridization solution containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (ScanArray slides) or 5× SSC (GenePix slides), 0.1% sodium dodecyl sulfate, and 0.1 mg/ml sonicated salmon sperm DNA. Two or three hybridizations were performed for each strain.
Genomic DNA from the Houston-1Ref subculture was used as the reference in the microarray hybridizations against genomic DNA from the 38 B. henselae strains as well as in hybridizations against DNA isolated from different subcultures of the Houston-1 strain. These hybridizations consistently gave low M values for a 10-kb region located at kb 1060 and 1070 of the sequenced genome. PCR and sequencing revealed a 10-kb tandem duplication flanked by a short repeated sequence in the Houston-1Ref strain. We also discovered a 10-kb deletion in the Houston-1Ref strain in a segment upstream and within the badA gene, which codes for an adhesin required for the induction of vasculoproliferative disorders (57, 70). The microarray data further suggested that Houston-1980517 was identical in gene content to Houston-1Seq, whereas the Houston-1ATCC isolate seems to be an intermediate that contains the deletion upstream of badA but not the tandem duplication of the kb 1060 to 1070 region. Additionally, Houston-1Ref differed from Houston-1Seq by a recombination event across the tuf genes, as verified by PCR and sequence analysis. Finally, the pulsed-field gel electrophoresis (PFGE) analysis suggested translocations across HGIa and HGIb in the different subcultures of the Houston-1 strain.
For the comparative microarray hybridizations of genomic DNA isolated after 5 and 10 days of culture, genomic DNA from the Houston-1Ref strain was used as the reference in one set of experiments, whereas DNA from the Houston-1ATCC subculture isolated after 5 days of growth was used in a second hybridization. The median M values in the second experiment were corrected by adding the M values from a separate hybridization of genomic DNA from the Houston-1ATCC strain against Houston-1Ref subculture before computing the mean of the replicate hybridizations.
Analysis of CGH data.
Scanning and image analysis were performed with a ScanArray 4000 scanner and the ScanArray Express software (Perkin-Elmer, Inc.) as reported previously or with a GenePix 4100A scanner and the GenePix Pro 5.1 software (Axon Instruments, Molecular Devices) at a resolution of 10 μm. The channel used for the reference strain is referred to as Ch1, and the test strain is referred to as Ch2. Photomultiplier tube voltage was manually adjusted to balance intensities in the two channels while avoiding a high number of saturated spots. Spots meeting any of the following criteria were considered bad and removed from analysis: spots flagged as bad or not found during quantification; Ch1 spot median below 5 (ScanArray slides) or 40 (GenePix slides) times the Ch1 background median; more than 10% saturated pixels in either channel; less than 95% of spot pixels having intensity higher than background intensity plus 1 standard deviation; less than 90% of spot pixels having an intensity higher than background intensity plus 2 standard deviations; background-corrected Ch1 spot median intensity below 0.2 times the overall slide median Ch1 intensity; or the PCR used to generate the probe had failed (no visible band on gel). Median background intensities were subtracted from the median spot intensities, and M values (log2 ratios) were computed as log2(Ch2/Ch1). Normalization was performed as described in reference 43, with the exception that lowess normalization was performed when plots of M versus log2Ch1 or M versus A [where A = 0.5 · (log2Ch1 + log2Ch2)] indicated density-dependent dye bias. Median M values from all replicate spots and arrays were computed for each strain and used to classify each probe as absent (M ≤ −2), uncertain (−2 < M ≤ −1), or present (M > −1). Status of genes and intergenic regions was similarly decided by median M values of all corresponding probes.
Probes were ordered by their position in the Houston-1 genome and sorted into regions of contiguous probes with the same pattern of absence/presence in the strains. Probes with uncertain status were considered compatible with both absence and presence, depending on the status of the surrounding probes, and if a region contained both uncertain probes and probes with known status for a certain strain, the status of the region was set to that of the known probes. For phylogenetic analysis, only regions comprising at least two probes, absent in at least one strain and for which results were retrieved for all strains, were used. Maximum parsimony analyses of the CGH data were performed using the PHYLIP programs pars, seqboot, and consense (24) with 1,000 bootstrap replicates and 100 random orderings of strains on a matrix containing absence (0), presence (1), or uncertain (?) status scores of the selected regions.
PFGE hybridization.
PFGE-restriction fragment length polymorphism analysis was performed using NotI, AscI, and SgfI restriction endonucleases separately, as well as in a combination of NotI and AscI in a double digest. Experimental procedures for cultivation, digestion, and gel electrophoresis were as described previously (43). PFGE gels were depurinated in 0.2 N HCl for 10 min, and then blotting was performed using a Hybond N+ membrane (GE Healthcare) following the alkaline transfer protocol according to instructions of the manufacturer.
DNA fragments serving as probes were amplified using the primers listed in our Table 1 of our supplementary information at the website http://www.egs.uu.se/molev/suppl.data/J.Bacteriol. PCR conditions were as follows: enzyme activation step of 95°C for 10 min with subsequent 34 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 90 s, and extension at 72°C for 120 s, followed by the final extension at 72°C for 10 min and a holding step at 8°C. Obtained PCR products were first purified using a QiaQuick PCR purification kit (QiaGen) and then labeled and probed onto membranes using the Gen Images AlkPhos direct labeling and detection system with chemiluminescence detection with CDP-Star (GE Healthcare). Hybridizations were performed overnight at 60 to 70°C followed by washes at 60 to 72°C with the manufacturer's recommended buffer, which contains 50 mM NaH2PO4. The signal was detected by exposure using Hyperfilm ECL.
Analysis of PFGE data.
The sizes of the PFGE bands were estimated manually by comparison to a low-range λ ladder and yeast chromosome PFGE markers (New England Biolabs). The observed PFGE blot pattern was manually edited for double bands, incomplete restriction, and cross-hybridization to multiple bands in individual digests. If probes in the tested strain hybridized to the same band as neighboring probes in the sequenced genome, the fragment was considered to be syntenic with that of the sequenced genome; if the hybridization pattern of neighboring probes differed, a translocation, inversion, or gain or loss of restriction site was inferred. When a single probe or a consecutive stretch of probes differed in hybridization pattern, a translocation was inferred, although mechanistically the same pattern would also be explained by two inversions. When two probes had switched location, two inversions were inferred.
To infer the putative genome structures and identify rearrangement events, an interactive program (XPulSee) was written in Perl/Tk. The program displays the structure (including positions of probes and restriction sites) of the sequenced genome, as well as the hypothetical structure of the tested strain. The observed PFGE blot patterns obtained with the different enzymes are shown alongside the predicted pattern for the sequenced Houston-1 strain and the hypothetical structure of the tested strain. By using the program to interactively generate different rearrangements (inversions, translocations, and gains or losses of restriction sites) of the hypothetical test strain structure and compare the expected PFGE profile to the observed one, we identified putative rearrangements that would explain the observed PFGE pattern. Although the data were not always sufficient to distinguish between slightly different hypotheses or determine the exact number or types of events, the approach helped identify regions of rearrangements.
Nucleotide sequence and microarray accession numbers.
The sequence of the novel rpoB allele has been submitted to GenBank under the accession number AM295003, and the spacer sequences have been submitted under accession numbers AM295307 to AM295311. The microarray data have been deposited in the ArrayExpress database of the European Bioinformatics Institute under the accession number E-TABM-88.
RESULTS
We have analyzed 38 B. henselae strains isolated from 11 humans and 27 felines of different geographic origins (Table 1). Five of the human isolates were obtained from HIV-infected individuals (CA-1, Houston-1, Houston-2, MO-2, and SD-2), and four were isolated from patients with cat scratch disease (GA-1, Marseille, SA-1, and Tx-4). The remaining 71% of strains include 26 isolates from cats and one strain from a cheetah. For this set of isolates, we identified single-nucleotide variants using MLST, characterized indels by microarray, and estimated genome rearrangements with the aid of probe hybridization to PFGE blots.
Population structure inferred by MLST analysis.
An analysis of the distribution of polymorphisms in nine selected core genes (batR, eno, ftsZ, gltA, groEL, nlpD, ribC, rpoB, and rrs) showed that the selected strains represent six (Tables 2 and 3) of the seven previously identified sequence types (35). Like the sequenced Houston-1 strain, 23 of the 38 isolates belonged to ST1, including 7 of the 11 human isolates. The rpoB allele of the Marseille URLLY8 strain displayed an extra single-nucleotide polymorphism (here called C1733T) not previously recorded and was classified into a novel type, ST8.
TABLE 3.
Characteristics of the nine loci in the 38 B. henselae strains analyzed
| Locus | Variable sites (%) | No. of alleles | Allelic polymorphismsa | Corresponding Houston-1 locus |
|---|---|---|---|---|
| rrs | 3 (0.59) | 2 | 2, TAG 173-175 ATTT | BH13790 |
| batR | 4 (0.75) | 4 | 2, T390C | BH00620 |
| 3, C153T, C156A, A306G, T390C | ||||
| 4, C153T, A306G, T390C | ||||
| eno | 0 | 1 | BH05720 | |
| ftsZ | 4 (0.77) | 3 | 2, A1074T, G1293A, G1356T | BH11180 |
| 3, A1074T, G1356T, C1426T | ||||
| gltA | 1 (0.26) | 2 | 2, C1026Tb | BH06380 |
| groEL | 1 (0.25) | 2 | 2, G1344A | BH13530 |
| nlpD | 1 (0.19) | 2 | 2, A606Gc | BH05630 |
| ribC | 4 (1.25) | 2 | 2, A235G, G285A, A419G, G424A | BH07570 |
| rpoB | 2 (0.40) | 3 | 2, G1779A | BH06100 |
| 3, C1733T, G1779A |
To further examine the internal relationships of the ST1 isolates, we compared the sequences of two spacer regions located in between BH03830-BH38840 and BH15070-BH15080. Differences were observed in the spacer sequence downstream of BH15070, where the Houston-1 isolate as well as the feline strains UGA-3, UGA-7, UGA-9, UGA-13, and UGA-26 contain a stretch of eight Ts (subtype 1a), compared to nine Ts in the other ST1 isolates (subtype 1b). Additionally, the strains IndoCat-2 and IndoCat-11 differ by two substitutions of A to G in the spacer downstream of BH03830 (subtype 1c). Interestingly, UGA-23 seems to be a recombinant strain (subtype 1d); it shares the loss of a T with isolates of subtype 1a as well as the two A-to-G substitutions with strains of subtype 1b, and it has three additional, unique mutations in the spacer region downstream of BH15070.
Microarray comparative genome hybridization.
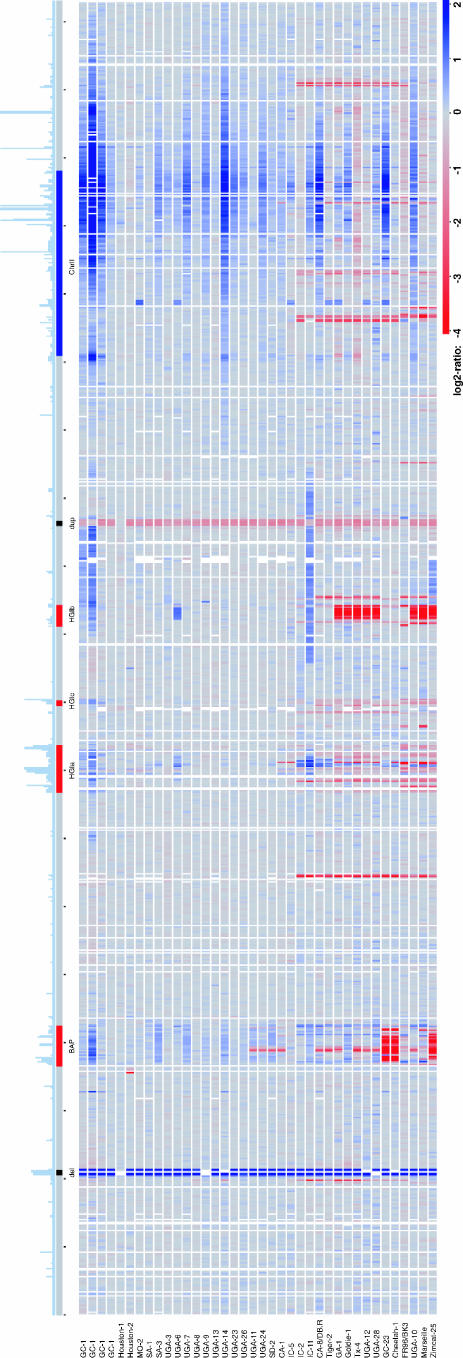
The variability in gene content within the B. henselae population was estimated by hybridizing genomic DNA from each isolate to a B. henselae microarray containing probes for 1,367 genes and 112 noncoding regions of the sequenced Houston-1 strain (Fig. 1). Neighboring probes with the same pattern of absence/presence across the strains were sorted into regions, each of which represents a putative insertion/deletion event. From the 1,269 genes (1,645 probes and 1,443 regions including pseudogenes and spacers) for which results were retrieved, a total of 105 different B. henselae genes were reported absent (145 probes and 111 regions including pseudogenes and spacers). The number of missing probes per strain ranged from 0 to 86 (Table 4). Up to 34 indel events per strain relative to the Houston-1 strain were estimated, of which 22 included at least two probes and gave results in all strains (Table 4).
FIG. 1.
Linear representation of the microarray CGH data ordered according to position in the Houston-1Seq genome. The frequency of repeats in the Houston-1Seq genome is shown to scale above the genome at the top (the number at which each sequence occurs in the genome with more than 80% sequence identity over 100 bp). The positions of the prophage region (BAP) and the genomic islands (HGIa, HGIb, and HGIc) in the Houston-1Seq genome are shown in red, and the chromosome II-like region (ChrII) is shown in blue. The regions deleted (del) and duplicated (dup) in the Houston-1Ref strain, used as a reference in the hybridization experiments, are indicated in black. Each horizontal line below the Houston-1Seq genome represents an isolate, with the color corresponding to the hybridization signal for the isolate relative to the Houston-1Ref strain (red for lower and blue for higher signal).
TABLE 4.
Summary of strain differences in gene content assessed by microarray comparative genome hybridization and genome structure inferred by PFGE hybridization
| Strain | No. of probes absenta | Total no. of regions absentb | No. of regions absent (≥2 absent probes)c | No. of rearrangementsd |
|---|---|---|---|---|
| CA-1 | 4 | 4 | 2 | 1 |
| CA-8 | 16 | 14 | 5 | 2 |
| Cheetah | 75 | 34 | 20 | 1 |
| FR96/BK3 | 36 | 22 | 14 | 2 |
| GA-1 | 40 | 21 | 9 | 2 |
| Goldie-1 | 31 | 13 | 7 | 2 |
| GreekCat-1 | 0 | 0 | 0 | 1 |
| GreekCat-9 | 0 | 0 | 0 | 1 |
| GreekCat-23 | 67 | 29 | 17 | 1 |
| GreekCat-25 | 0 | 0 | 0 | 2 |
| GreekCat-34 | 0 | 0 | 0 | 2 |
| Houston-1980517 | 0 | 0 | 0 | 1 |
| Houston-1Ref | 0 | 0 | 0 | 2 |
| Houston-2 | 1 | 1 | 1 | 1 |
| IndoCat-2 | 19 | 13 | 7 | 1 |
| IndoCat-5 | 2 | 2 | 2 | 1 |
| IndoCat-11 | 14 | 10 | 6 | 2 |
| Marseille | 60 | 28 | 16 | 3 |
| MO-2 | 0 | 0 | 0 | 1 |
| SA-1 | 0 | 0 | 0 | 1 |
| SA-3 | 0 | 0 | 0 | 2 |
| SD-2 | 2 | 2 | 1 | 3 |
| Tiger-2 | 24 | 19 | 9 | 2 |
| Tx-4 | 50 | 25 | 11 | 2 |
| UGA-3 | 0 | 0 | 0 | NDe |
| UGA-6 | 0 | 0 | 0 | 2 |
| UGA-7 | 0 | 0 | 0 | 1 |
| UGA-8 | 1 | 1 | 0 | 1 |
| UGA-9 | 0 | 0 | 0 | NDe |
| UGA-10 | 36 | 16 | 12 | 2 |
| UGA-11 | 3 | 3 | 1 | 2 |
| UGA-12 | 36 | 17 | 8 | 3 |
| UGA-13 | 0 | 0 | 0 | 2 |
| UGA-14 | 1 | 1 | 0 | 2 |
| UGA-23 | 0 | 0 | 0 | 1 |
| UGA-24 | 2 | 2 | 1 | 2 |
| UGA-26 | 0 | 0 | 0 | NDe |
| UGA-28 | 35 | 17 | 8 | MIf |
| ZimCat | 86 | 31 | 22 | 3 |
Total number of probes inferred to be absent compared to the Houston-1Ref strain.
Total number of regions inferred to be absent compared to the Houston-1Ref strain.
Number of regions inferred to be absent compared to the Houston-1Ref strain, counting only regions comprising at least two absent probes in at least one strain and for which results were retrieved for all strains.
Total number of rearrangements inferred by comparison to the Houston-1Seq genome.
ND, not determined. The PFGE banding pattern of UGA-3 is similar to that of UGA-6, and the patterns of UGA-9 and UGA-26 resemble those of UGA-7 and UGA-23.
MI, multiple inversions. The number of rearrangements could not be estimated due to complex hybridization patterns.
Of the 105 variably present genes, the majority (81%) are located in the prophage region (51 genes) or the genomic islands HGIa (9 genes) and HGIb (25 genes) (Fig. 1). Most of these putatively absent genes are of unknown function or encode phage functions. Additionally, variability was recorded in regions containing genes for adhesins, cytotoxins, and type IV secretion systems. Universally present in all strains are 1,164 genes, which suggests that the core B. henselae genome contains less than 92% of the genes present in the sequenced Houston-1 strain.
Clustering by MLST and CGH data.
To place the variability in gene content within the population structure of the species, we first compared the number of genes and regions inferred to be absent from strains of different sequence types. No or only minor differences were observed among the various ST1 and ST2 strains; only 1.9 and 2.5 probes, on average, are missing in these two clades relative to the Houston-1 isolate (Table 4). Interesting exceptions are the ST1c isolates IndoCat-2 and IndoCat-11, which have lost 19 and 14 probes, respectively, in up to 13 deletion events. Members of the other clades are even more divergent, with an inferred loss of 16 to 86 probes in circa 13 to 34 deletions per strain relative to the Houston-1 isolate, or 5 to 22 indels if only counting regions covered by at least two probes and for which data are available for all strains.
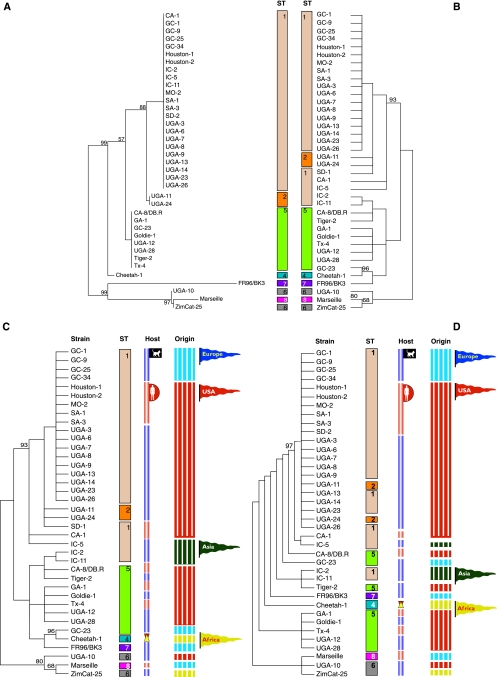
Next, we performed a clustering analysis based on the MLST data using the neighbor-joining method (Fig. 2A), which was broadly similar to a clustering based on the collected CGH data using the parsimony method (Fig. 2B), and counting as the unit of mutation segments that cover two or more neighboring probes with the same status. Both analyses distinguished the ST1 and ST2 isolates from the ST4 to ST7 isolates with bootstrap support values above 88% (Fig. 2A and B), with the exception of the ST1 isolates IndoCat-2 and IndoCat-11, which clustered with the ST5 group in the gene content analysis (Fig. 2B). Another highly supported group in the gene content analysis (96% bootstrap) not seen in the MLST clustering was that of two feline isolates of ST4 (GreekCat-23) and ST5 (Cheetah) (Fig. 2B). Both of these two isolates have lost the prophage region (Fig. 1).
FIG. 2.
Clustering structure according to MLST and gene content data collected from 38 strains of B. henselae. Results shown are from neighbor-joining clustering based on MLST data (A) and maximum parsimony clustering of the strains based on microarray CGH data (B to D), with phage sequences included (B and C) and excluded (D) from the analysis. Numbers show bootstrap support values. Vertical rows labeled with numbers are color coded to illustrate ST, host, and geographic origin of isolation of the strains. Colors used under the STs are as follows: beige, ST1; orange, ST2; blue, ST4; green, ST5; gray, ST6; purple, ST7; red, ST8. Colors used under hosts are as follows: blue, cat; red, human; yellow-red, cheetah. Colors used under the origin are as follows: blue, Europe; red, United States; green, Asia; yellow; Africa. Abbreviations of strain designations: GC, GreekCat; IC, IndoCat.
To test the influence of phage sequences on the clustering of the CGH data, the analysis was repeated with the prophage regions excluded (Fig. 2D). As before, the ST1 and ST2 isolates clustered with strong support (97%) also in this case, with the notable exceptions of IndoCat-2 and IndoCat-11. However, the clustering of GC-23 and Cheetah-1 was no longer supported, suggesting that the grouping of these two strains was partially influenced by the shared loss of several regions in the prophage. Part of the prophage is also absent from the ST6 strain ZimCat-25, but not from the ST6 strain UGA-10 nor from the ST8 strain Marseille URLLY8. These three strains are otherwise similar and grouped together in both analyses. Irrespective of whether the phage genes were included or excluded, the clustering analyses revealed no correlation with host or geographic origin of isolation, as visually depicted in Fig. 2C and D.
DNA amplification.
A few regions of the chromosome displayed an increased relative hybridization signal in several strains (Fig. 1). One of the amplified regions matched perfectly the prophage (BAP), and another matched the phage genes of HGIa. The amplifications at these two sites were located within or confined to the borders of the phage elements. We also observed a stronger hybridization signal of a segment in the vicinity of kb 1090 in GreekCat-9 and IndoCat-11 and to a lesser extent in GreekCat-1 and ZimCat-25. However, the most dramatic amplification was observed within the chromosome II-like region, which gave increased hybridization signals in half of the strains (Fig. 1). The amplified regions varied in size from 10 to 300 kb but were always centered at a position corresponding to kb 1660 in the sequenced Houston-1 genome. The amplitude showed a maximal M value of approximately 3.5 at the peak, which corresponds to a 10-fold-higher copy number relative to the reference strain, and gradually decreased in both directions.
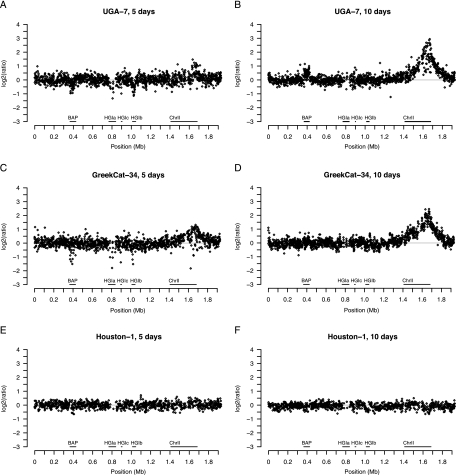
A comparison of the sizes of the amplified fragment following DNA isolation after growth on plates for 5 and 10 days showed that the extent of amplification was dependent on the time at which the cells were harvested (Fig. 3). In this comparison, we included strains that in the first study showed weak, intermediate, and very strong amplification (GreekCat-9, GreekCat-34, IndoCat-11, and UGA-7). In all cases, we observed a minor or weak amplification after 5 days of cultivation, which increased markedly in amplitude and size after 10 days. In effect, all strains showed a similar level of amplification after prolonged growth, as illustrated here with UGA-7 and GreekCat-34 (Fig. 3B and D). In comparison, only minor variations were observed for the different Houston-1 subcultures (Fig. 3E and F and data not shown). No correlation was observed between the extent of amplification and genotype; the amplifications were observed in strains of all sequence types. Nor was the amplification related to the host or geographic origin of isolation.
FIG. 3.
DNA amplification in B. henselae strains after 5 and 10 days of cultivation. The x axes show the sequence order of the Houston-1Seq strain. The y axes indicate the ratio of the hybridization signal in the tested strain relative to the Houston-1 strain used as a reference in the hybridization experiments, estimated as detailed in Materials and Methods. The extent of DNA amplification in strains UGA-7 (A and B), GreekCat-34 (C and D), and Houston-1Ref (E and F) after 5 days (A, B, and C) and 10 days (C, D, and E) of growth is shown.
Genome rearrangements.
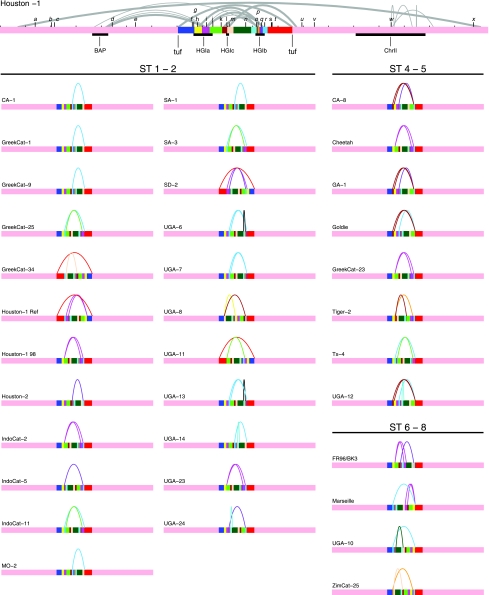
Our PFGE analyses showed that the B. henselae genomes are relatively homogeneous in size, in the range of 1.9 to 2.0 Mb (data not shown), although the genome size estimates obtained individually from the three different restriction enzymes used were occasionally inconsistent due to the presence of double bands and incomplete restriction. We also used PFGE data to infer the overall architecture of the investigated genomes, as was done previously (48, 49). To this end, we selected 11 probes from various locations and hybridized these to blots of restriction-digested DNA separated by PFGE (Fig. 4; see also Fig. 1 in our supplementary information at the website http://www.egs.uu.se/molev/suppl.data/J.Bacteriol). The hybridization pattern of each strain was compared to that of the sequenced strain using the program XPulSee (see Fig. 1 in our supplementary information). Although several equally possible rearrangement scenarios were often compatible with the hybridization data, a clear trend was that the rearrangements occurred near the terminus of replication (Fig. 4). The analysis was further refined by hybridizations with an extended set of 11 probes targeted against sequences near the ter region, and two probes were targeted to investigate the presence of a putative novel NotI site. In total, we inferred 106 rearrangement events, encompassing at least 29 different events, 8 of which comprised the gain or loss of restriction sites (see Table 2 in our supplementary information at the website http://www.egs.uu.se/molev/suppl.data/J.Bacteriol). These are minimal estimates, since smaller rearrangements in other areas of the chromosome may have gone undetected.
FIG. 4.
Schematic illustration of rearrangements in 35 B. henselae strains inferred by probe hybridizations to PFGE blots. Linear representation of the Houston-1Seq genome is at the top, with the positions of the hybridized probes shown in black letters (a to x). The locations of the prophage (BAP), the GEIs (HGIa, HGIb, and HGIc), and the chromosome II-like region (ChrII) are indicated as black boxes. Inverted repeats in the B. henselae genome are indicated as half-circles, with the length of the repeat proportional to the thickness of the line. The inverted repeats were identified by REPuter (41); only repeats over 200 bp and with a maximal edit distance of 20 are shown. When there are several repeats flanked by the same probes or restriction sites, only the longest repeat is shown. The Houston-1Seq genome is divided into colored segments by the breakpoints of the inferred rearrangements with end points at the duplicated tuf genes. Below, the inferred order of the segments is shown for each isolate (grouped by sequence type), with curved lines indicating the inferred inversions and translocations. Detailed information of restriction sites and inference of rearrangement scenarios are shown in Fig. 1 in our supplementary information at the website http://www.egs.uu.se/molev/suppl.data/J.Bacteriol.
The hybridization patterns suggested that the breakpoint regions were in the vicinity of the GEIs in 88% of cases and the remainder were inversions across the duplicated tuf genes (Fig. 4; see also Fig. 1 in our supplementary information at the website http://www.egs.uu.se/molev/suppl.data/J.Bacteriol). The inferred rearrangements, which represent our best attempt to explain the data, include smaller inversions across neighboring GEIs and larger inversions that span the entire set of GEIs. We were unable to pinpoint the exact borders of the inversions in the various strains. However, it seems likely that the inversions were mediated by inverted repeats, since the GEIs in the sequenced strain contain numerous inverted repeats (Fig. 4, top), with almost 50% of the GEI sequences in total belonging to repeat families.
We observed no correlation between genome structures and sequence types. Rather, isolates of the same sequence type often presented radically different structures, and similar rearrangement scenarios were in several cases inferred for strains of different sequence types. For example, several genomes showed an overall genome structure similar to that of the Houston-1980517 strain, including isolates as diverse as the American ST1 strain UGA-23, the Indonesian ST1 strains IndoCat-2 and IndoCat-11, and the ST5 isolates Cheetah and GreekCat-23. Five isolates of three different sequence types contained a nonsymmetrical inversion event across the terminus of replication, with inversion breakpoints at the duplicated tuf genes, as verified by PCR analysis and sequencing.
DISCUSSION
Accidental human infections with B. henselae occur frequently via contacts with domestic cats, with obvious risks for the emergence of novel pathogenic strains. It is therefore of interest to examine the natural population of B. henselae isolated from feline hosts and its association with human infections. In this study, we have analyzed the genome contents and structures of 38 B. henselae strains, 29% of which are human isolates. This collection of strains includes six of the seven previously described genotypes (35) and seems therefore representative of the global B. henselae population. Only one sequence type (ST3) was not recovered here; however, this class is so far represented by only a single isolate (35). In addition, we identified a new variant of the rpoB allele in the Marseille URLLY8 strain, which was classified into a novel sequence type, ST8.
Our results show that human infections have been induced by feline strains of diverse geographic origins and sequence types, in accordance with a high genotypic heterogeneity of B. henselae isolated from humans with disease symptoms (69). However, we failed to identify genes in the sequenced Houston-1 strain that were uniquely shared with the other human isolates, suggesting that the ability to infect humans is not simply due to the acquisition of novel virulence traits. Rather, strains of most sequence types appear capable of causing incidental human infections. In comparison, permanent host switches are rare, with B. quintana as the only known example. It is interesting that neither B. koehlerae (43) nor any of the B. henselae strains here examined are as reduced in size as B. quintana (2). This suggests that the reduction in size of the B. quintana genome was perhaps associated with the permanent switch to the human host population.
The CGH profiles of the B. henselae strains were broadly consistent with the identified MLST variants, although with a few notable exceptions in which B. henselae isolates of different STs clustered in the CGH analysis. One such anomaly was the clustering of the strain Cheetah (ST4) with GreekCat-23 (ST5). However, since the support for this clade was lost upon the removal of phage sequences from the clustering analysis, part of the explanation may be independent excision of the prophage regions in the two strains. Another possibility is that replication of phage genes in the reference strain makes it appear as if isolates with no or less replication of phage genes are missing the prophage. However, attempts to detect phage genes by PCR analyses using primers from various prophage genes consistently failed with Cheetah and GreekCat-23 (data not shown), which together with extremely low hybridization signals suggest that the prophage is truly missing from these strains.
Independent excision of prophages and genomic islands in unrelated strains or recombination between strains yield homoplasies in the CGH data, making it unreliable for phylogenetic analysis. Indeed, a microarray CGH analysis of Helicobacter pylori, a species associated with high recombination frequencies, also revealed similar gene losses in genotypically unrelated strains (30). In effect, the phylogenetic histories of H. pylori (30) and B. henselae (this study) can only partially be predicted from gene content data. Microarray CGH data are perhaps most useful for the classification of isolates from recently emerged clonal pathogens, such as Mycobacterium tuberculosis (39, 65), Yersinia pestis (71), and Staphylococcus aureus (44).
Nevertheless, CGH data are informative in that they reveal gene content variations among strains. The results of this study have shown that the difference between the two most frequently isolated genotypes, the Houston-1 and the Marseille strains, resides in the absence of approximately 15 to 20 kb from the central part of HGIb in the Marseille strain. This region encodes mostly Bartonella-specific genes and is also absent in B. quintana (2) and B. koehlerae (43) as well as from a clade in the ST5 group that encompasses both human and feline isolates. Additionally, we observed variability in the ST2 to ST8 isolates in two clusters of genes containing autotransporter and passenger domains characteristic of type V secretion (50), one of which has been annotated as a cohemolysin and putative cytotoxin (45). This is interesting, since it has been shown that vasculoproliferative lesions of the liver and spleen occurred in three out of four HIV patients infected with strains of the 16S genotype I but in none of eight patients infected with the other genotype (12). Genes in HGIb and in the clusters putatively coding for autotransporters are thus our current best candidates to explain differences in clinical manifestations among human isolates of different genotypes.
The amplification of the chromosome II-like region may potentially also contribute to the creation of novel sequence variants, since genes with higher copy numbers have a higher probability of being transmitted to other cells. The amplified region is up to 300 kb in size in some strains and covers most of the 282-kb-long chromosome-II like segment, which is thought to have been acquired from the integration of another replicon (2). The increase in size of the amplified fragment with time suggests that the replication process is gradually induced and perhaps coordinated with the transition to the stationary phase, during which phage induction and lysis have been observed (13). If so, the differences in the amplification patterns observed among the various strains in the initial analysis (Fig. 1) may depend on the stage at which the cells were harvested.
Like the prophage and the GEIs, sequences in this area of the chromosome show deviations in GC content, and there is a small peak in GC skew near the center of the amplified region (see Fig. 1 in reference 2), which is located within a segment of about 30 kb that may represent an extensively degraded prophage. This region is flanked by two tRNA genes and contains mostly noncoding DNA but also three putative phage genes (an exonuclease, a DNA helicase/primase, and a lysozyme) and a few hypothetical genes of unknown function. A phylogenetic analysis of the helicase shows that it clusters most closely with prophage homologs in Escherichia coli, Salmonella enterica serovar Typhimurium, and Legionella pneumophila and more distantly with helicases from the lytic T3 and T7 phages (data not shown). However, no structural phage genes were identified, providing no evidence for a functional prophage in this region, at least not in the Houston-1Seq strain. The corresponding region in B. quintana is shorter in size and contains the yopP gene, which codes for a secreted protein that modulates the host immune response and is located on a 70-kb virulence plasmid in Yersinia pestis (32, 40).
Escape replication of prophage regions has previously been observed in E. coli (27, 34) and Salmonella enterica (26) and may result from runoff replication due to the loss of excisionase and/or integrase genes (27). Likewise, the observed amplification in some B. henselae strains may be initiated from a replication initiation site derived from an element that was previously replicating autonomously, such as a bacteriophage, a plasmid, or an auxiliary replicon. It remains to be determined whether additional prophage genes and/or plasmid genes are located at the corresponding site in strains with strong amplification.
In contrast to previous estimates (51), our PFGE results indicate that the B. henselae genomes are relatively homogeneous in size, in the range of 1.9 to 2.0 Mb (data not shown). The data also indicated high rearrangement frequencies within and across regions that include the GEIs. Different rearrangement patterns were observed in strains of the same sequence type as well as similar patterns in strains of different sequence types. This makes classification systems based on PFGE or other structural data unsuitable for typing purposes.
The observed variability makes it tempting to speculate that there is selection for shuffling of sequences within and across the islands. Two hypotheses have previously been suggested to explain selection for locally high rearrangement frequencies. The adopt-adapt model suggests that the insertion of phages and/or GEIs leads to an unbalancing of the genome and thereby to fixation of chromosomal rearrangements, as observed in Salmonella enterica (46, 47). According to the adopt-adapt hypothesis, rearrangements occur until the chromosome is partitioned equally on both sides of ori and ter. Here, the expectation is that rearrangement frequencies are high in bacteria that experience high rates of sequence influx and that selection increases the proportion of cells with rapid DNA replication until a balanced genome is restored in the population. In support of this hypothesis is the observation that Salmonella enterica serovar Typhi strains with more balanced genomes and shorter generation times out-compete strains with less balanced genomes (46).
Another theory is that selection promotes recombination so as to generate antigenic variability. An example of this strategy is found in Streptococcus pyogenes, which contains two prophages that are integrated equidistant from the ter region. Superantigens are exchanged between the prophages by homologous recombination, thereby contributing to the diversity of virulence factors in S. pyogenes (52). The streptococcal rearrangements were estimated to have occurred in many strains with different serotypes in the past and to have varied in frequency over time in the clinical strains. Likewise, genomic inversions have been suggested to contribute to antigenic variation in Bacteroides fragilis (10) and Mycobacterium avium (67). Another example is the PPE family in Mycobacterium tuberculosis, members of which are highly variable across strains by multiple insertion-deletions and hypothesized to be involved in antigenic variation (1, 29). Additionally, recombination dominates over mutations in Porphyromonas gingivalis and may help mediate long-term survival in the human host (18). High frequencies of recombination, which is the basis for gene conversion, combinatorial gene shuffling, and genomic inversions, also play a role in antigenic variation in human pathogens such as Neisseria, Borrelia, and Treponema (61).
The difference between the two hypotheses is that the adopt-adapt model predicts that genome rearrangements will eventually come to a halt, whereas the model suggesting diversification by recombination predicts that structural alterations will occur even with a balanced genome. Since the sequenced Houston-1 strain of B. henselae already has a symmetric genome, the adopt-adapt model may not be applicable to this species. Also, rapid replication may not necessarily be a selective force in the evolution in B. henselae and other slowly growing pathogens that establish long-term infections in their hosts. In the natural population of such species, diversification and immune evasion by intrachromosomal recombination seem likely to be a more dominant selective force, which could help explain the extensive rearrangements observed in areas of the GEIs and the terminus of replication.
Interestingly, B. henselae isolates taken from naturally infected cats during peaks of bacteremia showed marked differences in their PFGE patterns, suggesting that rearrangements play a role in persistent infections (38), although it could not be ruled out that coinfections of strains of different genotypes also occurred. Additionally, PFGE profiles derived from human strains and their corresponding cat source isolates were concordant but differed in size for one or more fragments, indicative of genomic rearrangements caused by antigenic variation (12). Furthermore, in vivo passage of B. henselae in immunocompetent and immunodeficient mice resulted in significant morphological changes, possibly due to different numbers and sizes of fimbriae and other adhesins (66), some of which may be encoded by the GEIs.
Indeed, flanking a long stretch of phage genes in the largest island, HGIa, in the sequenced Houston-1 strain are multiple copies of fhaB, which codes for filamentous hemagglutinin (FHA), and of fhaB/hecB, which codes for the corresponding transport protein. FHA is the dominant adhesin in Bordetella (36, 55), which is a pathogen that causes respiratory tract infections in humans and animals. FHA in Bordetella pertussis is highly immunogenic in humans and is also a protective antigen in animal models (4, 9, 62). As in Bordetella, FHA in B. henselae may serve as a major antigenic determinant and play a role in host receptor recognition. Immediately downstream of fhaB is a short hypothetical gene putatively coding for a protein of 138 amino acids that is repeated 20-fold in the genome. In total, almost 50% of the sequences in the GEIs are members of repeat families. The repeated structure of the GEIs and the vicinity of the FHA genes to prophage genes facilitate recombination events and spread among cells by horizontal gene transfer, thereby further contributing to variability within the B. henselae population. Thus, as in S. pyogenes, homologous recombination across repeated sequences may contribute to the shuffling of sequence segments in antigenic determinants.
B. henselae infections are normally self-limiting, but the symptoms and severity of the disease depend on the immune system of the host. An interesting question is whether B. henselae is able to sense the immune status of the host and respond appropriately. Experimental studies similar to the one performed in this study of B. henselae grown in the natural reservoir could help determine whether amplifications, deletions, and rearrangements occur during host colonization and what the implications of these alterations are for virulence and antigenic diversity.
In short, we suggest that recombination within and across GEIs in the B. henselae genome continuously generates subpopulations with different sets, sequences, and possibly even different expression patterns of major adhesions. This may alter host receptor recognition patterns and support evasion of the host immune response during long-term residence in the natural reservoir. High recombination frequencies among genes coding for outer membrane and secreted proteins could also offer an advantage to strains that have a wider host range. We hypothesize that this provides a selective constraint on the preservation of GEIs in the B. henselae population, which leads to the testable hypothesis that the surface-exposed proteome differs for strains with different genomic architectures, despite otherwise similar gene pools.
Acknowledgments
We are indebted to Scott Handley and Russ Regnery for the provision of B. henselae strains. We thank Alix Boulouis and Na Guan for assistance in the MLST and DNA amplification analyses, respectively. We thank Annelie Waldén at the Royal Institute of Technology, Stockholm, Sweden, and Niclas Olsson at Uppsala University, Uppsala, Sweden, for microarray printing.
This research was supported by grants from the Foundation for Strategic Research in Sweden (S.S.F.), the Swedish Research Council (V.R.), the Göran Gustafsson Foundation, the Knut and Alice Wallenberg Foundation (K.A.W.), and the Network of Excellence EuroPathogenomics (LSHB-CT-2005-512061).
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Adindla, S., and L. Guruprasad. 2003. Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. J. Biosci. 28:169-179. [DOI] [PubMed] [Google Scholar]
- 2.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. Legault, D. H. Ardell, B. Canback, A. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. E. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amsbaugh, D. F., Z. M. Li, and R. D. Shahin. 1993. Long-lived respiratory immune response to filamentous hemagglutinin following Bordetella pertussis infection. Infect. Immun. 61:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 6.Bergmans, A. M., J. F. Schellekens, J. D. van Embden, and L. M. Schouls. 1996. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J. Clin. Microbiol. 34:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulouis, H., C. Chang, J. B. Henn, R. W. Kasten, and B. B. Chomel. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383-410. [DOI] [PubMed] [Google Scholar]
- 8.Breitschwerdt, E. B., and D. L. Kordick. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahill, E. S., D. T. O'Hagan, L. Illum, and K. Redhead. 1993. Mice are protected against Bordetella pertussis infection by intra-nasal immunization with filamentous haemagglutinin. FEMS Microbiol. Lett. 107:211-216. [DOI] [PubMed] [Google Scholar]
- 10.Cerdeno-Tarraga, A. M., S. Patrick, L. C. Crossman, G. Blakely, V. Abratt, N. Lennard, I. Poxton, B. Duerden, B. Harris, M. A. Quail, A. Barron, L. Clark, C. Corton, J. Doggett, M. T. G. Holden, N. Larke, A. Line, A. Lord, H. Norbertczak, D. Ormond, C. Price, E. Rabbinowitsch, J. Woodward, B. Barrell, and J. Parkhill. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463-1465. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. C., B. B. Chomel, R. W. Kasten, R. M. Heller, H. Ueno, K. Yamamoto, V. C. Bleich, B. M. Pierce, B. J. Gonzales, P. K. Swift, W. M. Boyce, S. S. Jang, H. J. Boulouis, Y. Piemont, G. M. Rossolini, M. L. Riccio, G. Cornaglia, L. Pagani, C. Lagatolla, L. Selan, and R. Fontana. 2000. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg. Infect. Dis. 6:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, C., B. B. Chomel, R. W. Kasten, J. W. Tappero, M. A. Sanchez, and J. E. Koehler. 2002. Molecular epidemiology of Bartonella henselae infection in human immunodeficiency virus-infected patients and their cat contacts, using pulsed-field gel electrophoresis and genotyping. J. Infect. Dis. 186:1733-1739. [DOI] [PubMed] [Google Scholar]
- 13.Chenoweth, M. R., G. A. Somerville, D. C. Krause, K. L. O'Reilly, and F. C. Gherardini. 2004. Growth characteristics of Bartonella henselae in a novel liquid medium: primary isolation, growth-phase-dependent phage induction, and metabolic studies. Appl. Environ. Microbiol. 70:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomel, B. B., H. Boulouis, S. Maruyama, and E. B. Breitschwerdt. 2006. Bartonella spp. in pets and effect on human health. Emerg. Infect. Dis. 12:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarridge, J. E., III, T. J. Raich, D. Pirwani, B. Simon, L. Tsai, M. C. Rodriguez-Barradas, R. Regnery, A. Zollo, D. C. Jones, and C. Rambo. 1995. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J. Clin. Microbiol. 33:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehio, C. 2004. Molecular and cellular basis of Bartonella pathogenesis. Annu. Rev. Microbiol. 58:365-390. [DOI] [PubMed] [Google Scholar]
- 17.Dillon, B., J. Valenzuela, R. Don, D. Blanckenberg, D. I. Wigney, R. Malik, A. J. Morris, J. M. Robson, and J. Iredell. 2002. Limited diversity among human isolates of Bartonella henselae. J. Clin. Microbiol. 40:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong, H., T. Chen, F. E. Dewhirst, R. D. Fleischmann, C. M. Fraser, and M. J. Duncan. 1999. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect. Immun. 67:3416-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet 347:441-443. [DOI] [PubMed] [Google Scholar]
- 20.Ehrenborg, C., L. Wesslen, A. Jakobson, G. Friman, and M. Holmberg. 2000. Sequence variation in the ftsZ gene of Bartonella henselae isolates and clinical samples. J. Clin. Microbiol. 38:682-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 22.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 23.Fabbi, M., L. De Giuli, M. Tranquillo, R. Bragoni, M. Casiraghi, and C. Genchi. 2004. Prevalence of Bartonella henselae in Italian stray cats: evaluation of serology to assess the risk of transmission of Bartonella to humans. J. Clin. Microbiol. 42:264-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle.
- 25.Foucault, C., B. La Scola, H. Lindroos, S. G. E. Andersson, and D. Raoult. 2005. Multispacer typing technique for sequence-based typing of Bartonella quintana. J. Clin. Microbiol. 43:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frye, J. G., S. Porwollik, F. Blackmer, P. Cheng, and M. McClelland. 2005. Host gene expression changes and DNA amplification during temperate phage induction. J. Bacteriol. 187:1485-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukasawa, T., K. Hirai, T. Segawa, and K. Obonai. 1978. Regional replication of the bacterial chromosome induced by derepression of prophage lambda IV. Escape synthesis of gal operon in phage 82. Mol. Gen. Genet. 167:83-93. [DOI] [PubMed] [Google Scholar]
- 28.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 29.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 30.Gressmann, H., B. Linz, R. Ghai, K. Pleissner, R. Schlapbach, Y. Yamaoka, C. Kraft, S. Suerbaum, T. F. Meyer, and M. Achtman. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guptill, L., C. Wu, H. HogenEsch, L. N. Slater, N. Glickman, A. Dunham, H. Syme, and L. Glickman. 2004. Prevalence, risk factors, and genetic diversity of Bartonella henselae infections in pet cats in four regions of the United States. J. Clin. Microbiol. 42:652-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haase, R., K. Richter, G. Pfaffinger, G. Courtois, and K. Ruckdeschel. 2005. Yersinia outer protein P suppresses TGF-beta-activated kinase-1 activity to impair innate immune signaling in Yersinia enterocolitica-infected cells. J. Immunol. 175:8209-8217. [DOI] [PubMed] [Google Scholar]
- 33.Heller, R., M. Artois, V. Xemar, D. De Briel, H. Gehin, B. Jaulhac, H. Monteil, and Y. Piemont. 1997. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J. Clin. Microbiol. 35:1327-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imae, Y., and T. Fukasawa. 1970. Regional replication of the bacterial chromosome induced by derepression of prophage lambda. J. Mol. Biol. 54:585-597. [DOI] [PubMed] [Google Scholar]
- 35.Iredell, J., D. Blanckenberg, M. Arvand, S. Grauling, E. J. Feil, and R. J. Birtles. 2003. Characterization of the natural population of Bartonella henselae by multilocus sequence typing. J. Clin. Microbiol. 41:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob-Dubuisson, F., B. Kehoe, E. Willery, N. Reveneau, C. Locht, and D. A. Relman. 2000. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology 146:1211-1221. [DOI] [PubMed] [Google Scholar]
- 37.Jacomo, V., P. J. Kelly, and D. Raoult. 2002. Natural history of Bartonella infections (an exception to Koch's postulate). Clin. Diagn. Lab. Immunol. 9:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabeya, H., S. Maruyama, M. Irei, R. Takahashi, M. Yamashita, and T. Mikami. 2002. Genomic variations among Bartonella henselae isolates derived from naturally infected cats. Vet. Microbiol. 89:211-221. [DOI] [PubMed] [Google Scholar]
- 39.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer, U., and C. A. Wiedig. 2005. Y. enterocolitica translocated Yops impair stimulation of T-cells by antigen presenting cells. Immunol. Lett. 100:130-138. [DOI] [PubMed] [Google Scholar]
- 41.Kurtz, S., J. V. Choudhuri, E. Ohlebusch, C. Schleiermacher, J. Stoye, and R. Giegerich. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.La Scola, B., Z. Liang, Z. Zeaiter, P. Houpikian, P. A. D. Grimont, and D. Raoult. 2002. Genotypic characteristics of two serotypes of Bartonella henselae. J. Clin. Microbiol. 40:2002-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindroos, H. L., A. Mira, D. Repsilber, O. Vinnere, K. Naslund, M. Dehio, C. Dehio, and S. G. E. Andersson. 2005. Characterization of the genome composition of Bartonella koehlerae by microarray comparative genomic hybridization profiling. J. Bacteriol. 187:6155-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litwin, C. M., and J. M. Johnson. 2005. Identification, cloning, and expression of the CAMP-like factor autotransporter gene (cfa) of Bartonella henselae. Infect. Immun. 73:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, G., W. Liu, R. N. Johnston, K. E. Sanderson, S. Li, and S. Liu. 2006. Genome plasticity and ori-ter rebalancing in Salmonella typhi. Mol. Biol. Evol. 23:365-371. [DOI] [PubMed] [Google Scholar]
- 47.Liu, G., A. Rahn, W. Liu, K. E. Sanderson, R. N. Johnston, and S. Liu. 2002. The evolving genome of Salmonella enterica serovar Pullorum. J. Bacteriol. 184:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu, S. L., and K. E. Sanderson. 1995. Genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu, S. L., and K. E. Sanderson. 1995. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J. Bacteriol. 177:6585-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruyama, S., R. W. Kasten, H. J. Boulouis, N. A. Gurfield, Y. Katsube, and B. B. Chomel. 2001. Genomic diversity of Bartonella henselae isolates from domestic cats from Japan, the USA and France by pulsed-field gel electrophoresis. Vet. Microbiol. 79:337-349. [DOI] [PubMed] [Google Scholar]
- 52.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 54.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. G. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A, Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 55.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resto-Ruiz, S., A. Burgess, and B. E. Anderson. 2003. The role of the host immune response in pathogenesis of Bartonella henselae. DNA Cell Biol. 22:431-440. [DOI] [PubMed] [Google Scholar]
- 57.Riess, T., S. G. E. Andersson, A. Lupas, M. Schaller, A. Schafer, P. Kyme, J. Martin, J. Walzlein, U. Ehehalt, H. Lindroos, M. Schirle, A. Nordheim, I. B. Autenrieth, and V. A. J. Kempf. 2004. Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 200:1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez-Barradas, M. C., R. J. Hamill, E. D. Houston, P. R. Georghiou, J. E. Clarridge, R. L. Regnery, and J. E. Koehler. 1995. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J. Clin. Microbiol. 33:1089-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sander, A., M. Posselt, N. Bohm, M. Ruess, and M. Altwegg. 1999. Detection of Bartonella henselae DNA by two different PCR assays and determination of the genotypes of strains involved in histologically defined cat scratch disease. J. Clin. Microbiol. 37:993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sander, A., M. Ruess, S. Bereswill, M. Schuppler, and B. Steinbrueckner. 1998. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J. Clin. Microbiol. 36:2973-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 62.Shahin, R. D., D. F. Amsbaugh, and M. F. Leef. 1992. Mucosal immunization with filamentous hemagglutinin protects against Bordetella pertussis respiratory infection. Infect. Immun. 60:1482-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamas, I., L. Klasson, B. Canback, A. K. Nåslund, A. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. E. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 64.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. B. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. L. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Velho, P. E. N. F., A. M. de Moraes, A. M. Uthida-Tanaka, M. L. Cintra, and R. Gigliogi. 2002. Ultrastructural changes in a standard strain of Bartonella henselae after passages through BALB/cAn mice. Ultrastruct. Pathol. 26:161-169. [DOI] [PubMed] [Google Scholar]
- 67.Wu, C., J. Glasner, M. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto, K., B. B. Chomel, L. J. Lowenstine, Y. Kikuchi, L. G. Phillips, B. C. Barr, P. K. Swift, K. R. Jones, S. P. Riley, R. W. Kasten, J. E. Foley, and N. C. Pedersen. 1998. Bartonella henselae antibody prevalence in free-ranging and captive wild felids from California. J. Wildl. Dis. 34:56-63. [DOI] [PubMed] [Google Scholar]
- 69.Zeaiter, Z., P. Fournier, and D. Raoult. 2002. Genomic variation of Bartonella henselae strains detected in lymph nodes of patients with cat scratch disease. J. Clin. Microbiol. 40:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, P., B. B. Chomel, M. K. Schau, J. S. Goo, S. Droz, K. L. Kelminson, S. S. George, N. W. Lerche, and J. E. Koehler. 2004. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl. Acad. Sci. USA 101:13630-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, D., Y. Han, E. Dai, Y. Song, D. Pei, J. Zhai, Z. Du, J. Wang, Z. Guo, and R. Yang. 2004. Defining the genome content of live plague vaccines by use of whole-genome DNA microarray. Vaccine 22:3367-3374. [DOI] [PubMed] [Google Scholar]
- 72.Zwick, M. E., F. Mcafee, D. J. Cutler, T. D. Read, J. Ravel, G. R. Bowman, D. R. Galloway, and A. Mateczun. 2005. Microarray-based resequencing of multiple Bacillus anthracis isolates. Genome Biol. 6:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]