Abstract
PcrA is a chromosomally encoded DNA helicase of gram-positive bacteria involved in replication of rolling circle replicating plasmids. Efficient interaction between PcrA and the plasmid-encoded replication initiator (Rep) protein is considered a requirement for the plasmid to replicate in a given host, and thus, the ability of a Rep protein to interact with heterologous PcrA helicases has been invoked as a determinant of plasmid promiscuity. We characterized transcription of the Streptococcus pneumoniae pcrA gene in its genetic context and studied the biochemical properties of its product, the PcrASpn helicase. Transcription of the pneumococcal pcrA gene was directed by promoter Pa, consisting of an extended −10 box. Promoter Pa also accounted for expression of a second essential gene, radC, which was transcribed with much lower efficiency than pcrA, probably due to the presence of a terminator/attenuator sequence located between the two genes. PcrASpn displayed single-stranded DNA-dependent ATPase activity. PcrASpn showed 5′→3′ and 3′→5′ helicase activities and bound efficiently to partially duplex DNA containing a hairpin structure adjacent to a 6-nucleotide 5′ or 3′ single-stranded tail and one unpaired (flap) nucleotide in the complementary strand. PcrASpn interacted specifically with RepC, the initiator of staphylococcal plasmid pT181. Although the pneumococcal helicase was able to initiate unwinding of the RepC-nicked pT181 DNA, it was much less processive in this activity than the cognate staphylococcal PcrA protein. Accordingly, PcrASpn was inefficient in in vitro replication of pT181, and perhaps as a consequence, this plasmid could not be established in S. pneumoniae.
DNA helicases are essential enzymatic motors that unwind double-stranded DNA (dsDNA) during various metabolic processes, such as replication, recombination, repair, and transcription (for reviews, see references 6, 16, 29, and 32). Helicases have been classified into different groups based on amino acid sequence comparisons. The majority of the helicases belong to two superfamilies, designated superfamilies SF1 and SF2, both of which include proteins encoded by prokaryotic, eukaryotic, and viral genes (14, 15). Helicases belonging to these two families contain seven conserved motifs distributed along the sequence with similar structures and arrangements, encompassing amino acid residues important for the helicase functions. Three of the best-characterized bacterial helicases are the Escherichia coli Rep and UvrD proteins (belonging to SF1) and DnaB, which has been placed in a separate family (family F4) (14). DnaB is the main replicative helicase and operates during the theta-type replication of the E. coli chromosome and many bacterial plasmids (24). Although not essential for the bacterium, Rep is required for replication of the single-stranded coliphages φX174 and M13 (44), whereas UvrD participates in DNA repair and in various recombination pathways (5) and is the replicative helicase used by various rolling circle replicating (RCR) plasmids in E. coli (7). In the case of gram-positive bacteria, the PcrA helicase is the best-characterized helicase at the biochemical and structural levels (48). The pcrA gene has been identified in the chromosomes of several gram-positive microorganisms, such as Bacillus subtilis, Bacillus stearothermophilus, Lactococcus lactis, Staphylococcus aureus, and Streptococcus pyogenes, and its involvement in replication of RCR plasmids was demonstrated previously (18). The pcrA gene has been found to be essential for cell viability in S. aureus and B. subtilis (35) and is required for replication of the staphylococcal RCR plasmids pT181, pC194, and pE194 in both of these hosts (9, 17, 35, 41). PcrA belongs to the SF1 superfamily of helicases and was the first SF1 helicase whose structure was solved (6, 43). PcrA is one of the few helicases that act as a monomer, in contrast to the more common replicative helicases, which are hexameric (6, 16, 29, 42, 48). PcrA has been shown to bind to single-stranded DNAs (ssDNAs), as well as to dsDNAs (13, 42).
The S. aureus pcrA gene encodes a 730-amino-acid protein (17, 18). Purified staphylococcal PcrA protein (PcrASau) showed ATPase activity which was stimulated by the presence of ssDNA, and it had both 5′→3′ and 3′→5′ helicase activities (2). Although the precise role of PcrASau in cell viability is still unknown, the ability of this protein to recognize DNA substrates with specific structures, coupled with its bipolar helicase activity, may be a key to understanding its precise role in different cellular processes (2). Physical interaction between PcrASau and pT181-encoded replication initiator protein RepC has been postulated to be essential for the initiation of rolling circle replication (9). PcrASau protein was shown to be recruited to the plasmid origin through its interaction with the initiator protein (RepC for pT181 and RepD for pC221), and this interaction could initiate unwinding of the DNA at the Rep-generated nick (9, 41). As in other RCR plasmids, the RepC nick site is located in the loop of a hairpin structure within the double-strand origin of replication, dso (11, 19, 20, 23), and nicking of the DNA leads to opening of the strands and generation of a 5′ ssDNA region. The ability of PcrASau to recognize folded structures that are present in the pT181-dso and its 5′→3′ helicase activity are consistent with the role of PcrA in unwinding the DNA to facilitate progression of the replication fork during plasmid rolling circle replication (2).
We studied expression of the pneumococcal pcrA gene at the transcriptional level and found that (i) the pneumococcal pcrA gene is the first gene of a bicistronic operon and (ii) transcription of a second essential gene appears to be regulated by a transcription terminator located between the two genes. A well-known feature of many RCR plasmids is their ability to become established and to replicate in a number of different bacteria. One plausible reason for such promiscuity is the existence of a functional interaction between the plasmid initiators and the heterologous PcrA proteins. To examine this possibility, we purified the 763-amino-acid Streptococcus pneumoniae PcrA (PcrASpn) protein from strain R6 and studied its biochemical activities and its ability to replace PcrASau in in vitro replication of plasmid pT181 and in unwinding of RepC-nicked pT181 DNA. Purified PcrASpn protein was active as a 5′→3′ and 3′→5′ DNA helicase. In addition, the protein displayed maximal helicase activity with substrates containing folded structures along with 5′ or 3′ single-stranded tails. Since PcrASpn exhibited helicase activities similar to those of PcrASau, we tested whether the pneumococcal helicase was also able to interact with the pT181 initiator protein RepC. Pull-down assays revealed a direct physical interaction between the RepC and PcrASpn proteins. However, PcrASpn was unable to support in vitro replication of pT181 DNA to the same extent as the cognate PcrASau protein. Moreover, PcrASpn was unable to extensively unwind RepC-nicked pT181 DNA in vitro. The failure to establish pT181 in S. pneumoniae suggested that this staphylococcal plasmid was unable to replicate in this bacterium, in agreement with the apparent inability of the RepC-PcrASpn complex to efficiently unwind the pT181 DNA. In contrast to these results, heterologous PcrA helicases from Bacillus cereus PcrABcer and Bacillus anthracis PcrABan (two species in which the pT181 replicon could be established) are known to extensively unwind RepC-nicked pT181 DNA and support pT181 replication in vitro (3). We propose that a functional interaction between the plasmid-encoded replication initiator protein and the host PcrA helicase is essential for establishment and replication of RCR plasmids in gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmid construction.
Strains and plasmids used in this study are listed in Table 1. E. coli cultures were grown in TY medium (40) with selection for kanamycin resistance (25 μg/ml) and/or ampicillin resistance (50 μg/ml). S. aureus cells were grown in brain heart infusion medium (Difco).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli M15 | K-12 derivative | Qiagen |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S. pneumoniae R6 | Wild type | 27 |
| S. aureus RN4220 | Restriction-deficient mutant of strain 8325-4 | 25 |
| S. aureus RN4220 pcrA3 | pcrA3 mutant | 17 |
| Plasmids | ||
| pT181-cop608 | Copy mutant of pT181 | 38 |
| pCR2.1 | pUC Plac lacZα pT7 neo bla f1-ori | Invitrogen |
| pCR2.1C | pCR2.1 cat | This study |
| pCR2.11 | pCR2.1 Δ3′pcrA | This study |
| pCR2.12 | pCR2.1 Δ5′pcrA | This study |
| pCR2.13 | pCR2.1 Δ5′3′pcrA | This study |
| pCR2.1C1 | pCR2.1 cat Δ3′pcrA | This study |
| pCR2.1C2 | pCR2.1 cat Δ5′pcrA | This study |
| pCR2.1C3 | pCR2.1 cat Δ5′3′pcrA | This study |
| pCR2.1PN | pCR2.1 pcrASpn | This study |
| pQE30 | QIAGEN | |
| pQE30PA | pQE30 His-pcrASau | 9 |
| pQE30PN | pQE30 His-pcrASpn | This study |
| pSK1377 | MBP-wild-type RepC | 49 |
S. pneumoniae cells were grown in AGCH medium (26) supplemented with 0.3% sucrose and 0.2% yeast extract with selection for resistance to chloramphenicol (2 μg/ml) when required. All cultures were grown at 37°C. The procedures used for development of competence and transformation with plasmid DNAs have been described previously (27).
Plasmids used in this work were constructed as follows. For pCR2.1PN, a DNA fragment containing the pcrA gene, including the Pa promoter, was amplified by PCR using the chromosomal DNA from S. pneumoniae R6 as the template and primers INS1 (5′-TGCTACTCCCCGTAAAGTTT-3′) and pcrA2 (5′-CGGGGTACCCCGGTACATAAAAATCCTCCTCA-3′). The 2,347-bp PCR fragment was then ligated into the pCR2.1 vector by means of the TOPO-TA cloning technology (Invitrogen).
For pQE30PN, the pcrA gene was amplified by PCR from the pneumococcal R6 strain. The sequences of the primers were 5′-ACTGGGTACCAACGCATTATTAAATGGAATGAATGACCGT-3′ for the forward primer and 5′-ACTGGGTACCGATTTTTTTCTCAATTGGAGCCACACTGGC-3′ for the reverse primer. The PCR primers contained KpnI linkers at their ends. The 2,290-bp PCR fragment was digested with KpnI and ligated into the equivalent site of the pQE30 vector. This DNA was expected to encode a PcrA protein with His6 residues fused at its amino-terminal end.
For pCR2.1C, The Sau3AI DNA fragment from plasmid pJS3 (4), containing the cat gene with its own rho-independent transcriptional terminator, was ligated into the BamHI site of the pCR2.1 vector.
For pCR2.1C1, an S. pneumoniae PCR fragment that contained a deleted pcrA gene lacking 516 bp of its 3′ end was generated using primers pcrA1 (5′-CCGGAATTCCGGTATTGACTTCAAGAGTAAGG-3′) and INS3 (5′-CATCTGGATCTTCAGTCGCA-3′). The resulting 1,794-bp DNA fragment was ligated into the pCR2.1 vector using the TOPO-TA cloning technology (Invitrogen), resulting in pCR2.11. The Δ3′pcrA DNA fragment of this plasmid was cut out with EcoRI and inserted into pCR2.1C, yielding pCR2.1C1.
For pCR2.1C2, an S. pneumoniae PCR fragment containing a deleted pcrA gene lacking 526 bp of its 5′ end was generated using primers pcrAc (5′-CAGTGTTATACAGCCTATCA-3′) and pcrA2. The resulting 1,814-bp DNA fragment was inserted into the pCR2.1 vector, resulting in pCR2.12. The Δ5′pcrA DNA fragment of this plasmid was cut out with EcoRI and inserted into pCR2.1C, yielding pCR2.1C2.
For pCR2.1C3, an S. pneumoniae PCR fragment containing a deleted pcrA gene which lacked 526 bp of its 5′ end and 516 bp of its 3′ end was generated using primers pcrAc and INS3. The resulting 1,270-bp DNA fragment was inserted into the pCR2.1 vector, resulting in pCR2.13. The Sau3AI DNA fragment from plasmid pJS3, containing the cat gene, was inserted in pCR2.13 opened with BamHI, resulting in pCR2.1C3.
Mapping of transcription initiation start sites.
Total RNA was isolated from S. pneumoniae R6 or from E. coli TOP10 harboring plasmid pCR2.1PN, and primer extension assays were performed as described previously (10), using 32P-labeled primer pcrAext (5′-TTGCACCGCCTCAGCCTG-3′), whose 3′ end is located 33 nucleotides (nt) downstream from the A residue of the pcrASpn ATG initiation codon.
RT-PCR.
Reverse transcription (RT) reactions were performed as follows. Total RNA from S. pneumoniae (400 ng) or “in vitro RNA” (whose concentration was adjusted so that the preparation contained an amount of template similar to the amount in the total pneumococcal RNA preparation) and 10 μM gene-specific primer gspa (5′-GTTTAGGCTCGTTGATAATA-3′) or gspb (5′-CTCTTTGACTATGTCGTTAATG-3′) were mixed in a final volume of 12 μl (adjusted with diethyl pyrocarbonate-treated water), denatured by incubation for 5 min at 70°C, and then placed on ice. This mixture was used to prepare an RT reaction mixture (total volume, 20 μl); the reaction was performed in cDNA synthesis buffer (Invitrogen) containing 5 mM dithiothreitol (DTT), 40 U RNase OUT, 1 mM deoxynucleoside triphosphate mixture, and 15 U Thermo-Script reverse transcriptase (Invitrogen), which was incubated for 60 min at 50°C. Reverse transcription was terminated by incubation at 85°C for 5 min. The RNA template was removed by treatment with 2 U of RNase H for 20 min at 37°C. An aliquot (2 μl) of the cDNA synthesis reaction mixture was used for each PCR. PCRs were carried out using Taq DNA polymerase (Invitrogen) and primers pcrAc and gspa. The annealing temperature was 54°C, and the extension time was 2 min at 72°C. The amplified DNAs were tested after 20 and 23 cycles, and under these conditions DNA synthesis was found to be exponential.
ATPase assays.
The ATPase activity of the His-tagged PcrASpn protein was determined by measuring hydrolysis of [α-32P]ATP or [α-32P]dATP as described previously (9). The products of the reaction were subjected to thin-layer chromatography, followed by autoradiography (9).
DNA binding assays.
The binding of His-PcrASpn to various DNA substrates was studied by performing electrophoretic mobility assays (EMSA) as described previously (2). Various DNA substrates were prepared by labeling one strand of the oligonucleotides with 32P at the 5′ end using T4 polynucleotide kinase (40) and annealing the cold complementary strand at a threefold molar excess. Substrate A contained 5′-dGCCTCGCTGCCGTCGCCA-3′ as the top strand and 5′-dTGGCGACGGCAGCGAGGCTTTTTTTTTTTTTTTTTTTT-3′ as the bottom strand. Substrate B contained 5′-dTTTTTTTTTTTTTTTTTTTTTGGCGACGGCAGCGAGGC-3′ as the top strand and 5′-dGCCTCGCTGCCGTCGCCA-3′ as the bottom strand. The top strand sequence of substrate C was 5′-dCATATGCACACAGTATGTGCGTCCAG-3′, and the bottom strand sequence was 5′-dGATCCAACCGGCTACTCTAATAGCCGGTTGGACGCACATACTGTGTGCATATG-3′. Substrate D contained 5′-dGTGGACGCACATACTGTGTGCATATGGATC-3′ as the top strand and 5′-dTATGCACACAGTATGTGCGTCCAACCGGCTATTAGAGTAGCCGGTTTGATCC-3′ as the bottom strand. Purified PcrASpn was mixed at different concentrations with labeled DNA (4 nM) and 10 ng/μl of poly(dI-dC) in a buffer consisting of 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 1 mM EDTA, 5 mM DTT, and 10% ethylene glycol. The reaction mixtures were incubated at room temperature for 15 min, and the DNA-protein complexes were resolved by electrophoresis on 6% native polyacrylamide gels (40). The gels were dried and subjected to autoradiography.
DNA helicase assays.
DNA substrates were prepared by labeling one strand of the oligonucleotides with 32P as described above. Helicase reactions were performed at 37°C for 20 min in a buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 3 mM MgCl2, 3 mM ATP, 10 mM DTT, 10% glycerol, 4 nM labeled DNA substrate, and different amounts of the PcrASpn helicase. The reactions were stopped by addition of sodium dodecyl sulfate (SDS) dye, and the products were analyzed by 10% native polyacrylamide gel electrophoresis (PAGE) (9). The gels were subsequently dried and subjected to autoradiography.
Plasmid pT181 DNA unwinding assays.
A 0.5-μg portion of pT181cop608 DNA was incubated in 1× TEKEM buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM KCl, 10 mM magnesium acetate, 10% [vol/vol] ethylene glycol) (23) containing 5 mM ATP in the presence or absence of RepC (200 ng) and in the presence or absence of His-tagged PcrASau or PcrASpn protein (10 to 50 ng each) at 32°C for 30 min. The reaction products were separated by electrophoresis on 1% agarose gels with Tris-borate-EDTA buffer containing 0.5 μg/ml ethidium bromide.
Preparation of cell extracts and in vitro replication.
Cell-free replication extracts were prepared from S. aureus strain RN4220 and the pcrA3 mutant as described previously (8, 23). The replication reaction mixtures (30 μl) contained 600 μg of protein extract, 500 ng of pT181cop608 DNA, 200 ng of RepC protein, and different amounts of the His-tagged PcrASpn or PcrASau protein. Replication products were labeled with [α-32P]dATP. The reaction mixtures were incubated at 32°C for 1 h, treated with proteinase K, and extracted with phenol-chloroform, and the DNA was precipitated with ethanol. The reaction products were separated by electrophoresis on 1% agarose gels using Tris-borate-EDTA buffer containing 1 μg/ml ethidium bromide. The gels were dried and subjected to autoradiography.
RepC-PcrA pull-down assays.
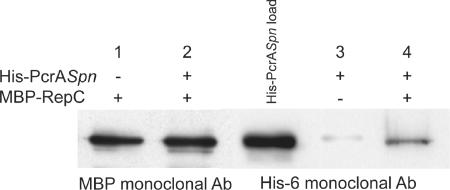
E. coli cell lysates (200 μl) containing the maltose-binding protein (MBP)-RepC fusion protein were adsorbed onto 50-μl amylose resin columns as described previously (8) and washed with buffer A (20 mM Tris-HCl [pH 8.0], 200 mM KCl, 10 mM 2-mercaptoethanol) containing 1% bovine serum albumin (BSA). Subsequently, 200 μl of 1× TEKEM buffer containing 1 μg of His-PcrASpn was mixed with MBP-RepC bound to the resin and incubated at 4°C for 1 h. The suspension was then washed three times with 1× TEKEM buffer, and the proteins were eluted directly in SDS-PAGE sample buffer (8). In control experiments, His-PcrASpn protein was loaded on amylose resin columns not containing any bound MBP-RepC. The eluted proteins were analyzed by 10% SDS-PAGE. Proteins were blotted onto membranes (3), hybridized with either an MBP monoclonal antibody (NEB) or His6 monoclonal antibody (QIAGEN), and visualized by using an ECL kit from Amersham according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Identification and sequence analysis of pcrASpn and its genomic context in gram-positive bacteria.
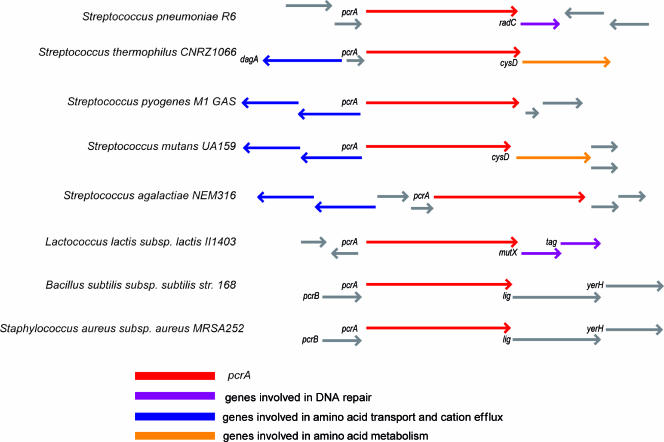
Using bioinformatic approaches, we identified the pcrA gene of S. pneumoniae in the genomes of two available sequenced strains, strains TIGR4 and R6 (SP1087 and spr0995, respectively) (http://genolist.pasteur.fr/StreptoPneumoList/index.html). To identify the genomic context of the pcrA gene in several gram-positive bacteria, comparative analyses were performed (Fig. 1) using the gene databases available at the NCBI web page (37, 45). The more conserved context was found in S. aureus and B. subtilis, where pcrA belongs to an operon formed by four homologous genes organized in the following order: pcrB-pcrA-lig-yerH. The lig gene encodes a DNA ligase, yerH encodes a putative lipoprotein, and pcrB encodes a protein with an unknown function. Downstream of pcrA, genes apparently involved in DNA repair, such as radC (S. pneumoniae), mutX, and tag (L. lactis), and genes involved in cysteine and methionine metabolism, such as cysD (Streptococcus mutans and Streptococcus thermophilus), were detected. However, in S. pyogenes, Streptococcus agalactiae, S. mutans, and S. thermophilus, we noted the presence of genes involved in amino acid transport and cation efflux located upstream from pcrA and in the opposite orientation.
FIG. 1.
Comparative analysis of the genomic context of the pcrA gene in several gram-positive bacteria. S. pneumoniae PcrA consists of 763 amino acids, while S. aureus PcrA consists of 730 amino acids. PcrA from S. pneumoniae is approximately 53% identical and 70% similar to PcrA proteins from other gram-positive organisms.
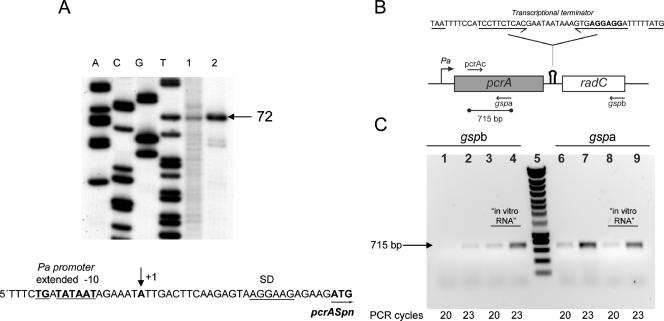
We also analyzed the sequence of the S. pneumoniae pcrA gene considering the known features of the pneumococcal genome (http://www.changbioscience.com/biotoolkit.html). A putative promoter (Pa) (Fig. 2A) was found to have a consensus extended −10 region but no recognizable −35 sequence, as is common in pneumococcal promoters (39). Pa is located 33 bp upstream of the pcrA ATG start codon. This codon was preceded by an optimal Shine-Dalgarno sequence, and the gene ended with a possible rho-independent transcriptional terminator sequence. The pcrA open reading frame (ORF) was 2,292 nucleotides long, and the corresponding protein consisted of 763 amino acids and had a theoretical molecular mass of 86,045 Da and an isoelectric point of 5.41. The radC gene is located downstream of pcrA (Fig. 1). The product of radC exhibits identity with the RadC protein of E. coli, whose function is unknown (30). The radC ORF was 681 nucleotides long, and its start codon was located 41 bp downstream of the pcrA stop codon, suggesting that the pcrA-radC locus could constitute a single operon. A putative Shine-Dalgarno sequence was found to overlap the proposed pcrA transcriptional terminator (Fig. 2B).
FIG. 2.
Transcriptional analysis of the pcrA-radC operon in S. pneumoniae. (A) Primer extension analysis for determination of the transcription start site of pcrA,. RNA samples from S. pneumoniae (lane 1) and E. coli harboring plasmid pCR2.1PN (lane 2) were reverse transcribed using 32P-labeled primer pcrAext (see Materials and Methods). A single extended 72-nt DNA was observed, which located the transcription start site at the A residue (+1) indicated by an arrow in the DNA sequence. Lanes A, C, G, and T contained DNA sequencing reaction products obtained using the same primer. The extended −10 sequence of the Pa promoter (underlined boldface type), the ribosome-binding site (SD) (underlined), and the first codon of pcrA (boldface type underlined with an arrow) are also indicated. (B) Schematic diagram of the genetic structure of the pcrA-radC locus, showing the positions of the Pa promoter and of the putative rho-independent transcriptional terminator (hairpin symbol). The DNA sequence encompassing the putative intrinsic terminator is shown above the schematic diagram, and the convergent arrows indicate the inverted repeat located upstream of the T tract. The pcrA stop codon and the radC start codon are underlined, and the putative radC Shine-Dalgarno sequence is indicated by boldface type. The primers used for RT (gspa and gspb) or PCR (pcrAc and gspa) are indicated by arrows. The location of the 715-bp PCR-amplified fragment is also shown. (C) RT-PCR analysis. Two cDNAs spanning pcrA-radC or pcrA were synthesized using total RNA prepared from S. pneumoniae R6 and oligonucleotide gspb or gspa, respectively. Amplified dsDNA was then obtained by PCR using oligonucleotides pcrAc and gspa as primers. Aliquots from the PCR mixtures were taken after 20 and 23 amplification cycles. Lanes 1 to 4, PCR products from pcrA-radC cDNA; lanes 6 to 9, PCR products from pcrA cDNA; lanes 3, 4, 8, and 9, control RT-PCRs using pcrA-radC “in vitro RNA” as the template; lane 5, DNA molecular weight standard (Smartladder; Eurogentec).
Pneumococcal pcrA gene belongs to an essential operon.
The proposed organization of S. pneumoniae pcrA-radC as an operon was tested by performing primer extension and RT-PCR assays using total RNA prepared from S. pneumoniae R6. The results of the primer extension analysis revealed the presence of a major 72-nt band, indicative of a single transcription initiation site (Fig. 2A, lane 1). When total RNA was prepared from E. coli TOP10 harboring plasmid pCR2.1PN (a construct that contains pcrA, including the Pa promoter), a major band that was the same size, albeit much stronger, was observed, which was indicative of the increase in gene dosage (Fig. 2A, lane 2). As expected for bacterial promoters, the initiation start site is a purine, which is located 7 nucleotides downstream of the 3′ end of the −10 box (28). These results demonstrated the existence of promoter Pa and showed that the “extended −10” region is efficiently recognized in both S. pneumoniae and E. coli. RT-PCR analysis was performed to determine whether pcrA and radC belong to the same operon, as well as to determine the functionality of the proposed transcriptional terminator. Two different cDNAs were synthesized by reverse transcription using total RNA from S. pneumoniae and a gene-specific primer for either pcrA (primer gspa) or radC (primer gspb). The cDNAs were PCR amplified by using two DNA primers (primers pcrAc and gspa) that annealed to the 5′ region and the 3′ region of pcrA, respectively. In both cases, the resulting amplified DNA was a single 715-bp DNA fragment (Fig. 2B and C) identical to the fragment obtained using the same primers but chromosomal DNA as the template, confirming the transcriptional linkage between pcrA and radC in an operon. No PCR DNA fragment was detected in negative control reactions in which the same RNA samples were added but reverse transcription was not performed (data not shown). This allowed us to eliminate the possibility that the presence of small amounts of genomic DNA in the RNA preparations could have accounted for generation of the PCR products observed.
A semiquantitative RT-PCR approach (33) was used to discriminate the transcripts that stopped at the putative terminator (pcrA mRNA) from the transcripts which passed through it (pcrA-radC mRNA). As a positive control for this approach, an in vitro-synthesized pcrA-radC RNA (“in vitro RNA”) was obtained by T7 RNA polymerase (RNAP)-mediated transcription from a DNA template that contained the entire pcrA-radC operon under the control of the φ10 promoter of phage T7. Since T7 RNAP does not recognize transcriptional terminators that are recognized by bacterial RNAPs, the “in vitro RNA” should have corresponded only to the bicistronic pcrA-radC transcript. This “in vitro RNA” was used to evaluate whether factors such as the processivity of the reverse transcriptase or the hybridization efficiency of each gene-specific primer contributed to the different yield of the 715-bp PCR product amplified from either gspa- or gspb-initiated cDNA. The results showed that the possibility of a contribution by these factors could be disregarded since, as expected for a bicistronic transcript, no significant differences were found in the yield of the amplified DNA when reverse transcription from the “in vitro RNA” control was initiated with gspa or with gspb (Fig. 2C). It should be noted that whereas both the pcrA and pcrA-radC mRNAs can yield the 715-bp amplified DNA fragment when reverse transcription is initiated from gspa, only the pcrA-radC mRNA can yield the PCR product if gspb is used as the primer for cDNA synthesis. The intracellular level of the pcrA mRNA was found to be approximately 20-fold higher than the intracellular level of the full-length pcrA-radC mRNA (Fig. 2C). These results confirmed that the expression of radC is regulated at the transcriptional level. Although we have not determined the size of the pcrA mRNA, we propose that the majority of transcription through pcrA ends at the putative transcriptional terminator located between the two genes.
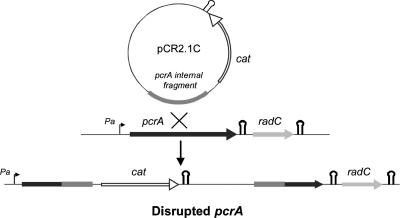
We next examined the essentiality of the pcrA-radC operon by means of a gene replacement strategy developed for S. pneumoniae (31). For this, we used plasmid pCR2.1C, an integrative vector unable to replicate in pneumococci, which introduced a selectable chloramphenicol resistance marker into the chromosome (Fig. 3). Two different pcrA fragments were separately cloned in this vector: an internal fragment (yielding plasmid pCR2.1C3, designed to perform gene disruption) and a fragment including the promoter Pa but lacking the 3′ end of pcrA (which generated plasmid pCR2.1C1, designed to reconstitute an intact copy of the gene and thus used as a positive control). No chloramphenicol-resistant (Cmr) colonies were found when pcrA was interrupted, indicating the essentiality of pcrA and/or radC (if pcrA disruption has a downstream polar effect). In the control experiment, in which the expression of the whole operon from promoter Pa was not affected and a copy of pcrA was reconstituted, ∼20,000 Cmr CFU per ml of competent culture and per μg of DNA was obtained. Furthermore, the polarity of the pcrA gene disruption was examined by cloning a pcrA fragment lacking the promoter and the 5′ end of the gene into the pCR2.1C vector (pCR2.1C2). The insertion-duplication event with this construct should have affected expression of radC from promoter Pa but should have left a functional copy of pcrA. Again, we did not detect any Cmr colonies, a result that points out the essentiality of radC for cell viability. While the polar effect of pcrA disruption was demonstrated, we could not rule out the possibility that pcrA is also essential for cell growth. Using a similar gene replacement strategy for the identification of conserved essential genes in S. pneumoniae Rx-1, pcrA was shown to be nonessential (46). PcrA from B. subtilis is thought to play an essential role in preventing untimely recombination events (36). Mutations in RecA modulator genes can suppress the lethality of pcrA mutants in this species. Thus, it is possible that suppressor mutations are present in the pneumococcal Rx-1 strain, making pcrA nonessential. Whether this could also be the case in strain R6 is unknown at present, since our results did not allow us to determine unambiguously the essentiality of pcrA in this strain. With respect to radC, the fact that interruption of pcrA in the Rx-1 strain did not reveal the essentiality of radC might have reflected a different organization affecting the pcrA genetic context in pneumococcal strains R6 and Rx-1. Alternatively, it could have been related to a difference in the magnitude of the polar effect caused by the pcrA gene disruption in the two approaches. Our gene replacement strategy maximized the polar effect since the cat gene, cloned with its own transcriptional terminator (Fig. 3), was inserted in the same direction as the pcrA gene fragment (Fig. 3). Thus, after the insertion-duplication event, transcription of genes downstream of pcrA was prevented unless the cloned pcrA fragment included promoter Pa. Our results demonstrate that transcription of radC from promoter Pa is essential for cell viability, showing again that pcrA and radC constitute a single operon in which expression of radC is downregulated at the transcriptional level.
FIG. 3.
Strategy for disruption of the S. pneumoniae pcrA gene. Plasmid pCR2.1C was used to test the essentiality of the pcrA-radC operon. This plasmid is an integrative vector that is unable to replicate in pneumococci, in which the Cmr (cat) gene from pJS3 (including its own transcriptional terminator) has been cloned. An internal pcrA gene fragment was cloned in this vector, and the insertion-duplication event was selected on chloramphenicol-containing medium. The transcriptional terminator of the cat gene and the transcriptional terminator proposed to attenuate transcription of radC are indicated by hairpin symbols.
ATPase activity of purified His-PcrASpn is stimulated by ssDNA.
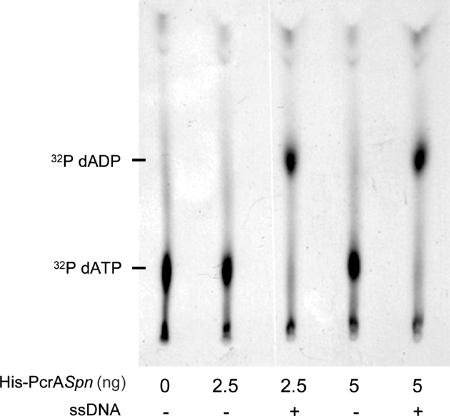
To purify PcrASpn, the pcrA ORF was amplified by PCR from S. pneumoniae R6 and cloned into the pQE30 expression vector. The resultant construct generated a translational fusion of PcrASpn with a His6 tag at its amino-terminal end. The His-PcrASpn protein was overproduced in E. coli and purified by affinity chromatography using nickel resin. His-PcrASpn protein bound to the resin was eluted with 0.2 M imidazole. Fractions were analyzed by SDS-PAGE, and the fractions in which the PcrA protein purity was greater than 90% were selected (not shown). The concentration of His-PcrASpn in these fractions was approximately 0.2 mg/ml. The protein had a molecular mass of approximately 86 kDa, which is consistent with the predicted size of the pcrA gene product (763 amino acids). To confirm that purified His-PcrASpn was enzymatically active, we measured its ATPase activity by examining hydrolysis of [α-32P]ATP and [α-32P]dATP as described previously (9). The results showed that His-PcrASpn had ATPase activity that efficiently hydrolyzed both dATP (Fig. 4) and ATP at similar levels (not shown). In addition, as observed for the PcrA helicases of S. aureus, B. anthracis, and B. cereus (3, 9, 34), the ATPase activity of His-PcrASpn was significantly stimulated by addition of ssDNA to the reaction mixtures (Fig. 4). The ATPase activities of PcrASpn and PcrASau were comparable (1).
FIG. 4.
ATPase activity of purified His-tagged PcrASpn. The dATPase activity of the pneumococcal helicase was measured by determining the hydrolysis of [α-32P]dATP. The reactions were performed in 1× TEKEM buffer containing 1 μCi of [α-32P]dATP and different amounts of His-PcrASpn. The ATPase activity of His-PcrASpn was activated by the presence of 100 ng of a 53-nt oligonucleotide (ssDNA). The products of [α-32P]dATP hydrolysis were analyzed by thin-layer chromatography.
Protein-protein interactions between His-PcrASpn and RepC.
Specific interactions between PcrASau and RepC were shown to be critical for pT181 DNA unwinding during the initiation of replication (9). Previous studies showed that PcrASau could physically interact with RepC (9). Therefore, we tested the ability of His-PcrASpn to physically interact with RepC. To do this, we performed pull-down assays (9), making use of the different epitope tags present on PcrA and RepC. Cell lysates from E. coli overproducing an MBP-RepC fusion protein (8) were absorbed to amylose resin and washed with buffer A. Then His-PcrASpn was mixed with MBP-RepC bound to the resin and incubated at 4°C for 1 h. The resin was then washed with buffer, and bound proteins were eluted directly in SDS-PAGE sample buffer. Western blot analysis using the anti-MBP monoclonal antibody showed that MBP-RepC was bound to the amylose resin, as expected (Fig. 5). While His-PcrASpn did not bind to the amylose resin incubated with BSA, it was retained on the resin to which MBP-RepC was bound (Fig. 5). These results indicate that His-PcrASpn and RepC can physically interact. Furthermore, the strength of the interaction was comparable to the strength of the interaction between PcrASau and RepC (1).
FIG. 5.
Interactions between His-PcrASpn and RepC. The physical interaction between PcrASpn and RepC proteins was analyzed by performing a pull-down assay as described in Materials and Methods. The fractions eluted from amylose resin columns (containing [+] or lacking [−] bound MBP-RepC) were probed with either anti-MBP or anti-His6 monoclonal antibodies (Ab). In a control experiment in which MBP-RepC was not adsorbed to the amylose columns (lane 3), 1% BSA was used instead. In lane His-PcrASpn load, 0.2 μg of purified pneumococcal protein was loaded directly onto the gel.
His-PcrASpn DNA binding and helicase activity.
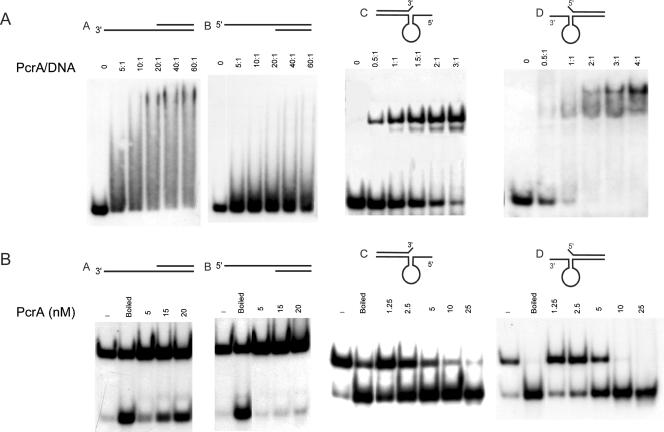
To characterize the pneumococcal PcrASpn helicase, we tested its DNA binding and helicase activities. To do this, several oligonucleotides with different conformations were used (Fig. 6). The substrates were dsDNAs with 3′- and 5′-end oligo(dT) overhangs (substrates A and B, respectively) and partial duplexes in which the single-stranded regions could form secondary structures, including (i) a hairpin with a 6-nt single-stranded overhang at the 5′ end and a 1-nt flap (unpaired nucleotide) at the 3′ end of the complementary strand (substrate C) and (ii) a hairpin with a 6-nt single-stranded overhang at the 3′ end and a 1-nt flap at the 5′ end of the complementary strand (substrate D). These oligonucleotides mimicked the dso of pT181, and their 5′ and 3′ single-stranded regions corresponded to the IRII element that can assume a hairpin structure (11, 19, 20). His-PcrASpn showed very little binding to substrate B, while incubation with substrate A generated detectable levels of DNA-protein complexes that exhibited diffuse migration on polyacrylamide gels, an indication of nonstable binding (Fig. 6A). We interpreted this result as an indication that the presence of an ssDNA-dsDNA junction is not enough to generate a stable His-PcrASpn-DNA complex, although binding to substrate A with the 3′ single-stranded tail was more efficient than binding to substrate B. Binding of His-PcrASpn to substrates C and D appeared to be much more stable, and the DNA-protein complexes migrated as discrete bands in both cases (Fig. 6A). His-PcrASpn showed comparable abilities to bind to these substrates harboring a single-stranded folded overhang at the 3′ or 5′ end (a slightly higher preference for binding to substrate D was observed). These results suggest that there is specific recognition of the hairpin formed at either the 3′ or 5′ regions of the probes. In the unwinding experiments (Fig. 6B), His-PcrASpn was active in unwinding duplex substrate A. However, the protein did not significantly unwind duplex substrate B, indicating that the pneumococcal helicase predominantly acts as a 3′→ 5′ helicase with tailed duplex substrates. When substrates C and D were used to test the helicase activity of His-PcrASpn, unwinding of both probes was observed, although the 5′ single-stranded tailed substrate was unwound slightly more efficiently. These results demonstrated that His-PcrASpn has bipolar 3′→ 5′ and 5′ → 3′ helicase activities, exhibiting similar efficiencies with partial duplex substrates in which the 3′ or 5′ single-stranded regions can fold into secondary structures. From this set of experiments, we concluded that PcrASpn showed maximal helicase activity with substrates to which it bound most efficiently in EMSA.
FIG. 6.
DNA binding and helicase activities of PcrASpn. (A) PcrASpn shows specific binding to DNA substrates containing a hairpin structure at their 3′ or 5′ end. EMSA were carried out by incubating His-PcrASpn at different molar ratios with four different 5′ 32P-labeled probes. The DNA-protein complexes were resolved on 6% polyacrylamide gels. (B) PcrASpn showed bipolar 5′→3′ and 3′→5′ helicase activities. His-PcrASpn was incubated with 32P-labeled partially duplex substrates containing either 3′ or 5′ oligo(dT) tails (substrates A and B) or with structured substrates containing 5′ or 3′ ssDNA regions (substrates C and D). The reaction products were resolved by native PAGE.
His-PcrASpn cannot extensively unwind RepC-nicked pT181 DNA.
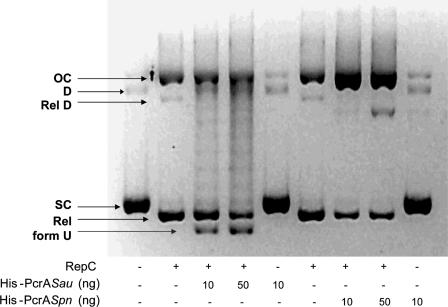
Rolling circle replication of pT181 is initiated by the plasmid-encoded initiator RepC, which introduces a sequence-specific nick at dso and becomes covalently attached to the 5′ end of the DNA (23, 47). The new 3′-OH end serves as the substrate for DNA polymerase-mediated DNA synthesis, a reaction that presumably involves DNA unwinding by PcrA (21, 22). Binding of RepC to dso is followed by recruitment of the PcrA helicase (among other host-encoded proteins of the replisome), which interacts directly with RepC (9). The specific interaction between the Rep proteins of RCR plasmids and the PcrA helicase might be a determinant of a broad host range for replication of these plasmids. To investigate further the interaction between heterologous PcrA helicases and the pT181-encoded RepC initiator, we performed a set of assays to functionally replace PcrASau with its pneumococcal counterpart. To do this, supercoiled (SC) pT181 DNA was incubated with RepC along with one of the helicases, and DNA unwinding was analyzed. The results (Fig. 7) showed that unwinding of RepC-nicked DNA by PcrASau resulted in generation of a faster-migrating band that corresponded to the unwound U form of the DNA (9). Although the U form does not correspond to the fully unwound pT181 DNA (9), it has been shown to contain extensive ssDNA regions that are sensitive to digestion with nuclease S1 (9). Incubation with both RepC and PcrASpn yielded a diffuse band migrating slightly faster than the open circular (OC) form (Fig. 7). This band was sensitive to nuclease S1 (not shown) and is thought to correspond to OC DNA with limited unwinding (9). In contrast to the results obtained with PcrASpn, incubation of SC pT181 DNA with RepC and the PcrA helicase of B. cereus or B. anthracis (3) resulted in generation of the U form, which reflected extensive unwinding of the DNA. We concluded that while PcrABcer and PcrABan were able to establish a functional interaction with RepC, the pneumococcal helicase failed to do this and was able to initiate unwinding only to a small extent. In this respect, it is worth pointing out that we have been unable to establish the staphylococcal pT181 plasmid in S. pneumoniae, even though the tet gene that it carries is highly homologous to the tet gene of plasmid pMV158 (27).
FIG. 7.
DNA unwinding activity of PcrASpn. DNA relaxation assays were performed as described in Materials and Methods. Plasmid pT181cop608 DNA was incubated in the presence (+) or absence (−) of RepC and/or different amounts of either His-tagged PcrASau or PcrASpn. The products were separated by 1% agarose gel electrophoresis in the presence of ethidium bromide. OC, nicked open circular DNA; D, supercoiled dimeric plasmid DNA; RelD, covalently closed relaxed dimeric DNA; SC, supercoiled monomeric plasmid DNA; Rel, covalently closed relaxed monomeric DNA; form U, unwound plasmid DNA.
His-tagged PcrASpn can support in vitro replication of plasmid pT181 to a limited extent.
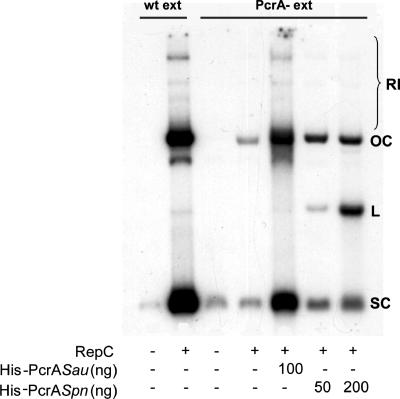
PcrASau has approximately 10-fold-stronger helicase activity with substrates containing hairpin structures along with a 5′ single-stranded tail than with similar substrates with a 3′ single-stranded tail (2). On the other hand, PcrASpn shows only a modest preference for the substrates with the 5′ single-stranded tail (Fig. 6B). Since PcrASpn is unable to promote extensive unwinding of the pT181 DNA, it is possible that unwinding may depend upon the processive movement of the helicase-initiator protein complex in the 5′-to-3′ direction. Future experiments should address this issue. Alternatively, interaction with the heterologous RepC initiator may not provide PcrASpn with an efficient unwinding capability. However, interaction of PcrASpn with other components of the replicative machinery of S. aureus might overcome this deficiency, allowing the helicase to promote replication of pT181. Thus, we examined whether the heterologous PcrASpn helicase could support replication of pT181 DNA in vitro. These experiments were performed with cell extracts prepared from S. aureus strain RN4220 and from the pcrA3 mutant, which is inactive in pT181 replication (8, 9). Replication of pT181 in wild-type extracts generated labeled DNA corresponding to SC and OC forms of the DNA, as well as replication intermediates (Fig. 8). The faint bands observed when pT181 DNA was incubated with the pcrA3 cell extract along with RepC may reflect the incorporation of a few labeled nucleotides due to the RepC nicking activity or other DNA-modifying activities present in the cell extract. As observed previously (9), addition of purified His-PcrASau to the pcrA3 extract restored replication of the pT181 DNA. Addition of purified His-PcrASpn to the pcrA3 extract resulted in limited replication of the pT181 DNA (Fig. 8). However, the levels of DNA synthesis were much lower than those obtained with the homologous PcrASau (Fig. 8).
FIG. 8.
In vitro replication of plasmid pT181. In vitro replication was carried out using the RepC protein and cell extracts from either wild-type S. aureus (wt ext) or the pcrA3 mutant of S. aureus (PcrA- ext) and different amounts of PcrA helicases. The positions of supercoiled pT181cop608 DNA (SC), linear DNA (L), open circular DNA (OC), and replication intermediates (RI) are indicated.
We concluded from our studies that the pneumococcal PcrA helicase has DNA binding specificity for substrates that have a folded structure (such as substrates C and D assayed here). This helicase also had a bipolar helicase activity, similar to the results obtained with PcrASau. Even though the pneumococcal helicase was able to initiate in vitro replication of the staphylococcal plasmid pT181 (Fig. 8), its efficiency was much lower than that of PcrASau. In this respect it is worth pointing out that molecular modeling of PcrASpn based on the known structure of the PcrA helicase of B. stearothermophilus was feasible only at the N-terminal region of the protein (not shown), indicating that interactions between these proteins and the initiators of RCR plasmids may be more complex than envisaged. In summary, PcrASpn, like the other PcrA helicases characterized so far (2, 3, 6, 35), was able to initiate unwinding of the RepC-nicked pT181 DNA to a limited extent, perhaps by direct interaction with the initiator, thus promoting synthesis of the leading strand by means of other host-encoded proteins, such as single-stranded DNA binding protein (SsbA), DNA polymerase I (12), and DNA polymerase III. A reconstituted system with purified proteins involved in replication by the rolling circle mechanism would shed light on these complex protein-protein and protein-DNA interactions. Future experiments using chimeric PcrA proteins and site-directed mutants of PcrA based on amino acid sequence divergence between PcrA proteins from different bacterial genera should shed light on the role of the directionality of PcrA helicases and their processivity in determining the host ranges of pT181 and other RCR plasmids in gram-positive bacteria.
Acknowledgments
We thank C. Nieto for help with the RT-PCR measurements and P. Burón for technical help.
J.A.R.M. was a recipient of an FPI fellowship from the Ministerio de Educación y Ciencia. This research was financed by grant BFU-2004-00687 from the Comisión Interministerial de Ciencia y Tecnología (to G.D.S.), by grant REIPI FIS CO3/14 from the Institute of Health Carlos III (to M.E.), and by grant GM31685 from the National Institutes of Health (to S.A.K.).
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Anand, S. P., A. Chattopadhyay, and S. A. Khan. 2005. The PcrA3 mutant binds DNA and interacts with the RepC initiator protein of plasmid pT181 but is defective in its DNA helicase and unwinding activities. Plasmid 54:104-113. [DOI] [PubMed] [Google Scholar]
- 2.Anand, S. P., and S. A. Khan. 2004b. Structure-specific DNA binding and bipolar helicase activities of PcrA. Nucleic Acids Res. 32:3190-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, S. P., P. Mitra, A. Naqvi, and S. A. Khan. 2004. Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J. Bacteriol. 186:2195-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballester, S., P. Lopez, J. C. Alonso, M. Espinosa, and S. A. Lacks. 1986. Selective advantage of deletions enhancing chloramphenicol acetyltransferase gene expression in Streptococcus pneumoniae plasmids. Gene 41:153-163. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H., M. Seigneur, S. D. Ehrlich, and B. Michel. 1997. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol. Microbiol. 26:557-567. [DOI] [PubMed] [Google Scholar]
- 6.Bird, L., J. Brannigan, H. Subramanya, and D. Wigley. 1998. Characterisation of Bacillus stearothermophilus PcrA helicase: evidence against an active rolling mechanism. Nucleic Acids Res. 26:2686-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruand, C., and S. D. Ehrlich. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 35:204-210. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T.-L., M. G. Kramer, R. A. Ansari, and S. A. Khan. 2000. Role of individual monomers of a dimeric initiator protein in the initiation and termination of plasmid rolling circle replication. J. Biol. Chem. 275:13529-13534. [DOI] [PubMed] [Google Scholar]
- 9.Chang, T.-L., A. Naqvi, S. P. Anand, M. G. Kramer, R. Munshi, and S. A. Khan. 2002. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J. Biol. Chem. 277:45880-45886. [DOI] [PubMed] [Google Scholar]
- 10.del Solar, G., P. Acebo, and M. Espinosa. 1995. Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 18:913-924. [DOI] [PubMed] [Google Scholar]
- 11.Dempsey, L. A., P. Birch, and S. A. Khan. 1992. Six amino acids determine the sequence-specific DNA binding and replication specificity of the initiator proteins of the pT181 family. J. Biol. Chem. 267:24538-24543. [PubMed] [Google Scholar]
- 12.Díaz, A., S. A. Lacks, and P. López. 1994. Multiple roles for DNA polymerase I in establishment and replication of the promiscuous plasmid pLS1. Mol. Microbiol. 14:773-783. [DOI] [PubMed] [Google Scholar]
- 13.Dillingham, M. S., P. Soultanas, P. Wiley, M. R. Webb, and D. B. Wigley. 2001. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. USA 98:8381-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 17.Iordanescu, S. 1993. Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet. 241:185-192. [DOI] [PubMed] [Google Scholar]
- 18.Iordanescu, S., and R. Basheer. 1991. The Staphylococcus aureus mutation pcrA3 leads to the accumulation of pT181 replication initiation complexes. J. Mol. Biol. 221:1183-1189. [DOI] [PubMed] [Google Scholar]
- 19.Jin, R., and R. P. Novick. 2001. Role of the double-strand origin cruciform in pT181 replication. Plasmid 46:95-105. [DOI] [PubMed] [Google Scholar]
- 20.Jin, R., A. Rasooly, and R. P. Novick. 1997. In vitro inhibitory activity of RepC/C*, the inactivated form of the pT181 plasmid initiation protein, RepC. J. Bacteriol. 179:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan, S. A. 2003. DNA-protein interactions during the initiation and termination of plasmid pT181 rolling-circle replication. Prog. Nucleic Acid Res. Mol. Biol. 75:113-137. [DOI] [PubMed] [Google Scholar]
- 22.Khan, S. A. 2005. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53:126-136. [DOI] [PubMed] [Google Scholar]
- 23.Koepsel, R. R., R. W. Murray, W. D. Rosenblum, and S. A. Khan. 1985. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc. Natl. Acad. Sci. USA 82:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg, A., and T. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Co., New York, N.Y.
- 25.Kreiswirth, B. N., M. J. S. Lofdahl, M. Betley, P. M. O'Reilly, M. S. Schlievert, and R. P. N. Bergdoll. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 26.Lacks, S. A. 1968. Genetic regulation of maltosaccharide utilization in pneumococcus. Genetics 60:685-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacks, S. A., P. López, B. Greenberg, and M. Espinosa. 1986. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 192:753-765. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, D. E. A., and S. Adhya. 2004. Axiom of determining transcription start points by RNA polymerase in Escherichia coli. Mol. Microbiol. 54:692-701. [DOI] [PubMed] [Google Scholar]
- 29.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 30.Lombardo, M.-J., and S. M. Rosenberg. 2000. radC102 of Escherichia coli is an allele of recG. J. Bacteriol. 182:6287-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López, P., M. Espinosa, D. L. Stassi, and S. A. Lacks. 1982. Facilitation of plasmid transfer in Streptococcus pneumoniae by chromosomal homology. J. Bacteriol. 150:692-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marians, K. J. 2000. Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Structure 8:R227-R235. [DOI] [PubMed] [Google Scholar]
- 33.McCluskey, J., J. Hinds, S. Husain, A. Witney, and T. J. Mitchell. 2004. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51:1661-1675. [DOI] [PubMed] [Google Scholar]
- 34.Naqvi, A., E. Tinsley, and S. A. Khan. 2003. Purification and characterization of the PcrA helicase of Bacillus anthracis. J. Bacteriol. 185:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petit, M.-A., E. Dervyn, M. Rose, K.-D. Entian, S. McGovern, S. D. Ehrlich, and C. Bruand. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 29:261-273. [DOI] [PubMed] [Google Scholar]
- 36.Petit, M.-A., and D. Ehrlich. 2002. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 21:3137-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33:D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasooly, A., P. Wang, and R. Novick. 1994. Replication-specific conversion of the Staphylococcus aureus pT181 initiator protein from an active homodimer to an inactive heterodimer. EMBO J. 13:5245-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144-155. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor N.Y.
- 41.Soultanas, P., M. Dillingham, F. Papadopoulos, S. Phillips, C. Thomas, and D. Wigley. 1999. Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res. 27:1421-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soultanas, P., M. S. Dillingham, P. Wiley, M. R. Webb, and D. B. Wigley. 2000. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 19:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanya, H. S., L. E. Bird, J. A. Brannigan, and D. B. Wigley. 1996. Crystal structure of a DExx box DNA helicase. Nature 384:379-383. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, S., C. Hours, M. Iwaya, H. E. D. Lane, and D. T. Denhardt. 1978. The Escherichia coli rep gene, p. 393-400. In D. T. Denhardt, D. H. Dressler, and D. S. Ray (ed.), The single-stranded DNA phages. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Tatusova, T. A., I. Karsch-Mizrachi, and J. A. Ostell. 1999. Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics 15:536-543. [DOI] [PubMed] [Google Scholar]
- 46.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, C. D., D. F. Balson, and W. Shaw. 1988. Identification of the tyrosine residue involved in bond formation between replication origin and the initiator protein of plasmid pC221. Biochem. Soc. Trans. 16:758-759. [Google Scholar]
- 48.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, A. C., R. A. Ansari, M. C. Schmidt, and S. A. Khan. 1998. An oligonucleotide inhibits oligomerization of a rolling circle initiator protein at the pT181 origin of replication. J. Biol. Chem. 273:16082-16089. [DOI] [PubMed] [Google Scholar]