Abstract
Phosphorylation of Escherichia coli CheY increases its affinity for its target, FliM, 20-fold. The interaction between BeF3−-CheY, a phosphorylated CheY (CheY∼P) analog, and the FliM sequence that it binds has been described previously in molecular detail. Although the conformation that unphosphorylated CheY adopts in complex with FliM was unknown, some evidence suggested that it is similar to that of CheY∼P. To resolve the issue, we have solved the crystallographic structure of unphosphorylated, magnesium(II)-bound CheY in complex with a synthetic peptide corresponding to the target region of FliM (the 16 N-terminal residues of FliM [FliM16]). While the peptide conformation and binding site are similar to those of the BeF3−-CheY-FliM16 complex, the inactive CheY conformation is largely retained in the unphosphorylated Mg2+-CheY-FliM16 complex. Communication between the target binding site and the phosphorylation site, observed previously in biochemical experiments, is enabled by a network of conserved side chain interactions that partially mimic those observed in BeF3−-activated CheY. This structure makes clear the active role that the β4-α4 loop plays in the Tyr87-Tyr106 coupling mechanism that enables allosteric communication between the phosphorylation site and the target binding surface. Additionally, this structure provides a high-resolution view of an intermediate conformation of a response regulator protein, which had been generally assumed to be two state.
Two-component systems, composed of an autohistidine kinase and a phosphorylatable response regulator, are widely used in bacterial signal transduction. In Escherichia coli, the response regulator CheY controls the sense of rotation of the flagellar motor by switching between two stable states (CheY and phosphorylated CheY [CheY∼P]), enabling bacterial chemotaxis. CheY∼P interacts strongly with the flagellar motor switch protein FliM to effect a change in the motor rotational bias from counterclockwise to clockwise. Unphosphorylated CheY also interacts with FliM, though with substantially reduced affinity. Because residues on the FliM binding surface of unphosphorylated CheY (e.g., Tyr106) would sterically block FliM from binding, the structural basis of the moderate affinity of unphosphorylated CheY for FliM remains unclear.
The structures of unphosphorylated, Mg2+-free CheY (apoCheY) (28) and CheY bound to the BeF3− phosphoryl analog (19) provide evidence that a network of conserved residues enables allosteric communication between the phosphorylation site (Asp57) and the target binding site. Like phosphorylation, activation by the BeF3− phosphoryl analog requires a bound Mg2+ ion and causes a coordinated rearrangement of Tyr106 and Thr87 toward the active site, resulting in the burial of the Tyr106 side chain and formation of an H bond between the Thr87 hydroxyl and the phosphoryl group, termed Y-T coupling (19). Another rearrangement that characterizes the activation process is the reorientation of Lys109 (which forms a salt bridge with Asp57 in unphosphorylated CheY) to form a salt bridge with Asp12 and one of the fluorine atoms of BeF3−, while Mg2+ binding alone also induces the Lys109-Asp12 salt bridge (4). The backbone conformation of CheY also changes in response to BeF3− activation, most notably at the β4-α4 and β5-α5 loops on the α4-β5-α5 surface (19), where FliM binds (20).
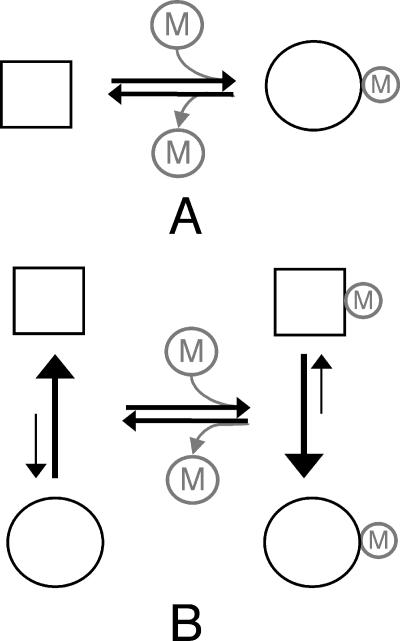
Dynamic excursions to the active state.
There exists a growing body of evidence that the unphosphorylated state of response regulators exists in dynamic equilibrium between inactive (unphosphorylated-like) and active (phosphorylated-like) conformations. Perturbations such as phosphorylation or mutation might then shift this equilibrium to favor the active conformation, giving rise to the notion of equilibrium shift activation (Fig. 1B). Here we imagine that the inactive conformation is favored in the absence of a ligand. The ligand binds more tightly to the active conformation and drives the equilibrium to favor the active conformation. Since the active and inactive conformations preexist, the idea that a bound ligand is not required to flip the conformational switch is fundamental to the equilibrium shift model. This is in contrast to the induced-fit model, where ligand binding and switching are concomitant (Fig. 1A). In this case the active conformation cannot be formed until the ligand is bound. It should be noted that although the original formulation of the Y-T coupling model includes no elements of equilibrium shift, the two models (Y-T coupling and equilibrium shift) are not mutually exclusive, as the equilibrating species could be coupled by the Y-T mechanism.
FIG. 1.
Two views of activation: In the induced-fit model (A), ligand (M) binding results directly in conformational change, while in the equilibrium shift model (B), ligand binding serves to stabilize the active conformation, which preexists in a small fraction of the population prior to phosphorylation. Squares, inactive conformation; circles, active conformation.
For the CheY homologs Spo0F and NtrC, nuclear magnetic resonance studies have shown that the regions that undergo phosphorylation-induced structural changes also experience dynamic excursions on the millisecond to microsecond time scale, consistent with coordinated conformational fluctuations (9, 27). In both studies the authors suggested that these fluctuations are due to conformational exchange between inactive and active conformations. These data have been interpreted in the context of a model whereby phosphorylation drives a preexistent equilibrium, rather than inducing a new conformation. There is some evidence that the CheY signaling surface experiences similar slow-time-scale fluctuations (23), and the concept of equilibrium shift activation was applied to CheY to rationalize the phosphorylation-independent activity of the variant CheYD13K, despite its paradoxically inactive conformation in the crystal (14). The structure of a related constitutively active mutant, CheYD13K/Y106W (CheY**), was also found in the inactive conformation when not bound to the 16 N-terminal residues of FliM (FliM16), yet this same mutant was able to switch to the active (BeF3−-CheY-like) conformation in complex with FliM16 (8). This result indicates that, at least for CheY**, phosphorylation is not necessary for conformational switching, and FliM16 binding is sufficient to throw the switch to the active conformation, consistent with the equilibrium shift model. Finally, the equilibrium shift model of activation has been used to rationalize the residual activity of unphosphorylated CheY at the flagellar motor (3, 6).
Intermediate CheY conformations.
In the atomic resolution structure of apoCheY (i.e., CheY in the absence of Mg2+, phosphoryl group, or peptide ligand; Protein Data Bank accession no. 1JBE.pdb), there exist two roughly equally populated conformations of the β4-α4 loop and the Tyr106 side chain (25). The authors of that study suggested that these sets of conformations are correlated and that one is the bona fide inactive conformation, which sterically blocks FliM, while the other, “meta-active” conformation is “responsible for the active properties seen in apoCheY” (25). As the authors point out, this structure does not support the equilibrium shift model, as the meta-active conformation is significantly different from the conformation of BeF3−-CheY. A physiological role for such intermediate species was recently demonstrated by the structures of CheY in complex with CheZ residues 200 to 214 (CheZC), in which CheZC-bound CheY is found in a partially switched conformation (11).
To more thoroughly map the regions of conformational space accessible to CheY in the absence of phosphorylation, we have solved the X-ray crystal structure of unphosphorylated Mg2+-bound CheY in complex with a 16-mer peptide corresponding to FliM16 to 2.4-Å resolution. Analysis of the structure indicates that unphosphorylated CheY accommodates bound FliM16 without switching to the canonical active conformation yet shares several features of the active conformation, perhaps most notably the complete burial of Tyr106. Combined with the observations that a functionally important region (the β4-α4 loop) is stabilized by an interaction network that is unique to this complex, while another region (the α1 helix) remains in a distinctly inactive conformation, these results indicate that the energy landscape between the inactive and active conformations of CheY is more complex than previously thought.
Finally, this structure provides a molecular view of how allostery in CheY works in reverse, whereby the binding of FliM is coupled to conformational changes in the active site that affect phosphorylation (24). Whereas neither the Y-T coupling nor the equilibrium shift model of activation is completely consistent with the details of the CheY-FliM16 complex, we discuss an alternate allosteric activation model that recognizes a role for the β4-α4 loop in signal propagation.
MATERIALS AND METHODS
Protein purification, crystallization, and data collection.
E. coli CheY was overexpressed and purified using a previously described protocol (13). The FliM peptide (MGDSILSQAEIDALLN) was purchased from Macromolecular Resources (Fort Collins, CO) at 90% purity. The Mg2+-CheY-FliM complex crystallized in space group P21. To form the complex, a solution of 0.5 mM CheY and 10 mM Tris (pH 7.0) was slowly saturated with solid FliM peptide (∼10 mM). Diffraction quality crystals were grown by mixing 3 μl of this solution with 3 μl of well solution containing 30% polyethylene glycol 6000, 50 mM MES (morpholineethanesulfonic acid), and 50 mM MgCl2, at pH 6.0 Crystals were grown in about 8 weeks at 4°C using the hanging-drop method. A single crystal was mounted in a 1-mm silanized glass capillary tube containing a reservoir of well solution to allow for room temperature data collection. Data collection took place at 24°C on an R-AXIS II (Molecular Structure Corporation) with CuKα radiation. A total of 149 nonoverlapping degrees of data were collected, with a 1.0° oscillation range and an exposure time of 10 min. Because we did not find suitable cryoconditions, data were collected on two sections of the same crystal to minimize the effects of radiation damage. In the first orientation, 103° were kept. In the second orientation, 46° were kept, giving a total of 149°. Data were processed to 2.4 Å in space group P21 with MOSFLM (21) and scaled using SCALA (7).
Structure determination.
Initial phases were determined by molecular replacement using the program EPMR (17) and Protein Data Bank entry 3CHY.pdb (28), with Tyr106 modeled as alanine, as the search model. As suggested by the Matthews coefficient, EPMR found a solution with two CheY molecules in the asymmetric unit (R value of 0.36 using data to 3.0 Å). Electron density generated following anisotropic B-factor and bulk solvent correction (used throughout refinement) clearly showed the bound peptide and the buried orientation of the Tyr106 side chain for both CheY molecules. Residues 5 to 12 of the two FliM16 peptides as well as the Tyr106 side chains were modeled into σA-weighted 2|Fo| − |fc| and |Fo| − |fc| maps. This model was refined against the complete range of data (40.7 to 2.40 Å) with multiple rounds of conjugate gradient least-squares minimization, torsion angle simulated annealing, and isotropic individual B-factor refinement using the program CNS (5) with noncrystallographic symmetry (NCS) restraints imposed on the two CheY molecules. Interspersed with rounds of refinement, manual adjustments to the model were performed using the program O (15), guided by σA-weighted 2|Fo| − |fc|, |Fo| − |fc|, and 2|Fo| − |fc| composite omit maps. During the later stages of refinement, residues 13 to 16 and 2 to 4 of the peptides, two magnesium(II) ions (one per CheY), and 144 water molecules were modeled. Neither NCS restraints nor simulated annealing was used in the later refinement cycles. Residues 14, 27, and 88 to 90 of CheY chain A and residues 14, 17, 88 to 90, and 100 of CheY chain B are modeled as two conformations. The R and Rfree values for the final model are 0.18 and 0.24, respectively. In Ramachandran space, 88.5% of residues are in the favored region, 9.5% are in the allowed region, 1.2% are in the generously allowed region, and 0.8% are in the disallowed region, due to the CheY Asn62 γ-turn (28). Data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter (unit) | Valuea |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 40.04, 62.16, 54.22 |
| α, β, γ (o) | 90.00, 96.77, 90.00 |
| Resolution (Å) | 40.7 (2.53)-2.40* |
| Completeness (%) | 97.3 (96.7) |
| I/σ I (σ) | 18.8 (2.1) |
| Rmerge | 0.126 (0.457) |
| Redundancy | 3.0 (3.1) |
| Mosaicity (°) | 0.4 |
| Refinement | |
| Resolution range (Å) | 40.7-2.4 |
| Number of reflections | 10,174 |
| Working set | 9,203 |
| Test set (10%) | 971 |
| R/Rfree | 0.181/0.240 |
| No. of atoms | |
| Protein | 2100 |
| Magnesium (Mg2+) | 2 |
| Water | 144 |
| Avg B-factor (Å2) | 47.0 |
| Protein | 46.5 |
| CheY | 42.6 |
| FliM peptide | 82.7 |
| Magnesium (Mg2+) | 29.3 |
| Water | 55.0 |
| Root mean square deviations | |
| Bond length (Å2) | 0.007 |
| Bond angle (o) | 1.3 |
Data for the highest-resolution shell are shown in parentheses.
Structure analysis.
Ramachandran statistics were generated using PROCHECK (18). Molecular images were generated using PyMOL (DeLano Scientific). Data plots were generated in SigmaPlot (Synergy Software, Reading, PA). Least-squares superposition of various CheY structures onto the structure of unphosphorylated CheY (Protein Data Bank accession no. 3CHY.pdb) (28) was performed using the program LSQKAB (16), which also calculates individual Cα displacements and overall root mean square deviation. All Cα atoms were used to generate the superpositions, with the exception of the nine C-terminal residues (121 to 129) of the BeF3−-CheY-FliM16 complex, which take on a conformation significantly different from that observed in the other CheY structures. Phi and psi angles were determined in the program O (15). Pseudodihedral angles were determined from the appropriate Protein Data Bank coordinate files by using an online program (www.biotechnology.uwc.ac.za/teaching/BTY226/Week5/dihed_calc.html) and confirmed manually with O (15). All references to previously solved crystallographic structures, unless stated otherwise, are a reference to the first chain found in the Protein Data Bank entry.
RESULTS
The refined structure contains two unphosphorylated Mg2+-bound CheY-FliM16 complexes per asymmetric unit (Protein Data Bank accession no. 2B1J.pdb), which will hereafter be referred to simply as CheY-FliM16. Electron density corresponding to residues 2 through 16 of the bound peptides (chains C and D, bound to CheY chains A and B, respectively) is unambiguous, though it is weak at the termini (Fig. 2). Met1 of FliM16 could not be located unambiguously, presumably due to disorder, and was not modeled. Met1 of CheY is cleaved during bacterial overexpression and so was not modeled. Several features of the CheY-FliM16 complex are reminiscent of the BeF3−-CheY-FliM16 complex: the Tyr106 side chain is exclusively buried, the Ile95 Cδ 1-methyl group is oriented toward the phenyl ring of Tyr106, Lys109 and Asp12 side chains form a salt bridge, and Asn59 is oriented toward the β4-α4 loop. Similarly, Thr87 is sequestered toward the active site, and the β4-α4 and β5-α5 loops have undergone conformational changes relative to apoCheY that are similar in direction to, yet smaller in magnitude than, those changes observed in the BeF3−-CheY-FliM complex (Fig. 3 and 4).
FIG. 2.
Electron density and structural model corresponding to FliM16 (chain C, green) bound to unphosphorylated Mg2+-CheY (chain A, tan). The displayed electron density is a|Fo| − |fc|-simulated annealing omit map generated with chain C omitted in the program CNS. In order to display the weak density at the peptide termini, the map has been contoured at 3.6 σ around the entire peptide (cyan, fine mesh, residues 2 to 16) and at 2.4 σ around the peptide termini (purple, coarse mesh, residues 2 to 5, 15, and 16).
FIG. 3.
Displacements of the Cα atoms of several CheY conformations following a least-squares overlay with inactive CheY (3CHY) are shown for all residues (A) and residues in and around the β4-α4 loop (B). The CheY conformation in the CheY-FliM16 complex (heavy line, Protein Data Bank accession no. 2B1J) is intermediate between that of apoCheY (lower line) and the BeF3−-CheY-FliM16 complex (light upper line, Protein Data Bank accession no. 1F4V). The mean displacements of chains A and B of the CheY-FliM16 complex are shown. Displacements associated with the meta-active apoCheY substructure are also shown (dashed black line, Protein Data Bank accession no. 1JBE).
FIG. 4.
The superimposed structures of the CheY-FliM16 complex (chain A is in dark blue and chain B is in light blue; Protein Data Bank accession no. 2B1J), apoCheY (red; Protein Data Bank accession no. 3CHY.pdb), and the BeF3−-CheY-FliM16 complex (green; Protein Data Bank accession no. 1F4V) are shown for the -α4 loop, Thr87, and Tyr106 (A), the β4-α4 loop and Asn59 (B), and the β5-α5 loop and the α1 helix (C). In panel B, the hydrogen bond formed between Gly89 and Asn59 (dashed line; chain B, subconformation b) is similar to the association between these residues in the BeF3−-CheY-FliM16 complex. In panel C, the α1 helix of the CheY-FliM16 complex, which remains in the inactive conformation (see Fig. 3), is omitted for clarity. (D) A previously unobserved conformation of Glu89 in the β4-α4 loop of the CheY-FliM16 complex (chain A) is stabilized by hydrogen bonds with Asn94 and Tyr106 (dashed lines). All structures were superimposed onto inactive CheY (3CHY).
Structure overview.
The FliM peptide is in a conformation similar to that of the previously determined CheY-FliM16 complexes (8, 20). Additionally, the Mg2+ binding site is arranged as in previous Mg2+-CheY structures: the side chain carboxyl oxygens of Asp13 and Asp57, the backbone carbonyl oxygen of Asn59, and three water molecules (one of which H bonds to the Asp12 carboxyl oxygen) form an octahedral coordinate sphere around the bound divalent magnesium ion (4, 19, 20, 26). In contrast, Mg2+-CheY bound to FliM16 is not found in the canonical active (BeF3−-CheY-like) or inactive (apoCheY- or Mg2+-bound CheY-like) conformation. Instead, CheY in this complex occupies a region of conformational space between inactive and active CheY.
The two CheY molecules in the asymmetric unit are NCS related yet are not identical; chain B has more “active character” than does chain A, as shown in Fig. 4. However, because the differences are small (<1 Å), occur primarily in loop regions, and may arise from different constraints placed on the two complexes within the crystal, they will not discussed in detail here.
Details of the CheY-FliM16 interface.
In complex with Mg2+-CheY, the conformation of FliM16 is similar to that observed previously in the BeF3−-CheY-FliM16 (20) and CheY**-FliM16 (8) complexes; residues 2 through 7 are in an extended conformation, and residues 8 to 14 form two helical turns. Additionally, the amount of surface area that is buried in the CheY-FliM16 interface is similar for the three complexes (1,075 Å2 for unphosphorylated CheY-FliM16, 1,110 Å2 for BeF3−-CheY-FliM16, and 991 Å2 for CheY**-FliM16). For residues 6 to 12, details of the interactions that stabilize the conformation of FliM16 and anchor it to CheY are very similar to those described previously (8, 20) and will not be described in detail here. In contrast, the termini (residues 2 to 5 and 13 to 16) of FliM16 in complex with Mg2+-CheY differ from those in the previously determined BeF3−-CheY-FliM16 and CheY**-FliM16 structures. The β-sheet-like interaction between residue 4 of FliM and residue 90 of CheY is not formed in one of the NCS-related complexes (chains B and D), and in the other complex (chains A and C) it takes place at a substantially farther distance (3.3 Å, versus 2.6 Å for BeF3−-CheY-FliM16). Additionally, the salt bridge between Asp3 of FliM16 and Lys91 of CheY is not formed in the unphosphorylated Mg2+-CheY-FliM16 complex. At the C terminus of FliM16, residues 13 to 16 are arranged much less tightly about the helical axis than has been seen in previous structures (i.e., the backbone unwinds somewhat), which displaces the bulk of Asn16 away from the CheY backbone at Tyr106, preventing the Asp16 side chain from contributing to the H bond network that anchors FliM16 to β-sheet 4 of CheY (see Fig. 4 in reference 8). Additionally, the salt bridge between Lys122 Nɛ and the FliM16 C-terminal carboxylate is replaced by an H bond between Lys122 Nɛ and the Asn16 side chain carbonyl. High B-factors for the C-terminal residues of FliM16 are consistent with the loss of previously observed stabilizing interactions.
The loss of several previously observed intermolecular contacts in the CheY-FliM16 interface is consistent with the reduced affinity of FliM for CheY relative to CheY∼P. The observed perturbation of contacts between CheY and the N-terminal region of FliM16 appears to result largely from the inability of the β4-α4 loop to adopt a fully active conformation in the CheY-FliM16 complex (Fig. 4A). Clearly this result does not support the equilibrium shift view of activation, which supposes that CheY and CheY∼P interact identically with FliM.
The β4-α4 loop.
The β4-α4 loop (Thr87 to Lys91) following Thr87 is found in a conformation that approximates that of BeF3−-CheY, though to different degrees for the two NCS-related complexes. Whereas BeF3− activation results in a ∼3.5-Å shift at the Glu89 Cα relative to apoCheY, in chain A of the CheY-FliM16 complex this position is displaced from its inactive position by ∼1 Å, while in chain B the shift is larger (∼2 Å) though still less than that observed for the BeF3−-CheY-FliM16-complex (Fig. 4A). Additionally, residues 88 to 90 are modeled as two equally populated conformations in both CheY-FliM16 complexes, largely to accommodate two conformations of the Glu89 side chain. Therefore, it appears that this loop has a significant degree of conformational freedom in the CheY-FliM16 complex. Besides being in a position intermediate to the that of apoCheY and BeF3−-CheY, it should be noted that the four loop subconformations we observe (two per CheY) are each distinct from that of meta-active CheY, where the largest displacement (relative to the 1.7-Å apoCheY structure, which did not model static disorder in the β4-α4 loop) are at Ala90 (Fig. 3B). Incidentally, two of the subconformations of the β4-α4 loop that we observe (present in chain A and chain B) are stabilized by H bonds between the Glu89 carboxylate side chain and the Asn94 side chain amide and Tyr106 side chain hydroxyl, giving the Glu89 side chain the previously unobserved ability to make van der Waal contact with Thr87 (Fig. 4D).
The α1 helix remains unswitched.
The α1 helix of unphosphorylated Mg2+-CheY in complex with FliM16 does not undergo conformational change relative to apoCheY (Fig. 3). Conversely, backbone atoms in the α1 helix experience some of the largest displacements (>2 Å) when active (e.g., BeF3−-CheY, BeF3−-CheY-FliM16, or CheY**-FliM16) and inactive (e.g., apoCheY, Mg2+-CheY, or CheY** alone) CheY structures are compared (Fig. 3; see also Fig. 1 in reference 8). In contrast to the case for wild-type CheY, FliM16 binding (in the absence of Mg2+ or a phosphoryl analog) is sufficient to flip the conformational switch in this region of CheY**.
Thr87 is partially switched and disordered.
Due to its role in phosphorylation/dephosphorylation chemistry, position 87 (or its equivalent) is highly conserved as a small hydroxyl-containing residue (Ser or Thr) in the response regulator protein superfamily. One feature of the CheY-FliM16 complex that is particularly confounding is the intermediate position and weak electron density of the Thr87 side chain, since this residue is well ordered in other CheY structures. The disorder at this position is evidenced by high side chain B-factors, which are not observed for the Thr87 side chain in other CheY structures or for buried residues generally. Because static disorder in this region prompted us to model two conformations of the β4-α4 loop (Fig. 4A), at several points during refinement dual conformations of Thr87 (backbone and side chain) were modeled and subjected to rounds of refinement. However, because these efforts led to increased Rfree values and no apparent improvement in the quality of the Thr87 side chain electron density, they were abandoned in favor of a single Thr87 conformation. We imagine that, similar to the β4-α4 loop, Thr87 in the CheY-FliM16 complex has significant conformational freedom, though we cannot rule out the possibility that it takes on two closely spaced static conformations in different unit cells of the crystal (static disorder) which are not resolved. As discussed further in Discussion, the Y-T coupling model would have predicted Thr87 to be forced to occupy the active conformation due to the complete burial of Tyr106 in complex with FliM16, though this is clearly not the case (Fig. 4A).
DISCUSSION
Conformational coupling in unphosphorylated CheY.
Information flow between the phosphorylation site and the target binding site has been shown to be bidirectional for CheY and other response regulator proteins (1, 24). The notion that ligands can affect CheY autophosphorylation kinetics by inducing conformational changes at the active site, termed conformational coupling, was hypothesized previously to explain the ability of FliM16 to accelerate CheY autophosphorylation by nearly 30-fold (24). The “two-way street” notion of allostery assumes that allosteric communication in both directions (phosphorylation site to the target binding site and vice versa) happens by a flip of the same conformational switch, yet in complex with FliM16, unphosphorylated CheY remains in a substantially inactive conformation (11).
In addition to supporting or refuting allosteric models, this structure provides insight as to the relationship between structure and function (binding, catalysis, etc.) in CheY. In the structure of BeF3−-CheY, the Thr87 hydroxyl and Ala88 backbone amide coordinate the BeF3− fluorine atoms that are thought to be analogous to the aspartyl-phosphate phosphoryl oxygens (19). In complex with FliM16, Thr87 and Ala88 of unphosphorylated CheY are displaced toward the phosphoryl binding site relative to apoCheY (Fig. 4A) and Mg2+-CheY, despite the absence of BeF3−. However, in the CheY-FliM16 complex, the Thr87 side chain does not move so far as to overlay with Thr87 of the BeF3−-CheY complex but instead is in a position intermediate to those of unphosphorylated CheY and BeF3−-CheY (Fig. 4A). This repositioning of Thr87 and Ala88 in the direction of the phosphoryl-binding site may account for the increased rate of CheY autophosphorylation in the presence of FliM16. This structure-function relationship is also supported by structural and biochemical studies of the CheY-CheAP2 complex, which has both a decreased autophosphorylation rate (24) and a significantly increased distance between the Thr87 side chain and the phosphorylation site (22) relative to CheY alone,. Similarly, when this hydroxyl is made unavailable to the active site by mutation of the Thr87 side chain to a non-Ser amino acid, the CheY autophosphorylation rate also decreases dramatically (2). In summary, (i) Thr87 can take on a spectrum of conformations intermediate to the fully active and fully inactive conformations, (ii) allosteric ligands can affect the Thr87 position, and (iii) the proximity of the Thr87 hydroxyl to the active site correlates positively with the autophosphorylation rate. These data provide evidence in support of the conformational coupling hypothesis and highlight a role for Thr87 in this process.
The equilibrium shift allosteric model.
The equilibrium shift model of activation predicts the structure of unphosphorylated Mg2+-CheY-FliM16 to be essentially identical to that of CheY∼P. While a comparison of BeF3−-CheY with the CheY-FliM16 structure clearly indicates that this is not the case, the degree to which these results refute the equilibrium shift model of activation is a matter of perspective. The CheY-FliM16 structure presented shows that in wild-type CheY the network of conserved residues can achieve an active-like arrangement in the absence of phosphorylation, as predicted by the equilibrium shift model. In contrast, phosphorylation appears to be a prerequisite for complete conformational switching of wild-type CheY, especially in regions remote from the target binding site such as the α1 helix and the β5-α5 loop (Fig. 3A). This indicates that, unlike for CheY**, the equilibrium shift model is not sufficient to describe the allosteric activation process for wild-type CheY.
FliM16 affinity and the two-state assumption.
The inability of unphosphorylated CheY to form several stabilizing contacts when bound to FliM16 (relative to BeF3−-CheY-FliM16) provides a molecular basis for the reduced FliM affinity of unphosphorylated CheY relative to that of CheY∼P. While the equilibrium shift model rests on the simple assumption that the folded protein can access only two stable states, its proponents must invoke preequilibrating subpopulations that are difficult or impossible to detect. Further, it ascribes affinity changes that result from phosphorylation to a shift in the inactive-to-active preequilibrium. Instead, our results suggest that the affinity difference has straightforward structural origins involving polar interactions near the peptide termini and that phosphorylation is in fact a prerequisite for the formation of the high-affinity FliM16 interface.
The concurrence of active-like (resembling BeF3−-CheY) and inactive-like (resembling apoCheY) features in the same CheY conformation represents a clear though unanticipated departure from the two-state model of CheY activation. In the structures of several Salmonella enterica serovar Typhimurium CheY-CheZC peptide complexes (solved in both the presence and absence of BeF3−), CheY is found to occupy an intermediate region of activation space (11), offering another example of CheY plasticity and reinforcing the notion that the two-state assumption it is not adequate for describing the spectrum of conformations that are accessible to CheY.
The Y-T coupling allosteric model.
The intermediate position of Thr87 relative to apoCheY and BeF3−-CheY was not anticipated by either the equilibrium shift model or the Y-T coupling model of activation. Because Tyr106 is buried in the CheY-FliM16 complex, yet the “coupled” Thr87 is not in the fully active position (Fig. 4A), the correlation between the conformations of these two residues does not appear to be exclusive and may involve the influence of another region of the protein. An alternate model to Y-T coupling that is consistent with the structure presented here would recognize the participation of the β4-α4 loop, in addition to steric occlusion, to explain the conformational correlation between Tyr106 and Thr87.
The structures of meta-active CheY (25), the CheY-CheZC complex (11), and the CheY-FliM16 complex indicate that Tyr106 can be buried without inducing a fully active conformation of the β4-α4 loop, while the structures of BeF3−-CheY (19) and the BeF3−-CheY-FliM16 (20), CheY**-FliM16 (8), and BeF3−-CheY-CheZC (11) complexes suggest that Thr87 adopts the fully active conformation only when both the Tyr106 side chain and β4-α4 loop are in their fully active conformations. Because the Thr87 Cα of the CheY-FliM16 complex is essentially in the active position, yet the Cα-Cβ bond vector is inclined away from the active site as in apoCheY (Fig. 4A and D), it appears that the fully active orientation of the β4-α4 loop is related to a twist in the backbone that makes the Thr87 hydroxyl available to the phosphorylation site. In a past study, the pseudodihedral angle defined by the Cα atoms of residues 87, 88, 89, and 90 was used to quantify the β4-α4 loop activation state (10). For unphosphorylated (FliM16-free) CheY structures, including meta-active CheY, the value of this pseudodihedral angle is in the range of 10° to 30°, while values of ∼110° characterize the BeF3−-CheY and BeF3−-CheY-FliM16 structures (Fig. 5A). In the CheY**-FliM16 complex, the only instance where complete switching to the active (BeF3−-CheY-like) conformation is achieved in the absence of a phosphoryl analog, the value of the 87:88:89:90 pseudodihedral angle is nearly identical to that of BeF3−-CheY (108° and 110°, respectively). Consistent with the intermediate conformations of this loop in the CheY-FliM16 structure (Fig. 4A), the values of the 87:88:89:90 pseudodihedral angle for the four observed subconformations (two per CheY) in the structure presented take on a spectrum of values intermediate to those for the canonical active and inactive conformations (Fig. 5A). The partially activated position of the β4-α4 loop in phosphono-CheY (12) is also evidenced by an intermediate value of the 87:88:89:90 pseudodihedral angle (Fig. 5A).
FIG. 5.
(A) A spectrum of values are observed for the 87:88:89:90 Cα pseudodihedral angle in different wild-type CheY structures. The values of this angle for the CheY-FliM16 complex (Protein Data Bank accession no. 2B1J) are intermediate those of apoCheY (Protein Data Bank accession no. 3CHY) and BeF3−-CheY (Protein Data Bank accession no. 1FQW). The remaining structures shown are the high-resolution apoCheY structure (Protein Data Bank accession no. 1JBE, 2 subconformations), phosphono-CheY (Protein Data Bank accession no. 1C4W), and the BeF3−CheY-FliM16 complex (Protein Data Bank accession no. 1F4V). In contrast to wild-type CheY, CheY** exhibits a switch-like response to FliM16-binding (CheY**, Protein Data Bank accession no. 1U8T, chain C; CheY**-FliM16, Protein Data Bank accession no. 1U8T, chain A). (B) There is not a strong correlation between the degree of active character in the β4-α4 loop and the rotamer state of the Tyr106 side chain; the buried conformation of Tyr106 is consistent with the entire spectrum of observed β4-α4 loop conformations. However, the combination of an active β4-α4 loop and the solvent-exposed rotamer of Tyr106 is unoccupied. Structures having two subconformations of either Tyr106 or the β4-α4 loop are represented by two points connected with a dashed line. (C) The degree of active character in the β4-α4 loop (as measured by the 87:88:89:90 Cα pseudodihedral angle) does correlate well with the phi and psi angles of Val86, which position theThr87 side chain. Data derived from the structures of Mg2+-CheY (Protein Data Bank accession no. 2CHE), as well as the activating mutants CheYD13K (Protein Data Bank accession no. 1EHC), CheY!95V (Protein Data Bank accession no. 1D4Z), and CheYY106W (Protein Data Bank accession no. 5CHY) are additionally shown in panels B, C, and D. To avoid redundancy, the lower-resolution apoCheY structure (Protein Data Bank accession no. 3CHY) is not included in panel B, C, or D, and only the high-resolution apoCheY structure (Protein Data Bank accession no. 1JBE) is shown.
By devising metrics for local conformational change and plotting the values of these metrics as pairwise combinations for a family of related structures, one can get a sense of the degree of coupling between two regions of a protein. First, the focus here will be on the β4-α4 loop and its degree of coupling to Tyr106 and Thr87, after which the apparently weak coupling between Tyr106 and Thr87 will be revisited in a quantitative manner. Analysis of a number of CheY structures indicates that the solvent-exposed (inactive) rotamer position of Tyr106 (as measured by the Tyr106 chi1 dihedral angle) is observed only when the β4-α4 loop is in the inactive conformation but that the buried (active) Tyr106 rotamer can be found in conjunction with the entire spectrum of possible β4-α4 loop conformations (Fig. 5B). From the perspective of the loop, the inactive (apoCheY-like) β4-α4 conformation is found in conjunction with both the buried and exposed Tyr106 conformations, while the active loop conformation is found only when Tyr106 is buried (Fig. 5B). From these data it is clear that the conformations of Tyr106 and the β4-α4 loop are not exclusively coupled, although the combination of Tyr106 being exposed and β4-α4 being active appears to be forbidden. Note that the combination of Tyr106 being buried and β4-α4 being inactive, which was reported previously to be sterically discouraged (25), is occupied by both meta-active CheY and, to a lesser degree, the CheY-FliM16 complex. There appear to be at least two reasons for this: (i) the pseudodihedral angle defined by the Cα positions of residues 87, 88, 89, and 90 of meta-active CheY is essentially the same as that of inactive CheY, and (ii) Tyr106 has some flexibility in the buried conformation and can move slightly to alleviate potential steric clashes with inactive-like conformations of the β4-α4 loop, as observed in the CheY-FliM16 complex. The later observation was also made by the authors of the report on the CheY-CheZC complex structure, where the buried (active) conformation of Tyr106 is also found in conjunction with a substantially inactive β4-α4 loop (11).
Further analysis of this set of CheY structures suggests that the conformation of the β4-α4 loop does correlate with the orientation of the Thr87 side chain. Because the phosphorylation-dependent reorientation of the Thr87 side chain is accomplished largely by changes in the Val86 phi and psi angles (−111° and 136°, respectively, in apoCheY and −137° and 100°, respectively, in BeF3−-CheY), these values were used as a metric to describe Thr87 orientation, providing a quantitative measure of the Thr87 activation state. By plotting the 87:88:89:90 pseudodihedral angle versus either the phi (Fig. 5C) or psi (Fig. 5D) angle of Val86 for the same family of CheY structures as in Fig. 5B, it is clear that aside from the CheY-FliM16 complex, there exist two well resolved clusters that correspond to the inactive and active forms of CheY. Whereas the BeF3−-CheY-FliM16 complex is situated firmly in the active region of pseudodihedral-phi and pseudodihedral-psi space, the CheY-FliM16 complex is found in an intermediate region, between the inactive and active clusters of CheY structures (Fig. 5C and D). This result supports the notion that Thr87 and the β4-α4 loop are coupled (“T-loop coupling”) and is consistent with the partially switched character of this complex that is evident in structure superpositions (Fig. 3 and 4). When the set of structures in Fig. 5B (except CheY-FliM16) are considered, the Val86 phi and psi angles correlate well with the 87:88:89:90 pseudodihedral angle (r values of 0.94 and 0.95, respectively, for a linear fit to Fig. 5C and D). Conversely, for this same set of structures the Val86 phi and psi angles are correlated poorly with the chi1 angle of Tyr106 (r values of 0.58 and 0.53, respectively, for a linear fit) (data not shown).
The T-loop-Y allosteric model.
In contrast with the central tenet of the Y-T coupling model, our analysis indicates that the conformation of Thr87 and the Tyr106 rotamer state are not strongly coupled (Fig. 5B), suggesting the existence of a more robust pathway along which the allosteric information can “flow.” Strong coupling does exist between the Thr87 side chain orientation and the conformation of the β4-α4 loop (Fig. 5C and D), highlighting an alternate pathway along which the phosphorylation signal could potentially be transmitted from the active site to the target binding site. Molecular dynamics simulations offer a similar picture of CheY allostery, wherein Thr87 and the phosphorylation signal communicate with Tyr106 through the β4-α4 loop, rather than by steric occlusion as suggested in the original Y-T coupling model. To supplant the Y-T coupling model, these data support a T-loop model whereby conformational changes at Thr87 (e.g., phosphorylation) are strongly coupled to the conformation of the β4-α4 loop. The conformation of the β4-α4 loop is likely to impact target binding directly, as evidenced by the inability of FliM16 to form previously observed contacts with the partially active loop of unphosphorylated CheY. Therefore, in regard to target binding, inducing the active conformation of the β4-α4 loop is an end in itself. However, we imagine that the more important role of this loop in CheY allostery is to facilitate communication between Thr87 and Tyr106.
Analysis of Fig. 5B highlights a potential route for allosteric communication from the β4-α4 loop to the bulky Tyr106 side chain on the ligand binding surface. Although our analysis has already concluded that the β4-α4 loop and Tyr106 are not strongly coupled in all cases, the apparent presence of an forbidden combination (Tyr106 exposed and the loop active [Fig. 5B]) means that the conformation of one appears to restrict, or gate, the conformation of the other, in certain cases. Specifically, an active loop specifies burial of Tyr106, and solvent exposure of Tyr106 specifies an inactive loop, while the presence of an inactive loop or a buried Tyr106 specifies nothing about the partner (Fig. 5B). Note that these relationships are directional; although an active loop specifies burial of Tyr106, the burial of Tyr106 does not specify an active loop.
The T-loop-Y model refers simply to an extension of the T-loop model that acknowledges the mutually restrictive relationship that exists between the β4-α4 loop and Tyr106. According to this model, in apoCheY the default inactive conformation of Thr87 is directly coupled to a substantially or fully inactive conformation of the β4-α4 loop. The notion that the inactive conformation(s) of the β4-α4 loop places little or no restriction on the Tyr106 (though it responds slightly to the conformation of Tyr106) agrees with the dual conformations of the aromatic ring in apoCheY. Any attempt to force the ring entirely into the buried conformation will be poorly or incompletely transmitted to the active site, since burial of Tyr106 places few, if any, restrictions on the β4-α4 loop, consistent with the CheY-FliM16 and CheY-CheZC (11) complex structures where BeF3− is absent. In contrast, forcing Tyr106 to be entirely exposed, as is the case when CheY is bound to CheA, effectively gates the β4-α4 loop in the fully inactive conformation, consistent with the CheY-CheAP2 complex structure (22). Similarly, locking Thr87 in the active conformation (by phosphorylation) results directly in an activated conformation of the β4-α4 loop, which efficiently gates Tyr106 to take on only the buried conformation, though it should be noted that in several of the recently published BeF3−-CheY-CheZC structures (11), the binding of BeF3− does not necessarily result in complete switching of Thr87 (or, incidentally, of the β4-α4 loop, consistent with our model). One of these structures (Protein Data Bank accession no. 2FMH.pdb) provides particularly compelling support for the T-loop coupling notion, since there exist dual conformations of Thr87 and the β4-α4 loop which have corresponding activation states (i.e., active-active and inactive-inactive). In summary, the T-loop-Y model (i) reconciles the complete switching of BeF3−-CheY and CheY-CheAP2 with the incomplete switching of the FliM16- or CheZC-bound CheY complexes and (ii) provides a rationale for the conformational heterogeneity of both Tyr106 in apoCheY and Thr87 in theCheY-FliM16 complex.
Tyr106 gating.
The predicted ability of the inactive β4-α4 loop to discourage the buried orientation of Tyr106, which was hypothesized on the basis of the meta-active CheY substructure (25) and described using the term “gating” in a more recent study (10), is apparent only as a weak trend in our analysis. Instead, our analysis indicates that restricting the loop in the active conformation very effectively gates the Tyr106 side chain in the buried orientation and that restricting the Tyr106 side chain in the solvent-exposed conformation effectively gates the loop to the inactive conformation. While the structural basis of the latter relationship is unclear, the ability of the active loop to gate Tyr106 is likely due to the hydrogen bond formed between the Tyr106 side chain hydroxyl and the Glu89 backbone carbonyl observed in activated forms of CheY (e.g., BeF3−-CheY, BeF3−-CheY-FliM16, BeF3−-CheY-CheZC, and even CheY**-FliM16, where the indole nitrogen on Trp106 serves as the proton donor). Therefore, these data encourage use of the term gating to acknowledge the more efficient Tyr106 gating ability of the active, rather than the inactive, loop conformation (Fig. 5B), though the two mechanisms are not mutually exclusive. The T-loop-Y model also recognizes the ability of Tyr106, when solvent exposed, to gate the β4-α4 loop in the inactive conformation, as is the case when CheY binds to CheAP2.
Relevance to CheZC peptide binding.
As mentioned previously, the recently published CheY-CheZC complex (11) also agrees well with this formulation of the T-loop-Y model, since the burial of Tyr106 by bound CheZ is not sufficient to cause switching of Thr87 and results in only modest displacement of the β4-α4 loop in the absence of BeF3−. Additionally, in all seven of the CheY-CheZ structures (with and without BeF3−) and the two substructures, differing degrees of Thr87 activation correlate with the β4-α4 loop activation state.
In contrast, the structure-function trend that we observe for Thr87 of CheY complexed with FliM16 and CheAP2 (positive correlation between the Thr87 hydroxyl-to-phosphorylation site distance and observed autophosphorylation rate) is not apparent in the CheY-CheZC complexes. Instead, in the CheY-CheZC structures that lack BeF3−, Thr87 retains a distinctly inactive conformation despite biochemical studies that shows CheZC binding to accelerate CheY phosphorylation kinetics (24). This may indicate that that the mechanism by which events at the peptide ligand binding surface are conformationally coupled to the active site may be subtly different for different ligands.
In conclusion, the crystallographic CheY-FliM16 complex demonstrates that the structural changes associated with CheY phosphorylation are not exclusively coupled and in some regions are capable of adopting conformations intermediate to those of the inactive and active states. Additionally, the partially switched conformation that we observe for the crystalline CheY-FliM16 complex agrees well with the nuclear magnetic resonance peak positions of this complex, which are typically intermediate to those of inactive CheY (apo or Mg2+-bound) and active (BeF3−-bound) forms of CheY (C. M. Dyer and F. W. Dahlquist, unpublished observations). In light of these results, it is clear that neither the equilibrium shift model, the Y-T coupling model, nor the two-state assumption upon which both models are based is a fully valid descriptor of allostery in CheY. Rather, the T-loop-Y model, which was anticipated on the basis of recent molecular dynamics simulations and acknowledges the active role of the β4-α4 loop in allosteric signaling (10), seems more consistent with the experimental results.
.
Acknowledgments
We thank John Perona for the use of the R-AXIS II. We also thank Brian Matthews for helpful suggestions regarding the manuscript prior to submission.
This work was funded by NIH grant GM059544 (to F.W.D.).
REFERENCES
- 1.Ames, S. K., N. Frankeema, and L. J. Kenney. 1999. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc. Natl. Acad. Sci. USA 96:11792-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleby, J., and R. Bourret. 1998. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J. Bacteriol. 180:3563-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak, R., and M. Eisenbach. 1992. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31:1821-1826. [DOI] [PubMed] [Google Scholar]
- 4.Bellsolell, L., J. Prieto, L. Serrano, and M. Coll. 1994. Magnesium binding to the bacterial chemotaxis protein CheY results in large conformational changes involving its functional surface. J. Mol. Biol. 238:489-495. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, N. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR systems (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Clegg, D. O., and D. E. J. Koshland. 1984. The role of a signaling protein in bacterial sensing: behavioral effects of increased gene expression. Proc. Natl. Acad. Sci. USA 81:5056-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 8.Dyer, C. M., M. L. Quillin, A. Campos, J. Lu, E. M. Westbrook, P. Matsumura, B. W. Matthews, and F. W. Dahlquist. 2004. Structure of the constitutively active double mutant CheYD13KY106W alone and in complex with a FliM peptide. J. Mol. Biol. 342:1325-1335. [DOI] [PubMed] [Google Scholar]
- 9.Feher, V. A., and J. Cavanagh. 1999. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature 400:289-293. [DOI] [PubMed] [Google Scholar]
- 10.Formaneck, M., L. Ma, and Q. Cui. 2006. Reconciling the “old” and “new” views of protein allostery: a molecular simulation study of chemotaxis Y protein (CheY). Proteins 63:846-867. [DOI] [PubMed] [Google Scholar]
- 11.Guhaniyogi, J., V. L. Robinson, and A. M. Stock. 2006. Crystal structures of beryllium fluoride-free and beryllium bound-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J. Mol. Biol. 359:624-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halkides, C. J., M. M. McEvoy, E. Casper, P. Matsumura, K. Volz, and F. W. Dahlquist. 2000. The 1.9 Å resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY. Biochemistry 39:5280-5286. [DOI] [PubMed] [Google Scholar]
- 13.Hess, J. F., R. B. Bourret, and M. I. Simon. 1991. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol. 200:188-204. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, M., R. B. Bourret, M. I. Simon, and K. Volz. 1997. Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY. J. Biol. Chem. 272:11850-11855. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. A., J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch, W. 1976. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32:922-923. [Google Scholar]
- 17.Kissinger, C. R., D. K. Gehlhaar, and D. B. Fogel. 1999. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D 55:484-491. [DOI] [PubMed] [Google Scholar]
- 18.Lakowski, R. A., M. W. Macarthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 19.Lee, S.-Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S.-Y., H. S. Cho, J. G. Pelton, D. Yan, R. K. Henderson, D. S. King, L.-s. Huang, S. Kustu, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of an activated response regulator bound to its target. Nat. Struct. Biol. 8:52-56. [DOI] [PubMed] [Google Scholar]
- 21.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB newsletter on protein crystallography, no. 26. Daresbury Laboratories, Warrington, United Kingdom.
- 22.McEvoy, M., A. C. Hausrath, G. B. Randolph, S. J. Remington, and F. W. Dahlquist. 1998. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc. Natl. Acad. Sci. USA 95:7333-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moy, F. J., D. F. Lowry, P. Matsumura, F. W. Dahlquist, J. E. Krywko, and P. J. Domaille. 1994. Assignments, secondary structure, global fold, and dynamics of chemotaxis Y protein using three- and four-dimensional heteronuclear (13C,15N) NMR spectroscopy. Biochemistry 33:10731-10742. [DOI] [PubMed] [Google Scholar]
- 24.Schuster, M., R. E. Silversmith, and R. B. Bourret. 2001. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA 98:6003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonovic, M., and K. Volz. 2001. A distinct meta-active conformation in the 1.1-Å resolution structure of wild-type apo CheY. J. Biol. Chem. 276:28637-28640. [DOI] [PubMed] [Google Scholar]
- 26.Stock, A., E. Martinez-Hackert, B. Rasmussen, A. West, J. Stock, D. Ringe, and G. Petsko. 1993. Structure of the Mg(2+) bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry 32:13375-13380. [DOI] [PubMed] [Google Scholar]
- 27.Volkman, B. F., D. Lipson, D. E. Wemmer, and D. Kern. 2001. Two-state allosteric behavior in a single-domain signaling protein. Science 291:2429-2433. [DOI] [PubMed] [Google Scholar]
- 28.Volz, K., and P. Matsumura. 1991. Crystal structure of Escherichia coli refined at 1.7-Å resolution. J. Biol. Chem. 266:15511-15519. [DOI] [PubMed] [Google Scholar]