Abstract
A mutant strain of Bradyrhizobium japonicum USDA110 lacking isocitrate dehydrogenase activity was created to determine whether this enzyme was required for symbiotic nitrogen fixation with soybean (Glycine max cv. Williams 82). The isocitrate dehydrogenase mutant, strain 5051, was constructed by insertion of a streptomycin resistance gene cassette. The mutant was devoid of isocitrate dehydrogenase activity and of immunologically detectable protein, indicating there is only one copy in the genome. Strain 5051 grew well on a variety of carbon sources, including arabinose, pyruvate, succinate, and malate, but, unlike many microorganisms, was a glutamate auxotroph. Although the formation of nodules was slightly delayed, the mutant was able to form nodules on soybean and reduce atmospheric dinitrogen as well as the wild type, indicating that the plant was able to supply sufficient glutamate to permit infection. Combined with the results of other citric acid cycle mutants, these results suggest a role for the citric acid cycle in the infection and colonization stage of nodule development but not in the actual fixation of atmospheric dinitrogen.
Symbiotic nitrogen fixation, the primary pathway by which inorganic nitrogen is made available for living organisms, requires complex communication and exchange of nutrients between the bacterial microsymbiont and the host plant. The complexity extends from preinfection and continues throughout the lifetime of the symbiosis. Specific rhizobial species infect particular leguminous host plants, for example, Bradyrhizobium japonicum infects soybean but not other leguminous plants. The termination of the infection and root colonization process takes the form of tumor-like growths on the roots called nodules. The bacteria within the nodule differentiate into a form known as a bacteroid, which is retained within a plant-derived membrane referred to as a symbiosome. The bacteroids derepress nitrogenase, the enzyme complex that reduces atmospheric N2 to ammonium. Nitrogenase requires a minimum of 16 mol of ATP per mole of N2, but estimates of the energy needed for the complete nitrogen fixation process indicate that around 40 mol of ATP is required per mole of N2 or, in terms of carbon, 6 g of carbon is required for every gram of N reduced (9, 39, 40).
Bacteroids repress the glycolytic and other carbohydrate catabolic pathways (9, 26). Respirometric and enzymatic analyses have suggested the operation of the citric acid cycle in bacteroids. Malate is the primary carbon source provided to the bacteroids from the plant. Recently, proteomic analysis of B. japonicum bacteroids showed the presence of many citric acid and poly-β-hydroxybutyrate (PHB) cycle enzymes but only a few in glycolysis or the pentose phosphate pathway (34). The operation of the citric acid cycle in bacteroids has been an apparent anomaly since α-ketoglutarate dehydrogenase is strongly inhibited and down-regulated under low-oxygen conditions within the nodule (33). Analysis of a B. japonicum mutant lacking α-ketoglutarate dehydrogenase (13, 14) indicated that a classical citric acid cycle was not needed to supply energy or electrons for the actual reduction of atmospheric dinitrogen, as the symbiotic nitrogen fixation activity of this mutant was normal. However, the mutant demonstrated an unusual nodule colonization phenotype, suggesting that a classical citric acid cycle was required for proper infection and colonization but a different malate-utilizing pathway was necessary for providing energy and electrons to nitrogenase. In a search for this novel malate-utilizing pathway, Green et al. discovered a coenzyme A (CoA)-independent α-ketoglutarate decarboxylase activity in B. japonicum bacteroids, which, in concert with succinate semialdehyde dehydrogenase, may be components of a pathway that provides the energy and electrons for symbiotic nitrogen fixation (17). In addition, Ebenau-Jehle et al. reported that three adjacent open reading frames (ORFs) in B. japonicum should be reannotated as the three subunits of 2-oxoglutarate:acceptor oxidoreductase and proposed that it replaces α-ketoglutarate dehydrogenase and provides the energy for symbiotic nitrogen fixation (10). These reports provide alternative routes of the metabolism of α-ketoglutarate in bacteroids.
The source of α-ketoglutarate in bacteroids is presumably NADP-dependent isocitrate dehydrogenase. A search of the B. japonicum genome revealed the presence of only a single gene with homology to isocitrate dehydrogenases (20). Previously, we had purified NADP-dependent isocitrate dehydrogenase from both cultured cells and bacteroids and demonstrated they were the same protein via N-terminal amino acid sequencing and immunological cross-reactivity (22). The N-terminal amino acid sequence matched that of the translated B. japonicum isocitrate dehydrogenase gene. Thus, B. japonicum possesses only a single isocitrate dehydrogenase that is expressed both symbiotically and in culture and is an enzymatic source of α-ketoglutarate.
To further characterize bacteroid metabolism, the isocitrate dehydrogenase gene from B. japonicum wild-type USDA110 was cloned with the goal of generating an idh null mutant and studying the effect of this mutation on the symbiotic relationship between B. japonicum and soybean.
MATERIALS AND METHODS
Bacterial strains and media.
The B. japonicum and Escherichia coli strains used as hosts for cloning and for donors in conjugation are listed in Table 1. B. japonicum was routinely cultured at 28°C on Tully's defined medium, pH 6.8, with 25 mM arabinose as a carbon source (13). HM medium was used in triparental conjugations (6). MB medium was used to plate transconjugants (12). The antibiotic concentrations used for B. japonicum were 100 μg streptomycin/ml, 100 μg kanamycin/ml, 20 μg nystatin/ml, and 30 μg chloramphenicol/ml. E. coli cultures were grown at 37°C at pH 7.5 in LB medium (28). The antibiotic concentrations used for E. coli were 30 μg streptomycin/ml, 50 μg kanamycin/ml, 100 μg ampicillin/ml, and 30 chloramphenicol μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid/vector | Relevant feature(s) | Source or reference |
|---|---|---|
| Bradyrhizobium japonicum strains | ||
| USDA110 | Cmr (wild type) | 28 |
| 5051 | 110 (idhA::str), Cmr Strr | This study |
| 5051(pRS7) | 5051, Cmr Strr Tetr | This study |
| Escherichia coli strains | ||
| DH5α | supE44 Δ(lacZYA-argF)U169 deoR (Φ80lacZΔM15) gyrA98 endA1 hsdR17(rK− mK+) recA1 thi-1 relA1 | 19 |
| HB101 | thi-1 hsdR20(rB− mB+) supE44 lacY1 ara14 recA13 leuB6 proA2 xly5 mtl-1rpsL20 (Strr) | 4 |
| INVαF′ | F′ endA1 recA1 supE44 deoR Δ(lacZYA-argF)U169 λ−relA1 (Φ80lacZΔM15) hsdR17(rK− mK+) recA1 endA1 gyrA96 | Invitrogen |
| Plasmids/vectors | ||
| pTZ18U | Ampr, lacZ′, f1 ori, MCS, Mob−, cloning vector | 42 |
| pCR 2.1 | Ampr Kmr, lacZ′, f1 ori, MCS, Mob−, T cloning vector | Invitrogen |
| pGEM-T Easy | Ampr, lacZ′, f1 ori, MCS, Mob−, T cloning vector | Promega |
| pLO1 | Kmr, sacB, RP4 oriT, ColE1 ori | 30 |
| pHP45Ω | Ampr Strr in pP45Ω replicon | 42 |
| pRK2013 | Kmr, Mob+, Tra+, ColE1 ori, helper plasmid | 11 |
| pLAFR5 | Tcr, cosmid vector | 8 |
| pRS2 | Ampr, 2.01-kb PCR-amplified idhA region in pGEM-T Easy vector | This study |
| pRS3 | Ampr Strr, 2.01-kb PCR-amplified idhA region with a 270-bp XmaI deletion and a 2-kb XmaI Ω cassette insertion in pGEM-T Easy vector | This study |
| pRS4 | Ampr Strr, 3.73-kb PCR-amplified idhA::str insert with XbaI ends in pGEM-T Easy vector | This study |
| pRS5 | Kanr Strr, 3.73-kb PCR-amplified idhA::str insert in pLO1 vector | This study |
| pRS6a | Ampr Kanr, 2.01-kb PCR-amplified idhA region in pCR 2.1 vector | This study |
| pRS6b | Ampr Kanr, 2.01-kb PCR-amplified idhA region in pGEM-T Easy vector | This study |
| pRS7 | Tetr Kanr, 4.1-kb PCR-amplified idhA region and kan cassette with HindIII ends in pLAFR5 | This study |
Growth measurements.
For growth on different carbon substrates, B. japonicum cells were first grown to late exponential phase (A630, ∼1.5) in Tully's medium with arabinose. Cells were then collected by centrifugation at 7,600 × g for 10 min, washed with 0.2 M NaCl, and resuspended in Tully's medium with the carbon source omitted. Cells were then inoculated into Tully's medium with the appropriate carbon source (20 mM l-malate, pyruvate, acetate, or succinate) to an A630 of 0.02 and incubated at 28°C with moderate shaking.
DNA manipulations.
Genomic DNA from B. japonicum was prepared as described by Hahn and Hennecke (18). The plasmids used are listed in Table 1. Plasmids were prepared on a small scale with Promega Wizard minipurification (Madison, WI) according to the manufacturer's instructions. Plasmids on a larger scale were prepared by a QIAGEN midi plasmid purification kit (Valencia, CA). Restriction enzyme digests, agarose gel electrophoresis, and ligations were performed according to the manufacturer's instructions (Promega, Madison, WI). E. coli conjugations were done either by electroporation or by heat shock, as outlined by Ausubel et al. (2). Conjugation of plasmid pRS7 into B. japonicum was performed by the triparental method of Ditta et al. (8). The electroporation was performed with a Gene Pulser (Bio-Rad, Hercules, CA) at 1.5 V, 25 μF, and 200 Ω in a 0.1-cm-gap-width pulser cuvette.
Southern blot analyses were performed in a sodium dodecyl sulfate-phosphate buffer as described previously (13). All probes were labeled by the random priming method with [α-32P]dCTP (NEN Life Sciences), using the Prime-a-Gene labeling system (Promega, Madison, WI). Probes were labeled to a specific activity of 0.9 × 109 dpm/μg.
Cloning and sequencing of NADP-dependent isocitrate dehydrogenase.
The B. japonicum NADP-dependent isocitrate dehydrogenase gene was cloned from genomic DNA using PCR. The oligonucleotides used for PCR of NADP-dependent isocitrate dehydrogenase, idhfor and idhrev, are listed in Table 2. The PCR conditions were as follows: an initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 98°C for 1 min, annealing at 55°C for 1 min, and elongation at 72°C for 4 min. A 50-μl reaction mixture contained 1 U Pfu Turbo DNA polymerase (Stratagene) and 0.5 mM of each deoxynucleoside triphosphate, 2 mM MgCl2, 10 pmol of each primer, and 1 μg genomic DNA. The reactions were spiked midway through the cycling conditions with 0.5 U Pfu Turbo DNA polymerase (Stratagene). After the addition of 3′ A-overhangs by incubation with Taq DNA polymerase (Promega), the amplified 2.01-kb DNA fragment was cloned into pGEM-T Easy vector (Promega), giving plasmid pRS2. Oligonucleotides idhhindf and idhhindr were used as described above for generating pRS7, which would generate a clone 423 bp upstream of the translational start and 377 bp downstream of the stop codon containing the native promoter and terminator elements. All constructs were sequenced using ABI BigDye Terminator chemistry on an ABI 377 automated sequencer (Applied Biosystems Inc., Foster City, CA) at the University of Missouri—Columbia DNA Core Facility. The resulting sequences were analyzed with the tBLASTx and tBLASTn programs (National Center for Biotechnology Information, Bethesda, MD) and SDSC Biology WorkBench (San Diego Supercomputer Center, UCSD, La Jolla, CA). Protein sequence analysis was done by using the ScanProsite (http://ca.expasy.org/tools/scanprosite/) against the PROSITE database. Amino acid alignments were done using the CLUSTALW multiple sequence alignment program in the SDSC Biology Workbench (http://workbench.sdsc.edu/).
TABLE 2.
Primers used in this study
| Primer | Description | Sequence (5′ to 3′) |
|---|---|---|
| idhfor | N-terminal forward for idh | AGCTGCGGCTCGACCAGGAT |
| idhrev | C-terminal reverse for idh | AGCTCCCAGTTCGGCCCGTA |
| idhxf | N-terminal forward for idh with added XbaI site | TCTAGAAGCTGCGGCTCGACCAGGAT |
| idhxr | C-terminal reverse for idh with added XbaI site | AGCTCCCAGTTCGGCCCGTATCTAGA |
| strfor | N-terminal forward for streptomycin | ACATTTGTACGGCTCCGCA |
| strrev | C-terminal reverse for streptomycin | TTAAGGTTTCATTTAGCGCCTCAAATAGA |
| idhfor2 | N-terminal forward for genomic DNA flanking idh | AATCGGCTGAGCGCAAAATT |
| strrev2 | C-terminal reverse for streptomycin | ATGCGGATCAGTGAGGGTTT |
| idhhindf | N-terminal forward for idh with added HindIII site | TAGGCACCCCAGGCTTTACACTTTATGCTTCC |
| idhhindr | C-terminal reverse for kanamycin with added HindIII site | CCCAAGCTTGGGTGAGCAAAAACAGGACAAAA |
Construction of the NADP-dependent isocitrate dehydrogenase null mutant.
An internal 270-bp XmaI-delineated region within the cloned idh-containing fragment in plasmid pRS2 was deleted by restriction digestion and replaced by a 2-kb XmaI Ω cassette (encoding resistance to both spectinomycin and streptomycin) from vector pHP45Ω (30), resulting in vector pRS3. PCR of pRS3 with oligonucleotide primers idhxf and idhxr (Table 2) amplified a 3.7-kb insert with flanking XbaI sites. PCR cycling conditions were as described previously except that the annealing temperature was 62°C for 1 min and extension was 72°C for 8 min. After the addition of 3′ A-overhangs by incubation with Taq DNA polymerase (Promega), the amplified 3.7-kb DNA fragment was cloned into pGEM-T Easy vector (Promega), giving plasmid pRS4. Plasmid pRS4 was digested with XbaI, and the 3.7-kb XbaI fragment carrying the disrupted idh was ligated to the unique XbaI site of plasmid pLO1 (24) to produce plasmid pRS5, which was transformed into B. japonicum by the triparental conjugation method of Ditta et al. (8). Recipient strain B. japonicum USDA110 (or, in the case of the complementing transconjugate, B. japonicum 5051) were grown to mid-log phase in HM medium, while mid-log E. coli donor and helper strains were grown to mid-log phase in LB medium with the appropriate antibiotics. One milliliter of each culture was then individually pelleted down, washed twice, and then resuspended in HM medium without antibiotics. The cultures were then mixed in a 1:1:1 ratio, spun, and finally resuspended in 50 μl total HM medium. The controls consisted of deleting each of the individual cultures. After mating for 2 days on a 0.45-μm-pore-sized filter (P/N 60172; Gelman Sciences, Ann Arbor, MI) at 30°C, the bacterial mix was resuspended in phosphate-buffered saline-Tween 20 and plated on a rich medium for 7 to 10 days, such as MB, with chloramphenicol as a counter selection for E. coli strains and streptomycin to select B. japonicum recipients. The streptomycin-resistant B. japonicum colonies were screened on MB medium containing 100 μg/ml kanamycin. The Strr Kans presumptive double recombination mutants were streaked for single-colony purification on MB medium containing chloramphenicol, nystatin, and streptomycin. Double-crossover events were confirmed by colony PCR amplifications and appropriate Southern blot hybridization analyses.
Enzyme assays.
Cultures of B. japonicum were grown in Tully's medium with arabinose to mid-log phase. Cells were harvested by centrifugation at 6,000 × g for 10 min, washed with 200 mM NaCl, and resuspended in breaking buffer {20 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], pH 7.0, 100 mM NaCl, 5 mM MgCl2, 0.4 mM EDTA, 1.5 mM dithiothreitol, 4% [wt/vol] glycerol}. Cells were broken by passing them twice through a French pressure cell at 16,000 lb/in2. The resulting extracts were centrifuged at 10,000 × g for 15 min, and the supernatant was desalted by dialysis against breaking buffer with the salt omitted. The protein content of this extract was determined by the Bradford method (5). Isocitrate dehydrogenase, malate dehydrogenase, α-ketoglutarate dehydrogenase, β-hydroxybutyrate dehydrogenase, and isocitrate lyase were assayed as previously described (13, 14, 16, 23). Isocitrate was purified by high-pressure liquid chromatography (0.014 N H2SO4 isocratic elution from Aminex HPX-87H ion exclusion organic acid analysis column; Bio-Rad Laboratories, Richmond, CA) before being used to remove malate and other compounds. Citrate synthase assays contained 0.3 mM DTNB [5,5′-dithiobis-(2-nitrobenzoate)], 0.3 mM acetyl-CoA, 25 to 50 μl enzyme extract, and 100 mM Tris buffer, pH 8.1, to a final total volume of 1 ml. Oxaloacetate (0.5 mM) was added to start the reaction, which was followed at 412 nm, where the mercaptide ion has a strong absorption. Glyceraldehyde-3-phosphate dehydrogenase assays contained 83 mM triethanolamine buffer, pH 7.6, 6.7 mM 3-phosphoglyceric acid, 3 mM l-cysteine, 2 mM MgSO4, 0.1 mM NADH, 1.1 mM ATP, and 10 units of 3-phosphoglyceric phosphokinase to a final volume of 3 ml. Each enzyme reaction was started by the addition of 25 to 50 μl of enzyme extract, and the change in absorbance at 340 nm was recorded. Background rates were established by a 2-min preincubation period of the complete reaction mixture minus the indicated substrate. Each enzyme activity was measured in sets of three replicates and at two different enzyme concentrations. The final pH of each assay mixture was within 0.1 unit of the buffer pH. All of the reagents used in the assays are from Sigma Inc., St. Louis, MO, or Fisher Scientific unless otherwise mentioned.
Western blot analyses.
The antibodies were prepared by the Laboratory Animal Medicine Office at the University of Missouri—Columbia School of Medicine and partially purified by ammonium sulfate fractionation and DEAE Affi-Gel Blue chromatography before use, and Western blot analysis was performed as described previously (22).
Plant growth and inoculation.
Soybean (Glycine max Merrill cv. Williams 82) seeds were surface sterilized and germinated as described by Oehrle et al. (29). After 48 h, seedlings were inoculated with Bradyrhizobium japonicum USDA110, mutant 5051, or complemented transconjugate 5051 (pRS7) for 30 min and then planted in autoclaved Leonard jars (23) containing a 3:3:1 mixture of vermiculite/perlite/sand supplied with a nitrogen-free plant nutrient solution. During growth, sterile nutrient solution was added to the lower reservoir as needed. Plants were grown in the greenhouse with a day/night temperature of 27°C (16 h)/24°C (8 h). Light intensity at the pot level was 450 μE · m−2 · s−1. Nodules were harvested from the crown area of the tap root 18 to 31 days after planting and used for further assays.
Nitrogen fixation activity.
The nitrogen fixation activity of intact root crown nodules was estimated using the acetylene reduction method of Schwinghamer et al. (35). Values were normalized to the total fresh weight of the nodules in each assay.
Bacteroid isolation and extract preparation.
Bacteroids were isolated aerobically from nodules as previously described (23). After the second sucrose gradient, the bacteroids were resuspended in 30 ml deionized, distilled water and collected by centrifugation at 800 × g for 15 min at 5°C. The pellet was resuspended in 5 ml breaking buffer (20 mM TES, pH 7.0, 100 mM NaCl, 5 mM MgCl2, 0.4 mM EDTA, 1.5 mM dithiothreitol, and 4% [wt/vol] glycerol) and ruptured with a French pressure cell at 16,000 lb/in2. Cell debris was removed by centrifugation at 30,000 × g for 20 min at 4°C.
Anaerobic bacteroid isolation was performed as described by Waters et al. (41).
Resuspended bacteroids were assayed for acetylene reduction as described by Karr and Emerich (21). Bacteroid viability for retention of streptomycin resistance was determined as described by Karr and Emerich (21).
Microscopy.
Light and transmission electron microscopies were performed as described by Green and Emerich (14, 15).
Infection and nodule development assays.
Soybean seeds were surface sterilized and germinated as described previously (29), inoculated and transplanted to sterile growth pouches (Mega International, Minneapolis, MN) containing nitrogen-free plant solution, and grown as described by Green and Emerich (14). Curled root hairs were stained with methylene blue, using the protocol described by Green and Emerich (15). Soybean root segments, ∼3 cm in length, were prepared as described for methylene blue staining and were stained with hematoxylin using a method modified by Green and Emerich (15). The roots used for hematoxylin staining were inoculated with 105 cells/ml.
Poly-β-hydroxybutyrate measurement.
PHB analysis was performed on bacteroids isolated from soybean nodules as described by Karr and Emerich (21).
RESULTS
Genomic analysis of B. japonicum NADP-dependent isocitrate dehydrogenase.
The B. japonicum genome has been sequenced and annotated (http://www.kazusa.or.jp/rhizobase/) by Kaneko et al. (20). The gene for NADP-dependent isocitrate dehydrogenase (EC 1.1.1.42), idh, designated blr5747, has a GC ratio of 64.3%, which is in the range of the overall GC ratio of B. japonicum (61 to 65%). The codon usage bias of B. japonicum idh places it in a category (group III) distinct from the nif and fix genes, consistent with the role of idh in central carbon metabolism (31). A presumptive purine-rich ribosome binding site, GAGG, was present 11 base pairs upstream of the ATG translation start site. A sequence resembling the −35 and −10 σ70 promoter sequence in B. japonicum was located 93 base pairs upstream of the ATG start site of idh, but it should be noted that promoter structures in bacteria can vary widely and the identification of promoters based on sequence information alone is speculative. Putative transcription termination signals were identified by the presence of inverted repeats 45 base pairs and 99 base pairs downstream from the last idh codon, and mRNA transcribed from this region can potentially form a characteristic stem and loop terminator structure.
The B. japonicum USDA110 idh gene is flanked upstream by a region with 74% identity to tRNA/rRNA methyl transferase of Rhodopseudomonas palustris. The putative metS ORF starts 339 base pairs upstream of the idh gene and is transcribed on the strand opposite idh. The second inverted repeat following the idh coding sequence centered at 99 base pairs downstream overlaps the start of an ORF that is 97 base pairs downstream of the idh stop codon. The ORF potentially encodes 166 amino acids transcribed in the same direction as idh. This ORF has no homology to any known sequences in the available databases. Considering that the upstream flanking gene is transcribed in an opposite direction from idh and that the gene encoding the hypothetical protein downstream from idh follows after the stem loops, idh is not predicted to be part of a polycistronic message. This gene arrangement seems to be unique to bradyrhizobia since an analogous gene arrangement was not observed for Mesorhizobium loti or Sinorhizobium meliloti.
Cloning and sequence analysis of B. japonicum NADP-dependent isocitrate dehydrogenase.
A 2,010-bp DNA fragment containing the NADP-dependent isocitrate dehydrogenase gene from B. japonicum strain USDA110 genomic DNA was obtained by PCR based on the published genomic sequence (20), using the oligonucleotide primers listed in Table 2. Sequencing of the entire PCR product showed it to be identical to the published genomic sequence (20).
The B. japonicum idh gene encodes a protein of 403 amino acids with a calculated molecular mass of 45.2 kDa. These values agree with those for B. japonicum isocitrate dehydrogenase purified from both cultured cells and bacteroids (22). To verify that the cloned idh gene encoded the B. japonicum NADP-dependent isocitrate dehydrogenase (EC 1.1.1.42), the translated idh gene sequence was compared to the N-terminal amino acid sequences of the NADP-dependent isocitrate dehydrogenases purified from cultured cells and from bacteroids isolated from infected soybean nodules (22). The N-terminal 20 amino acids from all three sources were identical, with the exception of the N-terminal methionine of the sequence translated from the DNA sequence. This confirmed that the cloned idh was indeed the B. japonicum USDA110 NADP-dependent isocitrate dehydrogenase. The absence of the N-terminal methionine from the mature protein indicated that only the methionine residue had been cleaved.
Based on the translated DNA sequence, B. japonicum isocitrate dehydrogenase was highly similar to Rhodopseudomonas palustris (91% identity), Mesorhizobium loti (84% identity), Sinorhizobium meliloti (83% identity), and Agrobacterium tumefaciens (82% identity), as might be expected. The sequence was also highly homologous to Glycine max (62% identity), Arabidopsis thaliana (62% identity), and Medicago sativa (60% identity). Furthermore, the alignment of the residues implicated in the binding of isocitrate and NADP+ is conserved in both the prokaryotic and the eukaryotic isocitrate dehydrogenase proteins. The idh gene contains the isocitrate and isopropylmalate dehydrogenase signature pattern, i.e., a glycine-rich stretch of residues located at the C terminus (269-NYDGDVQSDTVAQGFGSLGL-288) (36). Computer-aided protein sequence analysis did not detect any helix-turn-helix motifs or transmembrane domains interrupted by hydrophilic loops in the protein.
B. japonicum idh mutant.
A B. japonicum NADP-dependent isocitrate dehydrogenase null mutant was generated using homologous recombination as described in Materials and Methods. Southern blot analysis indicated that isolate 5051 had a banding pattern consistent with a double recombination event and insertion of the 2-kb streptomycin gene in the chromosomal DNA. The loss of the plasmid vector sequence was also confirmed by hybridization analysis. The marker exchange event was further verified in isolate 5051 by PCR amplification of genomic DNA isolated from strain 5051, using primers homologous to the streptomycin cassette and to B. japonicum genomic DNA outside the earlier-amplified 2.01-kb DNA region, allowing the amplification of the expected 1.1-kb PCR product from B. japonicum 5051, and confirmed exchange of the disrupted idh allele with wild-type B. japonicum USDA110 genomic idh and the gene arrangement predicted for an idh site-directed mutant.
Growth on different carbon substrates.
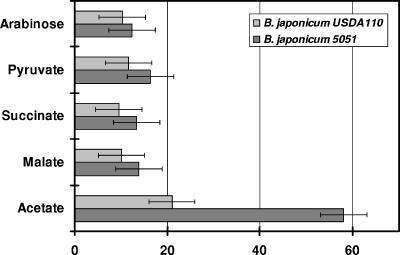
B. japonicum 5051 demonstrated auxotrophy for glutamate. Cultures grown in defined medium were supplemented with 0.125% (wt/vol) glutamate to compensate for the glutamate auxotrophy. Strain 5051 showed a slight increase in doubling time when arabinose, pyruvate, succinate, and malate were used as the sole carbon source but was significantly impaired when grown on acetate (Fig. 1). Complementation of strain 5051 with plasmid pRS7, which contained a full-length copy of the idh gene, including the presumptive promoter region, restored the glutamate auxotrophy and reduced the doubling time for growth on acetate to 25 ± 4 h (mean ± standard deviation), which is comparable to that for the wild type. This restoration of growth confirmed that the mutant phenotype is due to a mutation in the idh gene alone and not due to a polar effect on the transcription of its downstream genes. The restoration also suggested that the region upstream of the idh gene contained the native promoter.
FIG. 1.
Doubling times of B. japonicum strains on various carbon sources. B. japonicum USDA110 and strain 5051 were grown on defined Tully's medium with various carbon sources. Growth rates were determined as described in Materials and Methods. The carbon sources used were at a final concentration of 20 mM. Values are averages ± standard deviations of readings from three different cultures.
Biochemical analysis of strain 5051.
Cell-free protein extracts from USDA110 and strain 5051 cultured in Tully's arabinose defined medium harvested at mid-log phase were assayed spectrophotometrically for NADP-dependent isocitrate dehydrogenase activity by measuring the reduction of NADP+ at 340 nm (Table 3). The specific activity of isocitrate dehydrogenase in wild-type B. japonicum USDA110 was determined to be 290 ± 61 nmol/min/mg protein, a value similar to those obtained previously (22). Strain 5051 had no detectable isocitrate dehydrogenase activity when high-pressure liquid chromatography-purified isocitrate was used as the substrate. Neither NAD+ nor NADP+ as the cofactor resulted in isocitrate dehydrogenase activity in extracts from strain 5051. Complementation of strain 5051 with plasmid pRS7, containing the idh gene, restored enzyme activity levels to 276 ± 89 nmol/min/mg protein. The extracts were also assayed for activities of various enzymes of the citric acid cycle and other carbon metabolic pathways, such as the glycolytic and β-hydroxybutyrate cycle enzymes. The activity of α-ketoglutarate dehydrogenase activity of strain 5051 was 52% of B. japonicum USDA110 activity, whereas citrate synthase, malate dehydrogenase, β-hydroxybutyrate dehydrogenase, and glyceraldehyde-3-phosphate dehydrogenase activity levels were 122%, 92%, 91%, and 112%, respectively, of wild-type enzyme activity levels (Table 3). The levels of isocitrate lyase were found to be low in extracts from each of the cultures. The normal levels of the citric acid cycle and other enzymes indicate that there were no secondary suppressor mutations in strain 5051 that affected these enzymes.
TABLE 3.
Enzyme activities in cell extracts of B. japonicum USDA110, 5051, and 5051(pRS7) cultured on arabinose
| Enzyme | Enzyme activity (nmol/min/mg protein) of straina:
|
||
|---|---|---|---|
| USDA110 | 5051 | 5051(pRS7) | |
| Isocitrate dehydrogenase | 290 ± 61 | 0.4 ± 0.3 | 451 ± 267 |
| Citrate synthase | 429 ± 33 | 526 ± 37 | 625 ± 134 |
| α-Ketoglutarate dehydrogenase | 245 ± 33 | 109 ± 14 | 277 ± 89 |
| Malate dehydrogenase | 2,354 ± 204 | 2,184 ± 187 | 2,225 ± 332 |
| Hydroxybutyrate dehydrogenase | 84 ± 9 | 76 ± 11 | 84 ± 12 |
| Glyceraldehyde-3-phosphate dehydrogenase | 1,230 ± 57 | 1,303 ± 78 | 1,202 ± 111 |
| Isocitrate lyase | 0.7 ± 0.3 | 0.6 ± 0.4 | 0.7 ± 0.4 |
The values shown are the means ± standard deviations for three different cell extracts, each measured at two different enzyme levels.
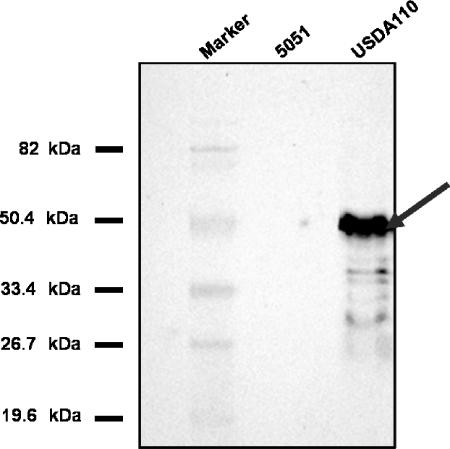
Rabbit polyclonal antibodies raised against purified B. japonicum NADP isocitrate dehydrogenase protein isolated from both bacteroids and cultured B. japonicum were used as probes in Western blot analyses to detect NADP-dependent isocitrate dehydrogenase protein in crude cell extracts of mutant 5051 and wild-type USDA110 (Fig. 2). The NADP-dependent isocitrate dehydrogenase antibody reacted with a peptide of approximately 43 kDa in B. japonicum USDA110 extracts but not in extracts obtained from strain 5051, indicating that isocitrate dehydrogenase protein was not produced in the mutated strain.
FIG. 2.
Immunodetection of Bradyrhizobium japonicum NADP-dependent isocitrate dehydrogenase. Soluble protein extracts from isolated B. japonicum USDA110 and strain 5051 were subjected to separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to membranes, and probed with rabbit polyclonal antibodies raised against B. japonicum NADP-dependent isocitrate dehydrogenase protein.
Symbiotic phenotype.
The symbiotic performance of strain 5051 was followed for 31 days after inoculation (DAI) onto soybean seedlings. Strain 5051 formed nodules on soybean plants, and the nodules appeared to be normal upon visual inspection. The uninoculated control did not form nodules. The nodules formed by mutant 5051 were able to reduce acetylene at rates lower, but not statistically different, than those of the wild type (Table 4). Earlier than 30 to 32 days after inoculation, the acetylene reduction rates of nodules formed by strain 5051 were lower than those of nodules formed by the wild type; after that, the reverse was generally true, but again, the differences were not statistically significant. In an experiment using the complemented strain, the acetylene reduction rates (means ± standard deviations) measured at day 18 were as follows: USDA110, 2.5 ± 0.2 μmol/g/h; strain 5051, 2.3 ± 0.2 μmol/g/h; and strain 5051(pRS7), 2.7 ± 0.1 μmol/g/h.
TABLE 4.
Acetylene reduction activity, nodule weight, and shoot weight of soybean plants inoculated with B. japonicum USDA110 and strain 5051a
| Strain | Acetylene reduction activity (μmol/g/hr) at:
|
Nodule wt (mg/nodule) at:
|
Shoot wt (g) at:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 18 DAI | 22 DAI | 31 DAI | 18 DAI | 22 DAI | 31 DAI | 18 DAI | 22 DAI | 31 DAI | |
| USDA110 | 2.8 ± 0.2 | 4.3 ± 0.3 | 3.9 ± 0.5 | 10.7 ± 0.7 | 9.8 ± 0.7 | 12.5 ± 0.9 | 0.9 ± 0.1 | 1.1 ± 0.1 | 1.5 ± 0.2 (0.8 ± 0.2) |
| 5051 | 2.4 ± 0.3 | 4.0 ± 0.1 | 3.8 ± 0.1 | 7.6 ± 0.7 | 9.5 ± 0.6 | 13.1 ± 0.5 | 1.1 ± 0.1 | 1.24 ± 0.2 | 1.2 ± 0.2 (0.6 ± 0.2) |
The values shown are the means ± standard deviations for three different cell extracts, each measured at two different enzyme levels. The values for uninoculated controls are shown in parentheses.
The average fresh weight of the nodules formed on the root crown of soybean plants inoculated with mutant 5051 lagged behind those induced by wild-type USDA110 at 18 days after inoculation, but no differences were observed at 22 days or later (Table 4). At 18 DAI, the average fresh weight of nodules formed by strain 5051 was only 71.4% that of the wild type; it was closer to normal at 22 DAI (96.9%) and slightly more than that of the wild type at 31 DAI (104.8%). At 31 DAI, soybean plants infected by strain 5051 had 9.67 ± 1.04 nodules while wild-type USDA110 had 12.34 ± 1.15 nodules. The average shoot dry weight of strain 5051-inoculated soybean plants was not significantly different from that of B. japonicum USDA110-infected plants throughout symbiosis (Table 4).
Bacteroids were isolated from nodules obtained at 22 days from nodules of plants inoculated with either USDA110 or strain 5051. The acetylene reduction activities of the isolated bacteroids were 4.3 ± 0.3 μmol/h/mg protein for bacteroids from USDA110 and 4.0 ± 0.8 μmol/h/mg protein for bacteroids from strain 5051. The ratio of acetylene reduction activity comparing nodules formed by USDA110 and strain 5051 was the same as the ratio of ex planta acetylene reduction activity of bacteroids isolated from nodules of USDA110 and strain 5051. Bacteroids were recovered from the nodules formed by inoculation with strain 5051, and the streptomycin resistance of the B. japonicum colonies recovered from the nodules was determined. More than 98% of the colonies were streptomycin resistant, indicating the stable retention of the antibiotic marker used to construct strain 5051.
The bacteroids recovered from nodules infected with strain 5051 had negligible NADP-dependent isocitrate dehydrogenase activity compared to those of the wild type. The complemented strain restored NADP-dependent isocitrate dehydrogenase enzyme activity in crude protein extracts of bacteroids. Isocitrate lyase activities were found to be similar in bacteroids formed from the wild type, the mutant, and the complemented mutant (Table 5). The nodules induced by mutant 5051 contained slightly less leghemoglobin than the root nodules induced by wild-type USDA110 (340 versus 304 mg/ml; 86%). A slight increase in PHB concentration in mutant nodules compared to that in wild-type nodules was observed (18.6 versus 16.0 mg/ml; 116%).
TABLE 5.
Enzyme activities of bacteroids isolated from nodules 18 days after inoculation with B. japonicum strains USDA110, 5051, and 5051(pRS7)
| Inoculum | Sp act (nmol/min/mg protein) fora:
|
|||
|---|---|---|---|---|
| NADP isocitrate dehydrogenase | α-Ketoglutarate dehydrogenase | Malate dehydrogenase | Isocitrate lyase | |
| USDA110 | 186.9 ± 3.6a | 169.5 ± 0.1 | 1,710 ± 65 | 1.1 ± 0.3 |
| 5051 | 0.4 ± 0.3 | 115.1 ± 11.7 | 2,193 ± 78 | 1.1 ± 0.4 |
| 5051(pRS7) | 227.5 ± 7.1 | 177.5 ± 13.5 | 1,784.1 ± 137 | 1.0 ± 0.4 |
The values shown are the means ± standard deviations for three different cell extracts, each measured at two different enzyme levels.
Western blot analyses were performed using polyclonal antibodies raised against purified B. japonicum NADP-dependent isocitrate dehydrogenase protein isolated from both bacteroids and cultured B. japonicum. An immunoreactive peptide of approximately 43 kDa was observed in bacteroid extracts from B. japonicum USDA110 but not in bacteroid extracts from strain 5051. The Western blot analyses and enzyme measurements confirm that the isocitrate dehydrogenase-deficient phenotype in the mutant was stable during symbiosis.
Nodule morphology.
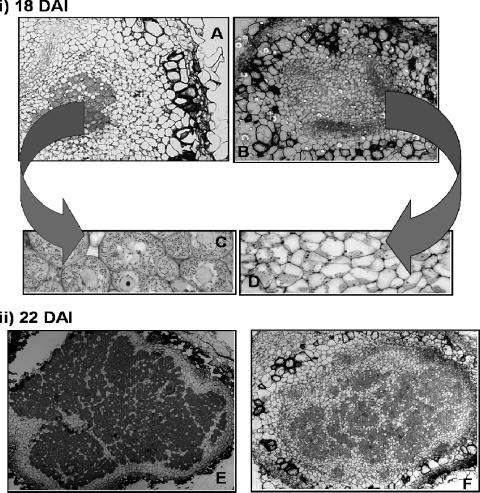
The symbiotic characterization of strain 5051 suggested that it had a delayed nodule development phenotype. Light micrographs of nodules formed by strain 5051 exhibited an unusual infection pattern during the early infection and colonization period of nodule development. At 18 days, only the peripheral plant cells of the central region of the developing nodule were infected, resulting in a region in the center of the nodule of uninfected cells surrounded by a ring of infected cells (Fig. 3A and B). In contrast, the wild-type nodules had a normal morphology, with the central region of the nodule uniformly infected with bacteria. Plant cells invaded by B. japonicum USDA110 showed a uniform distribution of the bacteria, whereas plant cells invaded by strain 5051 restricted them to only portions of the cell (Fig. 3C and D). Invading infection threads were also observed in transmission electron micrographs of nodules infected with strain 5051, indicating that at 18 days after inoculation, they were still in the process of being infected. The morphology of mature nodules formed by mutant 5051 was similar to the morphology of those induced by the wild type (Fig. 3E and F) but resembled the “patchy” phenotype of nodules produced by inoculation with the B. japonicum α-ketoglutarate null mutant (14). There were also more starch granules in 18-, 22-, and 31-day-old nodules formed by the mutant than in those formed by the wild type.
FIG. 3.
Light micrographs of nodules formed by the Bradyrhizobium japonicum strains USDA110 and 5051. Nodules were inoculated with either USDA110 or strain 5051 and harvested 18 or 22 days after inoculation. For panels A and B, nodules were fixed and embedded in paraffin and the sections were stained with fast green and safranin. For panels C and D, nodules were embedded in resin and the sections were stained with toluidine blue. Panels A to D are micrographs of 18-day-old nodules formed by USDA110 (A and C) and 18-day-old nodules of strain 5051 (B and D). Panels E and F are micrographs of 22-day-old nodules formed by USDA110 (E) and strain 5051 (F). (A) Magnification, ×20. (B, E, and F) Magnification, ×15. (C) Magnification, ×640. (D) Magnification, ×320.
The delayed nodulation phenotype of B. japonicum 5051 suggested a defect in infection thread initiation or elongation which affects the invasion of plant cells. Visual examination of methylene blue-stained roots indicated no apparent difference between the mutant and the wild type in the induction of curled root hairs. As determined by hematoxylin staining, the number of foci induced by mutant 5051 was consistently lower but not significantly different than that induced by the wild type. (Table 6).
TABLE 6.
Focus formation and nodule density of soybean seedlings inoculated with B. japonicum USDA110 or strain 5051a
| Inoculum | No. of foci per root segment at:
|
No. of nodules per root segment | |
|---|---|---|---|
| 3 DAI | 5 DAI | ||
| USDA110 | 9.2 ± 1.7 | 6.2 ± 1.4 | 2.3 ± 0.3 |
| Strain 5051 | 8.6 ± 1.6 | 5.5 ± 2.0 | 1.4 ± 0.4 |
The values shown are the means ± standard deviations from three replicate experiments.
DISCUSSION
The cloned idh gene was the only one annotated in the B. japonicum genome, and its translated N-terminal sequence was identical to that of the same enzyme purified from extracts of cultured cells and of bacteroids (22). The translated amino acid sequence has considerable homology to that of the NADP-dependent isocitrate dehydrogenase from related bacterial species but also to that of soybean. An idh mutant, strain 5051, was created via homologous recombination. Cell extracts of the idh mutant displayed no enzyme activity above background in cell extracts with either NAD+ or NADP+ as the added cofactor or immunological reactivity with antibody raised against purified isocitrate dehydrogenase, indicating that B. japonicum possesses only one isocitrate dehydrogenase-like gene under culture or symbiotic conditions.
The citric acid cycle performs two essential functions in prokaryotes: (i) it provides biosynthetic precursors, and (ii) it generates energy via production of NAD(P)H and FADH2. The loss of NADP-dependent isocitrate dehydrogenase affects these two functions differently in bacteria, typically leading to an inability to use carbon substrates requiring catabolism via the citric acid cycle as sole energy sources for growth. Among the exceptions to this rule are the S. meliloti isocitrate dehydrogenase mutant and B. japonicum citric acid cycle mutants. The S. meliloti isocitrate dehydrogenase mutant was able to utilize mannitol, succinate, and aspartate as sole carbon sources just as well as the wild type and utilizes α-ketoglutarate, malate, pyruvate, and arabinose only slightly less efficiently (27). This unusual growth characteristic was also observed in B. japonicum citric acid cycle mutants. The B. japonicum aconitase (37), α-ketoglutarate dehydrogenase (14), and fumarase (1) mutants were all able to grow at near-wild-type rates and achieve similar culture densities on a variety of citric acid cycle intermediates and four-carbon substrates. The authors of the papers on aconitase (37) and fumarase (1) mutants speculated that B. japonicum may have more than one copy of aconitase and fumarase genes. Subsequent analysis of the genome has not uncovered genuine homologs of acn, and although homologs of fumC are annotated, their function has not been demonstrated. B. japonicum strain 5051 is another exception to the rule that citric acid cycle mutants cannot use citric acid cycle intermediates as the sole source of carbon and energy, as it was able to use malate and succinate for growth, though not as efficiently as the wild type (20 to 30% increase in doubling time).
The citric acid cycle-independent utilization of dicarboxylates implies an alternative pathway for energy generation. One of the alternative metabolic pathways that would allow bypassing of the decarboxylating reactions of the citric acid cycle is the glyoxylate cycle. B. japonicum USDA110 is capable of growth on acetate in culture since it operates the glyoxylate cycle (16). However, strain 5051 grows very poorly on acetate as the sole carbon source, indicating that the glyoxylate cycle is not induced sufficiently to restore growth. Furthermore, the glyoxylate cycle is not induced in bacteroids (16). An isocitrate dehydrogenase mutant may increase the acetate pools and result in the increased PHB content observed in bacteroids.
The absence of isocitrate dehydrogenase means that the product of the reaction, α-ketoglutarate, is not formed. Strain 5051 is a glutamate auxotroph and thus requires a source of glutamate for growth in culture. During symbiosis, the plant may provide sufficient glutamate to permit growth and differentiation (3, 7, 33, 38). Indeed, amino acid uptake may be a part of nutrient cycles that is necessary for nitrogen fixation (25, 32). Plant-supplied glutamate could be metabolized via the γ-aminobutyric acid pathway (33) or transaminated to α-ketoglutarate and metabolized via the CoA-independent α-ketoglutarate decarboxylase activity (17) or the putative oxoglutarate:acceptor oxidoreductase (10). The NADP-dependent isocitrate dehydrogenase enzyme, encoded by gene blr5747, does not provide the substrate for these reactions.
Karr et al. (23) demonstrated that the specific activity of isocitrate dehydrogenase declines during symbiotic development, with the decline beginning at the initiation of nitrogenase activity. This is consistent with the observed phenotype of the isocitrate dehydrogenase mutant, in which nodule colonization is the most notable defect. This suggests that isocitrate dehydrogenase is required for efficient nodule formation but not for the nitrogen fixation process. In planta, the infection and nodule initiation steps proceed normally, albeit somewhat more slowly with strain 5051 than with USDA110, but the resulting nodules are capable of nitrogen fixation. This is in contrast to the isocitrate dehydrogenase mutant of S. meliloti, which forms indeterminate nodules on alfalfa but was ineffective (27). The phenotypes of both the B. japonicum α-ketoglutarate dehydrogenase (14) and the isocitrate dehydrogenase mutants are at the nodule colonization stage of symbiosis. The nodules which are formed have normal levels of acetylene reduction activity when the delayed nodule formation is taken into account. The negligible effect of each mutation on the other enzymes that were measured implies that the effect is “localized” to specific functions during colonization and not a general metabolic effect where cellular function is forced out of balance. The basis of the delay may simply be a nutritional limitation, that is, the availability of α-ketoglutarate. Insufficient α-ketoglutarate may retard nodule develop just enough to cause the discernible delay. The α-ketoglutarate dehydrogenase mutant yielded a more pronounced phenotype than the isocitrate dehydrogenase mutant, suggesting that provision of succinyl-CoA is more critical than the provision of α-ketoglutarate.
Biochemical and genetic evidence collected over the last 40 years has supported a role for organic acids in symbiotic nitrogen fixation. In recent years, site-directed mutations in the enzymes of the citric acid cycle of B. japonicum and other rhizobial species have shown that these enzymes are necessary for nodule development but not necessarily for nodule function, specifically the fixation of atmospheric dinitrogen.
Acknowledgments
We thank Dale Karr, Laura Green, and Nathan Oehrle for helpful discussions.
This work was supported by USDA NRICGP.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Acuna, G., S. Ebeling, and H. Hennecke. 1991. Cloning, sequencing and mutational analysis of the Bradyrhizobium japonicum fumC-like gene: evidence for the existence of two different fumarases. J. Gen. Microbiol. 137:991-1000. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Bergersen, F. J., and G. L. Turner. 1988. Glutamate as a carbon source for N2-fixing bacteroids prepared from soybean root nodules. J. Gen. Microbiol. 134:2441-2448. [Google Scholar]
- 4.Boyer, H. B., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cole, M. A., and G. H. Elkan. 1979. Multiple antibiotic resistance in Rhizobium japonicum. Appl. Environ. Microbiol. 37:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day, D. A., P. S. Poole, S. D. Tyerman, and L. Rosendahl. 2001. Ammonia and amino acid transport across symbiotic membranes in nitrogen-fixing legume nodules. Cell. Mol. Life Sci. 58:185-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, M. F. 1998. Tricarboxylic acid cycle and anapleurotic enzymes in rhizobia. FEMS Microbiol. Rev. 22:105-123. [DOI] [PubMed] [Google Scholar]
- 10.Ebenau-Jehle, C., M. Boll, and G. Fuchs. 2003. 2-Oxoglutarate:NADP+ oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J. Bacteriol. 185:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, C. L., and R. J. Maier. 1991. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 57:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green, L. S., and D. W. Emerich. 1997. Bradyrhizobium japonicum does not require α-ketoglutarate dehydrogenase for growth on succinate and malate. J. Bacteriol. 179:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, L. S., and D. W. Emerich. 1997. The formation of nitrogen-fixing bacteroids is delayed but not abolished in soybean infected by an α-ketoglutarate dehydrogenase-deficient mutant of Bradyrhizobium japonicum. Plant Physiol. 114:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, L. S., and D. W. Emerich. 1999. Light microscopy of early stages in the symbiosis of soybean with a delayed-nodulation mutant of Bradyrhizobium japonicum. J. Exp. Bot. 50:1577-1585. [Google Scholar]
- 16.Green, L. S., D. B. Karr, and D. W. Emerich. 1998. Isocitrate dehydrogenase and glyoxylate cycle enzyme activities in Bradyrhizobium japonicum under various growth conditions. Arch. Microbiol. 169:445-4451. [DOI] [PubMed] [Google Scholar]
- 17.Green, L. S., L. Youshong, D. W. Emerich, F. J. Bergersen, and D. A. Day. 2000. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 182:3298-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, M., and H. Hennecke. 1984. Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 193:46-52. [Google Scholar]
- 19.Hanahan, D. 1983. Studies on transformation of Escherichia coli. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 21.Karr, D. B., and D. W. Emerich. 1988. Uniformity of the microsymbiont population from soybean nodules with respect to buoyant density. Plant Physiol. 86:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karr, D. B., and D. W. Emerich. 2000. Bradyrhizobium japonicum isocitrate dehydrogenase exhibits calcium-dependent hysteresis. Arch. Microbiol. 376:101-108. [DOI] [PubMed] [Google Scholar]
- 23.Karr, D. B., J. K. Waters, F. Suzuki, and D. W. Emerich. 1984. Enyzmes of the poly-β-hydroxybutyrate and citric acid cycles of Rhizobium japonicum bacteroids. Plant Physiol. 75:1158-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenz, O., E. Schwartz, J. Dernedde, M. Eitinger, and B. Friedrich. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 176:4385-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodwig, E. M., A. H. F. Hosie, A. Bourdes, K. Findlay, D. Allaway, R. Karunakaran, J. A. Downie, and P. S. Poole. 2003. Amino-acid cycling drives nitrogen fixation in the legume—Rhizobium symbiosis. Nature 422:722-726. [DOI] [PubMed] [Google Scholar]
- 26.McDermott, T. R., S. M. Griffith, C. P. Vance, and P. H. Graham. 1989. Carbon metablolism in Bradyrhizobium japonicum bacteroids. FEMS Microbiol. Rev. 63:327-340. [Google Scholar]
- 27.McDermott, and M. L. Kahn. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. F. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Oehrle, N. W., D. B. Karr, R. J. Kramer, and D. W. Emerich. 2000. Enhanced attachment of Bradyrhizobium japonicum to soybean through reduced root colonization of internally-seedborne microorganisms. Can. J. Microbiol. 46:600-606. [DOI] [PubMed] [Google Scholar]
- 30.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Ramseier, T. M., and M. Gottfert. 1991. Codon usage and G+C content in Bradyrhizobium japonicum are not uniform. Arch. Microbiol. 156:270-276. [DOI] [PubMed] [Google Scholar]
- 32.Rosendahl, L., M. J. Dilworth, and A. R. Glenn. 1992. Exchange of metabolites across the peribacteroid membrane in pea root nodules. J. Plant Physiol. 139:635-638. [Google Scholar]
- 33.Salminen, S. O., and J. G. Streeter. 1990. Factors contributing to the accumulation of glutamate in Bradyrhizobium japonicum bacteroids under anaerobic conditions. J. Gen. Microbiol. 136:55-61. [Google Scholar]
- 34.Sarma, A. D., and D. W. Emerich. 2005. Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics 5:4170-4184. [DOI] [PubMed] [Google Scholar]
- 35.Schwinghamer, E. A., H. J. Evans, and M. D. Dawson. 1970. Evaluation of effectiveness in mutant strains of Rhizobium by acetylene reduction relative to other criteria of N2 fixation. Plant Soil 33:192-212. [Google Scholar]
- 36.Szewczyk, E., A. Andrianopoulos, M. A. Davis, and M. J. Hynes. 2001. A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADP-dependent isocitrate dehydrogenase in Aspergillus nidulans. J. Biol. Chem. 276:37722-37729. [DOI] [PubMed] [Google Scholar]
- 37.Thöny-Meyer, L., and P. Kunzler. 1996. The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for effective root nodule symbiosis. J. Bacteriol. 178:6166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Udvardi, M. K., C. L. Salom, and D. A. Day. 1988. Transport of l-glutamate across the bacteroid membrane but not the peribacteroid membrane from soybean root nodules. Mol. Plant-Microbe Interact. 1:250-254. [Google Scholar]
- 39.Udvardi, M. K., and D. A. Day. 1997. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:493-523. [DOI] [PubMed] [Google Scholar]
- 40.Vance, C. P., and G. H. Heichel. 1991. Carbon in N2 fixation: limitation or exquisite adaptation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42:373-392. [Google Scholar]
- 41.Waters, J. K., B. L. Hughes, L. C. Purcell, K. O. Gerhardt, T. P. Mawhinney, and D. W. Emerich. 1998. Alanine, not ammonia, is excreted from N2-fixing soybean nodule bacteroids. Proc. Natl. Acad. Sci. USA 95:12038-12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]