Abstract
Chromosomal rearrangements and base substitutions contribute to the large intraspecies genetic diversity of Helicobacter pylori. Here we explored the base excision repair pathway for the highly mutagenic 8-oxo-7,8-dihydroguanine (8-oxoG), a ubiquitous form of oxidized guanine. In most organisms, 8-oxoG is removed by a specific DNA glycosylase (Fpg in bacteria or OGG1 in eukaryotes). In the case where replication of the lesion yields an A/8-oxoG base pair, a second DNA glycosylase (MutY) can excise the adenine and thus avoid the fixation of the mutation in the next round of replication. In a genetic screen for H. pylori genes complementing the hypermutator phenotype of an Escherichia coli fpg mutY strain, open reading frame HP0142, a putative MutY coding gene, was isolated. Besides its capacity to complement E. coli mutY strains, HP0142 expression resulted in a strong adenine DNA glycosylase activity in E. coli mutY extracts. Consistently, the purified protein also exhibited such an activity. Inactivation of HP0142 in H. pylori resulted in an increase in spontaneous mutation frequencies. An Mg-dependent AP (abasic site) endonuclease activity, potentially allowing the processing of the abasic site resulting from H. pylori MutY activity, was detected in H. pylori cell extracts. Disruption of HP1526, a putative xth homolog, confirmed that this gene is responsible for the AP endonuclease activity. The lack of evidence for an Fpg/OGG1 functional homolog is also discussed.
One of the most striking characteristics of Helicobacter pylori, a pathogenic bacterium infecting over 50% of the human population and associated with gastritis, peptic ulcer, and gastric cancer (10, 12), is its enormous intraspecies genetic variability (39). At the origin of such diversity are both mutations and recombination events. During the course of infection of the gastric mucosa, H. pylori induces an oxidative stress that can affect not only the host tissue but also the bacteria. Therefore, H. pylori must cope with the reactive oxygen species (ROS) generated as part of the host inflammatory response. We have shown previously that H. pylori strains defective in the repair of oxidized pyrimidines are more sensitive to the mutagenic and lethal effects of activated macrophages. Furthermore, strains unable to repair oxidized pyrimidines show attenuated colonization capacity, demonstrating that, during gastric infection by H. pylori, the host effectively induces lethal and premutagenic oxidative lesions on the pathogen genome (26). Oxygen radicals are produced not only by the host inflammatory response but also as by-products of the normal metabolism of all cells. Among the main lesions produced in DNA by ROS is an oxidized form of guanine, 8-oxo-7,8-dihydroguanine (8-oxoG), with a strong mutagenic potential. Indeed, when present in the template strand, this modified base induces the incorporation of an adenine opposite it during DNA replication, leading to G:C-to-T:A transversions (13, 31). Confirmation for this mutational mechanism comes from bacterial genetics experiments that unveiled a sophisticated DNA repair system responsible for the avoidance of 8-oxoG-induced mutations. This so-called GO system, first characterized for Escherichia coli, involves base excision repair (BER) processes initiated by two distinct DNA glycosylases. If an 8-oxoG is formed on DNA, the oxidized base present in an 8-oxoG:C pair can be recognized and excised by Fpg. If, however, replication proceeds before removal of the 8-oxoG and adenine is incorporated instead of cytosine, the resulting A:8-oxoG base pair is the substrate for another DNA glycosylase, MutY, capable of removing the normal base when opposite 8-oxoG, avoiding in such a way the fixation of the mutation and, by recreating the 8-oxoG:C pair, allowing Fpg a new opportunity to remove the lesion (21, 22). The importance of such a system and therefore of the mutagenic potential of 8-oxoG is confirmed by the spontaneous mutator phenotypes, due exclusively to G:C-to-T:A transversions, of strains deficient in either DNA glycosylase, Fpg or MutY. Moreover, disabling both fpg and mutY genes has a synergistic effect on the spontaneous mutation frequency, again by exclusive increase of G:C-to-T:A changes, leading to one of the strongest mutator phenotypes described for E. coli.
The fpg gene was identified first as coding for a formamidopyrimidine (Fapy) DNA glycosylase from E. coli (3) and then as a mutator gene (mutM) (5, 23). Biochemical and genetic experiments showed that the main cellular substrate for Fpg is indeed the 8-oxoG:C pair (2, 20). The MutY (MicA) activity was first identified both genetically in a screen (29) and biochemically in cellular extracts (17, 18) as an activity capable of excising adenine from A:G mismatches independently of MutS, MutL, or MutH functions. At the same time, a new mutator gene, named mutY, was isolated by a genetic screen (24). This gene was shown to code for the activities described before (36). It was demonstrated later that the relevant substrate for MutY is the adenine in an A:8-oxoG pair (20).
Surprisingly, in neither of the Helicobacter pylori strains whose complete genomic sequences are available (1, 32) has a homolog of fpg or its eukaryote functional homolog OGG1 been found. With the aim of assessing the impact of 8-oxoG on Helicobacter pylori genetic stability, we investigated here, by biochemical and genetic means, the presence of the pathogen BER activities capable of avoiding the mutagenic effects of 8-oxoG.
(A preliminary description of the screen used in this work and the MutY protein was presented previously [27a].)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains CC104 (8) and its derivative PR180 (CC104 mutY::Kanr) (29) were grown in Luria-Bertani (LB) broth or agar supplemented with 200 μg/ml ampicillin and/or 100 μg/ml rifampin. H. pylori strains used were SS1 (a kind gift from Agnès Labigne, Institut Pasteur, Paris, France) and X47-2AL (16) and its derivatives X47-2AL nucT (27), X47-2AL mutY, and X47-2AL nucT xth (this work). Disruption mutants were generated by insertion of kanamycin or apramycin resistance cassettes. H. pylori strains were grown on blood or horse serum brucella agar plates at 37°C in a 5% CO2 and 95% humidity atmosphere. Disruption mutants of H. pylori strains were cultured with 20 μg/ml of kanamycin and/or 25 μg/ml of apramycin. Mutagenesis assays were done on horse serum brucella agar plates supplemented with 20 μg/ml rifampin, as described previously (26).
Plasmid and library construction.
The HP0142 gene, with its own start codon, was amplified by PCR from H. pylori genomic DNA by use of oligonucleotide primers deduced from the H. pylori strain 26695 genome sequence (5′-GAAGATCTCTGGAAACTTTACACAACGC and 5′-GGAATTCACCCCCAAATAAATTTTTTT). The amplified DNA product was cloned into pGEX-4T-1 previously digested with BamHI and EcoRI, resulting in plasmid pGEX-HP0142. In this system, the HP0142 polypeptide is produced as a fusion protein with glutathione S-transferase (GST) at its N terminus. In order to create an expression library, H. pylori genomic DNA digested with EcoRI was ligated into a Lambda ZapII vector (Stratagene).
Complementation assays with E. coli PR180.
E. coli PR180 was transformed with pGEX-4T-1 or pGEX-HP0142. For mutagenesis assays, overnight cultures were diluted in order to start 10 2-ml cultures from each genotype with 1,000 to 5,000 cells/ml inocula. Cultures were grown overnight at 37°C in the presence of 50 μM IPTG (isopropyl-β-d-thiogalactopyranoside). A portion (200 μl) of the cultures was plated onto LB-rifampin plates, and serial dilutions were plated onto LB plates to determine viable cells. Mutation rates were calculated using the median method (15).
Overproduction and purification of HP0142.
E. coli PR180 carrying plasmid pGEX-HP0142 was used to inoculate 500 ml of LB medium supplemented with ampicillin and was incubated at 37°C with shaking until the A600 reached 1.5. IPTG was added to 50 μM, and growth was continued overnight at 18°C with gentle shaking. Cells were harvested by centrifugation and lysed by lysozyme treatment and sonication. GST-HP0142 was purified by GST affinity chromatography. The eluted protein was dialyzed against 20 mM Tris-HCl, pH 8, 5 mM β-mercaptoethanol, and 10% glycerol.
DNA substrates and enzymatic assays.
Oligodeoxyribonucleotides (34-mer) containing at position 16 a single 8-oxoG residue (a kind gift of Jean Cadet, CEA-Grenoble), a tetrahydrofuranyl (THF) residue (Eurogentec), or an adenine were labeled at their 5′end with [γ-32P]ATP (3,000 Ci/mmol; Amersham) and T4 polynucleotide kinase (New England Biolabs). The 32P-labeled strands were hybridized with a complementary sequence containing a cytosine (C) opposite 8-oxoG or THF or containing an 8-oxoG opposite adenine 16, yielding duplexes 8-oxoG:C, THF:C, and A:8-oxoG:C, respectively.
In a standard reaction (10-μl final volume), 50 fmol of labeled duplex was incubated in reaction buffer (25 mM Tris-HCl, pH 7.6, 2 mM Na2EDTA, 50 mM NaCl) with the indicated protein fraction (either total cell extracts or purified proteins) at 37°C. Where stated, the reaction was stopped by adding NaOH to 0.2 N and the reaction mixture was incubated for another 10 min at 37°C. After addition of 6 μl of formamide dye, the products were separated by 7 M urea-20% polyacrylamide gel electrophoresis. Gels were analyzed using a Molecular Dynamics PhosphorImager.
RESULTS
Screen for H. pylori functions complementing the hypermutator phenotype of an E. coli fpg mutY strain.
In E. coli, inactivation of both fpg and mutY leads to a strong hypermutator phenotype due to the incapacity of the cells to avoid the mutagenic effects of 8-oxoG. We therefore screened an H. pylori genomic library for genes capable of reducing the spontaneous mutation frequencies of E. coli strain BH1190, a CC104 derivative deficient in Fpg, MutY, and RecA functions (BH1190). CC104 is a LacZ-deficient strain in which the lac-negative phenotype can be reverted only by a G-to-T transversion (8). Using a papillation assay (24), we looked for H. pylori genes whose expression lowered the reversion frequency of the CC104 lacZ allele. The recA mutation was introduced to lower the level of false positives in the screen (37). Ninety-eight clones with lower papillation frequencies were selected. Each of them was then tested individually for its lacZ reversion frequency. Three of them displayed consistently lower transversion rates. Sequencing of the plasmids revealed that one carried a putative thioredoxin reductase gene while the other two carried open reading frame (ORF) HP0142, coding for a putative MutY gene. Crude cell extracts were prepared from the other 95 E. coli clones, and their Fapy DNA glycosylase activities were tested. None of them displayed the capacity to excise Fapy residues, a specific substrate for an Fpg functional homolog.
HP0142 codes for a MutY functional homolog.
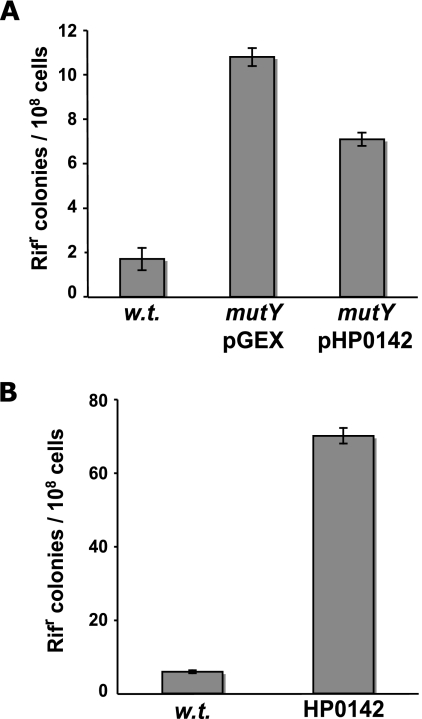
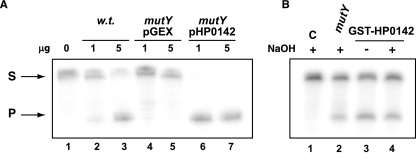
The protein encoded by HP0142, depending on the H. pylori strain analyzed, shares 34 to 39% identity and 53 to 58% similarity to the MutY protein from E. coli. In particular, the active site key residues as well as the iron-sulfur cluster are well conserved. To analyze whether this ORF codes for a protein with MutY activity, HP0142 fused to GST was expressed in E. coli strain PR180, where the mutY gene was disrupted. We analyzed the capacity of HP0142 to complement the mutator phenotype of MutY-deficient E. coli by comparing the spontaneous mutation rates of mutY strains expressing either GST or GST fused to the H. pylori HP0142 protein. As shown in Fig. 1A, expression of GST-HP0142 in this strain partially complemented the spontaneous mutator phenotype resulting from inactivation of mutY in E. coli. To confirm that HP0142 is indeed a mutY homolog, we prepared cellular extracts from PR180 expressing either GST or GST-HP0142 fusion proteins and tested them for adenine DNA glycosylase activity on an oligonucleotide substrate carrying an A/8-oxoG mismatch. Figure 2A confirms that the adenine DNA glycosylase activity present in wild-type extracts is completely eliminated by disruption of mutY (Fig. 2A, compare lanes 4 and 5 with lanes 2 and 3). However, expression of the GST-HP0142 fusion protein resulted in a strong A:8-oxoG adenine DNA glycosylase activity in the extracts (Fig. 2A, lanes 6 and 7).
FIG. 1.
HP0142 gene product codes for an antimutator activity. (A) Spontaneous mutation frequencies of E. coli wild-type (w.t.) or mutY strains carrying the indicated plasmids. (B) Spontaneous mutation frequencies of wild-type or HP0142 X47-2AL H. pylori strains. Shown are medians ± standard deviations.
FIG. 2.
Adenine DNA glycosylase activities of HP0142. DNA glycosylase activities were determined using a 34-mer oligonucleotide carrying an A/8-oxoG pair at position 16. The strand labeled is the one carrying the A. The products of the reaction (P) were separated from the substrates (S) by denaturing gel electrophoresis. (A) Activities of the indicated amounts of cell extracts from E. coli wild-type (w.t.) (lanes 2 and 3) or mutY strains (lanes 4 to 7) carrying the vector (lanes 4 and 5) or the HP0142-expressing plasmid (lanes 6 and 7). (B) Activities of the HP0142 purified protein. Reactions were stopped with (lanes 2 and 4) or without (lane 3) NaOH. C, control.
The GST-HP0142 fusion peptide was purified from E. coli strain PR180 pGEX-HP0142. The purified protein displayed an adenine DNA glycosylase activity on duplex DNA harboring an A:8-oxoG base pair (Fig. 2B, lane 4). The cleavage of the oligonucleotide was observed even when the chemical cleavage by NaOH at the end of the reaction was omitted (Fig. 2B, lane 3), suggesting that HP0142 possesses an AP lyase activity in addition to its DNA glycosylase activity. The biochemical results presented above are in agreement with HP0142 being the H. pylori MutY homolog.
H. pylori MutY is an antimutator.
To explore the in vivo function of H. pylori MutY, we created an H. pylori X47 strain where mutY was inactivated by disruption of HP0142 with a kanamycin resistance cassette. Inactivation of H. pylori mutY resulted in a 12-fold increase in the spontaneous mutation rate compared to that for the parental strain (Fig. 1B). Taken together, the biochemical and genetic evidence shows that HP0142 codes for a MutY homolog and participates in maintaining the genetic stability of H. pylori strains.
Search for Fpg-like enzymatic activities in H. pylori cell extracts.
As mentioned above, examination of the complete genomic sequences of H. pylori, as well as the results from our screen, failed to reveal ORFs potentially coding for an Fpg-like enzyme. To search for an Fpg activity, we prepared total cell extracts from strain X47-2AL nucT. NucT protein is a major DNA nuclease from H. pylori that impedes the use of DNA substrates for reactions involving extracts (27). A 32P-5′-end-labeled oligonucleotide with an 8-oxoG residue opposite a cytosine at position 16 was used as the substrate in X47-2AL nucT extracts. No significant 8-oxoG DNA glycosylase activity could be detected under standard reaction conditions, even at high protein concentrations (data not shown). Various salt concentrations did not reveal any activity on the 8-oxoG/C containing substrate (data not shown). Moreover, no DNA glycosylase activity was detected in cellular extracts on DNA substrates carrying Fapy modifications, another substrate for the Fpg and OGG1 family DNA glycosylases (data not shown). Fpg and OGG1 family 8-oxoG DNA glycosylases characterized so far form a Schiff base intermediate with the AP site generated by excision of 8-oxoG. This transient covalent complex can be stabilized by reducing agents. With the aim of identifying putative 8-oxoG DNA glycosylases/AP lyases, we therefore performed NaBH4 trapping experiments on H. pylori extracts by using DNA duplexes with 8-oxoG:C base pairs. Under conditions where both Fpg and OGG1 form covalent complexes, we could not detect covalent complexes in H. pylori total cell extracts (data not shown). Taken together, these results suggest that there are no enzymatic activities in H. pylori capable of excising 8-oxoG from DNA under standard reaction conditions used for the characterized 8-oxoG DNA glycosylases.
AP endonuclease activity.
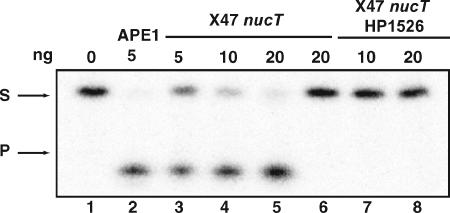
We then searched for the activity responsible for the cleavage of the AP site, the enzymatic step in BER that follows the excision of the base. For that purpose, we used an oligonucleotide substrate carrying an AP site analog, THF. Two main families of AP endonucleases have been described, corresponding to exonuclease III (Xth) and to endonuclease IV (Endo IV) (Nfo) from E. coli (9). The former requires Mg ions for activity, while the latter can cleave AP sites in the absence of the divalent cation. Because only an xth homolog was detected after sequencing of the H. pylori genome, we first tested the AP cleavage activity in cell extracts in the presence of Mg2+. As seen in Fig. 3, lanes 3 to 5, an efficient AP endonuclease activity is unveiled under such conditions. When parallel reactions were carried out in the presence of the Mg chelator EDTA, no detectable cleavage was observed, even at the highest extract concentration (Fig. 3, lane 6).
FIG. 3.
Mg-dependent AP endonuclease activities in H. pylori X47 nucT and X47 nucT HP1526. Activities were determined on the indicated amounts of total cell extracts from H. pylori X47 nucT (lanes 3 to 6) or X47 nucT HP1526 (lanes 7 and 8) by use of a 34-mer oligonucleotide harboring the AP site analog (F) at position 16 of the labeled strand. Reactions were carried out in the presence (lanes 1 to 5 and 7 to 8) or absence (lane 6) of Mg ions. Reaction products were analyzed as described in the legend for Fig. 2. Lane 2 corresponds to the activity of the human APE1 protein as a control.
ORF HP1526 codes for a 250-amino-acid protein with 28 to 30% identity and 47 to 49% similarity to the Xth protein from E. coli. To analyze whether this ORF codes for a protein with Xth activity, we created an H. pylori X47-2AL nucT strain where HP1526 was inactivated by disruption with an apramycin resistance cassette. As seen in Fig. 3, lanes 7 and 8, no AP endonuclease activity was detected in cell extracts from this strain, even in the presence of Mg2+. This result provides evidence for HP1526 being the H. pylori Xth homolog.
DISCUSSION
A large proportion of premutagenic DNA lesions in cells arise by the action of ROS generated either by the normal metabolism or by environmental agents. In particular, pathogenic bacteria are exposed during infection to ROS generated by the host as part of the defense response. It has been suggested previously that the induction of mutations observed with Salmonella enterica serovar Typhimurium cells exposed to phagocytes (40), as well as the reduced virulence of S. enterica serovar Typhimurium recombination mutants (4, 6), could be due to oxidative DNA damage in the bacteria. Consistent with the induced mutagenesis, experiments with E. coli showed that exposure to phagocytes induces oxidative DNA damage and, more specifically, 8-oxoG, a highly mutagenic lesion (30). More recently, by use of H. pylori defective in Nth, the DNA glycosylase responsible for the removal of oxidized pyrimidines, it was shown that DNA repair of oxidative DNA damage is necessary to remove macrophage-induced mutagenic and lethal lesions. Consistent with that observation, the ability to repair oxidative damage in H. pylori DNA was shown to be essential for bacterial persistence in the infected mouse stomach (26). In the present work, we explored the H. pylori DNA repair mechanisms involved in the avoidance of mutations induced by 8-oxoG. It is interesting to note that, from the genetic screen for an H. pylori antimutator activity in an E. coli fpg mutY doubly defective strain, we isolated a thioredoxin reductase gene, confirming the importance of the endogenously generated ROS in the formation of mutagenic DNA lesions. The other two candidates corresponded to vectors carrying ORF HP0142 coding for a putative MutY protein. Expression of HP0142 in E. coli was able to partially complement the mutator phenotype resulting from disabling the endogenous mutY gene. The biochemical experiments showing the adenine DNA glycosylase activity on A:8-oxoG pairs of the protein encoded by HP0142 confirmed that this open reading frame corresponds to the H. pylori mutY gene. More importantly, the disruption of H. pylori mutY resulted in a hypermutator phenotype, demonstrating the role of H. pylori MutY in a physiological mutation avoidance mechanism. A puzzling observation was the absence of a detectable H. pylori MutY activity in wild-type H. pylori extracts although H. pylori MutY expression in E. coli resulted in a very strong DNA glycosylase activity in extracts prepared under essentially the same conditions (data not shown). Some possible explanations are that MutY is present at very low levels under our culture conditions or that H. pylori possesses specific chemical or protein inhibitors. It would be interesting to determine the expression pattern of the gene at the mRNA or protein level.
In BER, once the base is removed by the specific DNA glycosylase, the resulting AP site is recognized and cleaved by an AP endonuclease. Here we show that such an activity is present in H. pylori extracts (Fig. 3). Consistent with the predicted presence of a gene coding for a homolog of exonuclease III (HP1526) (1, 32), the activity observed requires the presence of Mg ions in the reaction mixture. In the presence of EDTA no AP endonuclease activity was detected, suggesting that, unlike in E. coli, there is no member of the AP endonuclease Endo IV family. It would seem that for this enzymatic step, as for other DNA repair (25, 26) or metabolic (7) functions, H. pylori possesses a reduced set of proteins with little redundancy. Completion of BER could be assured by the predicted DNA polymerase I (HP1470) and DNA ligase (HP0615).
The experiments described in this work failed to detect a DNA glycosylase capable of eliminating 8-oxoG from H. pylori genomic DNA. Although this is consistent with the lack of putative Fpg or OGG1 coding sequences in the genome, it raises the question of the elimination of this ubiquitous mutagenic DNA lesion. The presence of a functional MutY homolog strongly suggests that H. pylori is no exception and that 8-oxoG is indeed formed in its DNA. A similar situation is found for the fission yeast Schizosaccharomyces pombe, where a homolog of MutY has been characterized previously (19) but no putative OGG1 or Fpg coding genes were present and neither was an 8-oxoG DNA glycosylase activity found in cell extracts (S. Boiteux, personal communication). It seems unlikely that 8-oxoG residues accumulate indefinitely and are transmitted to the progeny. Although the assays performed here and the analysis of the complete genomic sequences point to an absence of an H. pylori 8-oxoG DNA glycosylase, other, so-far-unknown enzymatic mechanisms could be responsible for the elimination of the oxidized guanine. A precedent is the recent determination of the AlkB activity capable of reversing alkylation DNA damage in E. coli (11, 35). In H. pylori, an interesting candidate to be involved in the repair of 8-oxoG is the protein MutS2. It was recently reported that, besides its role in recombination (28), this protein has affinity for 8-oxoG-containing DNA and that H. pylori deficient in this protein shows a strong induced mutagenesis with a bias towards G:C-to-T:A transversions when exposed to an oxidative stress and accumulates threefold-higher levels of 8-oxoG under such conditions (38). However, the mechanism by which MutS2 can initiate the repair of 8-oxoG remains to be established. An alternative for the elimination of 8-oxoG is that this base is further oxidized to yield a new DNA modification whose repair can be carried out by another DNA glycosylase (14, 33, 34).
Acknowledgments
We thank Serge Boiteux (CEA-Fontenay aux Roses) for his support and communication of unpublished data. We are grateful to Agnès Labigne and Eliette Touati (Institut Pasteur, Paris) for sharing bacterial strains and fruitful discussions, A-Lien Lu (University of Maryland) for analysis of the MutY sequence, and Stéphanie Marsin for her critical reading of the manuscript. We also thank Patricia Auffret van der Kemp (CEA-Fontenay aux Roses) for the Fpg and APE1 proteins. The CC104 strain was a kind gift from Claire Cupples.
This work was supported by the ECOS-Sud program, the CEA, the CNRS, and a grant from the Agence Nationale pour la Recherche (HPGENDIV to J.P.R.). E.J.O. was a recipient of a Fundación Antorchas fellowship.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Bessho, T., K. Tano, H. Kasai, and S. Nishimura. 1992. Deficiency of 8-hydroxyguanine DNA endonuclease activity and accumulation of the 8-hydroxyguanine in mutator mutant (mutM) of Escherichia coli. Biochem. Biophys. Res. Commun. 188:372-378. [DOI] [PubMed] [Google Scholar]
- 3.Boiteux, S., T. R. O'Connor, and J. Laval. 1987. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 6:3177-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., C. J. Lipps, M. Y. So, and F. Heffron. 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7:933-936. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera, M., Y. Nghiem, and J. H. Miller. 1988. mutM, a second mutator locus in Escherichia coli that generates G · C → T · A transversions. J. Bacteriol. 170:5405-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cano, D. A., M. G. Pucciarelli, F. Garcia-del Portillo, and J. Casadesus. 2002. Role of the RecBCD recombination pathway in Salmonella virulence. J. Bacteriol. 184:592-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 11.Falnes, P. O., R. F. Johansen, and E. Seeberg. 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419:178-182. [DOI] [PubMed] [Google Scholar]
- 12.Ge, Z., and D. E. Taylor. 1999. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu. Rev. Microbiol. 53:353-387. [DOI] [PubMed] [Google Scholar]
- 13.Grollman, A. P., and M. Moriya. 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9:246-249. [DOI] [PubMed] [Google Scholar]
- 14.Hazra, T. K., J. G. Muller, R. C. Manuel, C. J. Burrows, R. S. Lloyd, and S. Mitra. 2001. Repair of hydantoins, one electron oxidation product of 8- oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 29:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lea, D., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 16.Londono-Arcila, P., D. Freeman, H. Kleanthous, A. M. O'Dowd, S. Lewis, A. K. Turner, E. L. Rees, T. J. Tibbitts, J. Greenwood, T. P. Monath, and M. J. Darsley. 2002. Attenuated Salmonella enterica serovar Typhi expressing urease effectively immunizes mice against Helicobacter pylori challenge as part of a heterologous mucosal priming-parenteral boosting vaccination regimen. Infect. Immun. 70:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, A. L., and D. Y. Chang. 1988. Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics 118:593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, A. L., and D. Y. Chang. 1988. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell 54:805-812. [DOI] [PubMed] [Google Scholar]
- 19.Lu, A. L., and W. P. Fawcett. 1998. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J. Biol. Chem. 273:25098-25105. [DOI] [PubMed] [Google Scholar]
- 20.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 23.Michaels, M. L., L. Pham, C. Cruz, and J. H. Miller. 1991. MutM, a protein that prevents G.C—T.A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 19:3629-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nghiem, Y., M. Cabrera, C. G. Cupples, and J. H. Miller. 1988. The mutY gene: a mutator locus in Escherichia coli that generates G.C—T.A transversions. Proc. Natl. Acad. Sci. USA 85:2709-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Rourke, E. J., C. Chevalier, S. Boiteux, A. Labigne, L. Ielpi, and J. P. Radicella. 2000. A novel 3-methyladenine DNA glycosylase from Helicobacter pylori defines a new class within the endonuclease III family of base excision repair glycosylases. J. Biol. Chem. 275:20077-20083. [DOI] [PubMed] [Google Scholar]
- 26.O'Rourke, E. J., C. Chevalier, A. V. Pinto, J. M. Thiberge, L. Ielpi, A. Labigne, and J. P. Radicella. 2003. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. USA 100:2789-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Rourke, E. J., A. V. Pinto, E. A. Petroni, M. E. Tolmasky, and L. Ielpi. 2004. Evidence for the active role of a novel nuclease from Helicobacter pylori in the horizontal transfer of genetic information. J. Bacteriol. 186:2586-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.O'Rourke, E. J., A. Mathieu, L. Ielpi, and J. P. Radicella. 2003. Genetic variability and DNA repair: base excision repair activities in Helicobacter pylori. Genome Lett. 2:41-47. [Google Scholar]
- 28.Pinto, A. V., A. Mathieu, S. Marsin, X. Veaute, L. Ielpi, A. Labigne, and J. P. Radicella. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17:113-120. [DOI] [PubMed] [Google Scholar]
- 29.Radicella, J. P., E. A. Clark, and M. S. Fox. 1988. Some mismatch repair activities in Escherichia coli. Proc. Natl. Acad. Sci. USA 85:9674-9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlosser-Silverman, E., M. Elgrably-Weiss, I. Rosenshine, R. Kohen, and S. Altuvia. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 182:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibutani, S., M. Takeshita, and A. P. Grollman. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349:431-434. [DOI] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.Tretyakova, N. Y., J. C. Niles, S. Burney, J. S. Wishnok, and S. R. Tannenbaum. 1999. Peroxynitrite-induced reactions of synthetic oligonucleotides containing 8-oxoguanine. Chem. Res. Toxicol. 12:459-466. [DOI] [PubMed] [Google Scholar]
- 34.Tretyakova, N. Y., J. S. Wishnok, and S. R. Tannenbaum. 2000. Peroxynitrite-induced secondary oxidative lesions at guanine nucleobases: chemical stability and recognition by the Fpg DNA repair enzyme. Chem. Res. Toxicol. 13:658-664. [DOI] [PubMed] [Google Scholar]
- 35.Trewick, S. C., T. F. Henshaw, R. P. Hausinger, T. Lindahl, and B. Sedgwick. 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419:174-178. [DOI] [PubMed] [Google Scholar]
- 36.Tsai-Wu, J. J., J. P. Radicella, and A. L. Lu. 1991. Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY. J. Bacteriol. 173:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Kemp, P. A., D. Thomas, R. Barbey, R. de Oliveira, and S. Boiteux. 1996. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl. Acad. Sci. USA 93:5197-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, G., P. Alamuri, M. Z. Humayun, D. E. Taylor, and R. J. Maier. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 39.Wang, G., M. Z. Humayun, and D. E. Taylor. 1999. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 7:488-493. [DOI] [PubMed] [Google Scholar]
- 40.Weitzman, S. A., and T. P. Stossel. 1981. Mutation caused by human phagocytes. Science 212:546-547. [DOI] [PubMed] [Google Scholar]