Abstract
Escherichia coli cells with mutations in recBC genes are defective for the main RecBCD pathway of recombination and have severe reductions in conjugational and transductional recombination, as well as in recombinational repair of double-stranded DNA breaks. This phenotype can be corrected by suppressor mutations in sbcB and sbcC(D) genes, which activate an alternative RecF pathway of recombination. It was previously suggested that sbcB15 and ΔsbcB mutations, both of which inactivate exonuclease I, are equally efficient in suppressing the recBC phenotype. In the present work we reexamined the effects of sbcB15 and ΔsbcB mutations on DNA repair after UV and γ irradiation, on conjugational recombination, and on the viability of recBC (sbcC) cells. We found that the sbcB15 mutation is a stronger recBC suppressor than ΔsbcB, suggesting that some unspecified activity of the mutant SbcB15 protein may be favorable for recombination in the RecF pathway. We also showed that the xonA2 mutation, a member of another class of ExoI mutations, had the same effect on recombination as ΔsbcB, suggesting that it is an sbcB null mutation. In addition, we demonstrated that recombination in a recBC sbcB15 sbcC mutant is less affected by recF and recQ mutations than recombination in recBC ΔsbcB sbcC and recBC xonA2 sbcC strains is, indicating that SbcB15 alleviates the requirement for the RecFOR complex and RecQ helicase in recombination processes. Our results suggest that two types of sbcB-sensitive RecF pathways can be distinguished in E. coli, one that is activated by the sbcB15 mutation and one that is activated by sbcB null mutations. Possible roles of SbcB15 in recombination reactions in the RecF pathway are discussed.
Homologous genetic recombination is a fundamental process that has two major roles in living cells: first, to facilitate DNA repair, thus maintaining chromosome integrity; and second, to rearrange genes within and between chromosomes, thereby promoting genetic diversity. The right balance between these two roles of recombination contributes considerably to cell survival and evolution.
In wild-type Escherichia coli, a number of recombination events proceed via the RecBCD-mediated pathway (13, 36). RecBCD is a complex multifunctional enzyme composed of three subunits encoded by the recB, recC, and recD genes. It recognizes blunt or nearly blunt double-stranded DNA (dsDNA) ends (39), which can arise in the cell by spontaneous or induced chromosome breakage or by DNA transfer during conjugation, transduction, and transformation. Starting from a dsDNA end, RecBCD initiates recombination by unwinding and simultaneously degrading DNA. Upon encountering a specific sequence designated Chi, the 3′-5′ nuclease activity of the enzyme is attenuated, and weaker 5′-3′ activity is activated (3). This nuclease modification allows production of a long 3′ single-stranded DNA (ssDNA) tail, onto which RecBCD directs loading of RecA protein (4, 5). A nucleoprotein filament created in this way plays a crucial role in further reactions of homologous DNA pairing and strand exchange.
recB and/or recC mutants of E. coli lack all RecBCD activities and exhibit severe recombination deficiency during genetic crosses, as well as sensitivity to various DNA-damaging agents (UV, ionizing radiation, chemical agents, etc.) that produce dsDNA breaks (12, 21). The low residual level of recombination in recB(C) null mutants can be restored to a level close to the wild-type level by extragenic sbcB and sbcC(D) suppressor mutations that activate an alternative RecF recombination pathway (named after recF, the first gene discovered in this pathway) (12, 19, 23, 25). Initiation of recombination in the RecF pathway depends on several enzymes that substitute for missing activities of RecBCD. Recombinogenic 3′ ssDNA overhangs are produced by the combined action of the RecQ helicase (or the UvrD and HelD helicases) and RecJ 5′-3′ ssDNA exonuclease, whereas the RecFOR proteins facilitate the loading of RecA protein onto prepared ssDNA (for reviews, see references 20 and 24).
The exact mechanism of activation of the RecF pathway by sbc mutations is not completely understood. Mutations in sbcB were shown to inactivate exonuclease I (ExoI), the enzyme that digests ssDNA in the 3′-5′ direction (23). It was assumed that elimination of ExoI activity by an sbcB mutation preserves recombinogenic 3′ ssDNA tails formed by the RecBCD-independent mechanism (12, 23). The sbcC and sbcD mutations accumulate spontaneously during propagation of recBC sbcB strains and are required for full suppression of the RecBC− phenotype (18, 25). The sbcC and sbcD genes belong to the same operon and code for subunits of the SbcCD nuclease (18). It was shown previously that SbcCD acts both as an endonuclease that cleaves hairpin structures and as an exonuclease that degrades linear dsDNA molecules (14, 15). Genetic evidence suggests that the SbcB and SbcCD enzymes have redundant roles in blunting UV-generated DNA ends prior to RecBC(D) action, implying that SbcCD has an exonuclease activity with 3′ protruding ends (35). Such an activity might explain the antirecombinogenic effect of the SbcCD enzyme in recBC sbcB cells and the necessity for sbcC(D) mutations in order to obtain full activation of the RecF pathway.
Most previous genetic studies of the RecF pathway were performed with recBC sbcB sbcC(D) strains carrying the sbcB15 suppressor mutation. sbcB15 belongs to the first group of sbcB mutations isolated, which were shown to suppress efficiently both DNA repair and the homologous recombination deficiency of recBC strains (23). These mutations were isolated after treatment of recBC cells with the chemical mutagen ethyl methane sulfonate (23), and they were mapped within the gene coding for ExoI (40). In further genetic characterization of the sbcB locus, two sbcB deletion mutants were also isolated and tested for recBC suppression in parallel with the sbcB15 allele. It was found that the ΔsbcB and sbcB15 mutations had the same suppressive effect on the RecBC− phenotype, increasing conjugational recombination and UV resistance to nearly wild-type levels (40). Concomitant with characterization of sbcB mutations, another class of ExoI mutations was identified after treatment of recBC cells with nitrosoguanidine. Interestingly, this class, designated xonA mutations, efficiently suppressed UV sensitivity but had only a modest suppressive effect on the conjugational recombination defect in recBC cells (22). The failure of xonA mutations to restore conjugational recombination with the same efficiency as sbcB was first suggested to result from a small amount of residual ExoI activity left in xonA cells (22). Later characterization of xonA mutations argued against this hypothesis since it was shown that some xonA mutants (xonA2 and xonA6) are completely devoid of ExoI activity, whereas sbcB15 mutants possess traces of this activity (31). Therefore, it was proposed that mutant ExoI in xonA strains might have some unspecified activity (other than ssDNA degradation) which interferes with RecF pathway enzymes (31).
Although early findings suggested that the sbcB15 mutation was functionally equivalent to an sbcB deletion (and therefore could be considered a null mutation) (for a review, see reference 21), several later observations indicated that the two mutations had different effects on some recombination processes. A study of the role of ssDNA exonucleases in λ phage crosses revealed that the sbcB15 allele inhibits the nucleolytic processing of DNA ends much more strongly than ΔsbcB inhibits this processing (32). Bidnenko et al. (9) studied recombinational repair in rep mutants (deficient for an auxiliary replicative helicase, the Rep protein), which suffer from frequent breakage of the replication fork and are therefore not viable in the absence of RecBCD. They found that ΔsbcB sbcC mutations restore the viability of rep recBC mutants, while sbcB15 sbcC mutations do not. Recently, the sbcB15 mutation was found to increase the requirement for RuvABC proteins in recombinational repair after UV and γ irradiation, while an sbcB deletion had no such effect (43). Taking into account the finding that the sbcB15 allele is efficiently expressed as a stable full-length product (31), it has been suggested that the mutant SbcB15 protein, although inactive as ExoI, might have some other activity (possibly DNA binding) that affects the recombination process (9, 32, 43).
In the present work we reexamined the effects of the sbcB15, ΔsbcB, and xonA2 mutations on the RecBC− phenotype in different experimental systems by measuring DNA repair and recombination proficiency. The sbcB and xonA alleles were tested individually, as well as in combination with an sbcC mutation. We found that the effects of the two sbcB mutations differ significantly, and in the majority of tests the sbcB15 mutation proved to be a stronger recBC suppressor than ΔsbcB. Our results also showed that the xonA2 mutation has the same effect on recombination processes as ΔsbcB, suggesting that xonA2 is a null mutation. In addition, we demonstrated that recombination in a recBC sbcB15 sbcC mutant shows an alleviated requirement for the RecFOR complex and RecQ helicase compared with recBC ΔsbcB sbcC and recBC xonA2 sbcC strains.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The E. coli strains used in this study are listed in Table 1. New strains were constructed by P1 transduction, as described by Miller (28). Transductants were isolated on LB medium plates (28) supplemented with appropriate antibiotics (chloramphenicol, 15 μg/ml; kanamycin, 50 μg/ml; tetracycline, 10 μg/ml) or on M9 plates (28) supplemented with glucose (0.4%), vitamin B1 (1 μg/ml), and all required amino acids (100 μg/ml). The phenotype of sbcC201 transductants was confirmed by the increased efficiency of plating of λ phage carrying a 571-bp palindrome (18). λpal571 formed plaques on sbcC transductants with about 1,000-fold higher efficiency than it formed plaques on sbcC+ strains. Transfer of the xonA2 allele into the ΔsbcB recipient (Table 1) was additionally verified by PCR using primers 5′GACATGATCTGTTGCCACTC3′ (upstream) and 5′CCATCACCGATTATCAGCAG3′ (downstream).
TABLE 1.
E. coli strains
| Straina | Relevant genotype | Reference or source |
|---|---|---|
| AB1157 | rec+sbc+b | 6 |
| JC5519 | recB21 recC22 | 6 |
| JC7623 | recB21 recC22 sbcB15 sbcC201 | 23 |
| JC8260 | recB21 recC22 xonA2 hisG+ | 22 |
| JJC260 | sbcD300::kan | B. Michel |
| JJC889 | ΔsbcB::cam | 9 |
| N2364 | sbcC201 phoR79::Tn10 | 25 |
| BW6156 | Hfr proAB+ (PO3 of P4X) | 42 |
| K797 | phoR79::Tn10 | CGSC 6456c |
| KL742 | hisG+thyA748::Tn10 | CGSC 6212c |
| V330 | Δ(recC-argA)234 | 2 |
| WA576 | recF400::Tn5 | B. Micheld |
| SWM1003 | ΔrecQ::kan | 25 |
| LMM981 | recB21 recC22 sbcB15 sbcC201 hisG+ | P1.KL742 × JC7623 to His+ |
| LMM1124 | recB21 recC22 ΔsbcB::cam | P1.JJC889 × JC5519 to Cmr |
| LMM1128 | recB21 recC22 sbcB15 hisG+ | P1.LMM981 × LMM1124 to His+ Cms UVr |
| LMM1298 | recB21 recC22 sbcC201 phoR79::Tn10 | P1.N2364 × JC5519 to Tcr λpals |
| LMM1329 | recB21 recC22 ΔsbcB::cam sbcC201 phoR79::Tn10 | P1.JJC889 × LMM1298 to Cmr UVr |
| LMM1330 | recB21 recC22 sbcB15 hisG+sbcC201 phoR79::Tn10 | P1.LMM981 × LMM1329 to His+ Cms |
| LMM1331 | recB21 recC22 ΔsbcB::cam sbcC201 phoR79::Tn10 recF400::Tn5 | P1.WA576 × LMM1329 to Kmr UVs |
| LMM1332 | recB21 recC22 sbcB15 hisG+sbcC201 phoR79::Tn10 recF400::Tn5 | P1.WA576 × LMM1330 to Kmr UVs |
| LMM1362 | recB21 recC22 ΔsbcB::cam sbcC201 phoR79::Tn10 ΔrecQ::kan | P1.SWM1003 × LMM1329 to Kmr UVs |
| LMM1363 | recB21 recC22 sbcB15 hisG+sbcC201 phoR79::Tn10 ΔrecQ::kan | P1.SWM1003 × LMM1330 to Kmr UVs |
| LMM1745 | recB21 recC22 xonA2 hisG+sbcC201 phoR79::Tn10 | P1.JC8260 × LMM1329 to His+ Cms |
| LMM1746 | recB21 recC22 xonA2 hisG+sbcC201 phoR79::Tn10 recF400::Tn5 | P1.WA576 × LMM1745 to Kmr UVs |
| LMM1747 | recB21 recC22 xonA2 hisG+sbcC201 phoR79::Tn10 ΔrecQ::kan | P1.SWM1003 × LMM1745 to Kmr UVs |
| LMM1748 | recB21 recC22 xonA2 hisG+sbcD300::kan phoR+ | P1.JJC260 × LMM1745 to Kmr Tcs |
| LMM1749 | recB21 recC22 xonA2 hisG+sbcD+phoR79::Tn10 | P1.K797 × LMM1748 to Tcr Kms UVs λpalr |
| LMM1721 | thyA748::Tn10 | P1.KL742 × AB1157 to Tcrthy |
| LMM1724 | Δ(recC-argA)234 thyA+ | P1.V330 × LMM1721 to thy+ UVs T42s |
| LMM1725 | Δ(recC-argA)234 thyA+ ΔsbcB::cam | P1.JJC889 × LMM1724 to Cmr |
| LMM1726 | Δ(recC-argA)234 thyA+sbcB15 hisG+ | P1.LMM981 × LMM1725 to His+ UVr |
| LMM1732 | Δ(recC-argA)234 thyA+ ΔsbcB::cam sbcC201 phoR79::Tn10 | P1.N2364 × LMM1725 to Tcr UVr λpals |
| LMM1733 | Δ(recC-argA)234 thyA+sbcB15 hisG+sbcC201 phoR79::Tn10 | P1.LMM981 × LMM1732 to His+ Cms |
All strains except BW6156, K797, KL742, and V330 are derivatives of AB1157.
Other markers are F− thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 λ− rac− hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 qsr′.
Strain supplied by M. Berlyn of the Escherichia coli Genetic Stock Center.
Strain originated from the laboratory of W. Wackernagel.
Bacterial cultures were grown in LB medium (28) at 37°C with shaking. Cell growth was monitored by measuring the optical density at 600 nm (OD600). To determine the colony-forming ability of the strains, cells were appropriately diluted in phosphate buffer and plated on LB medium plates.
Irradiation experiments.
Bacteria were grown from a single colony in LB medium at 37°C until the OD600 was 0.2. For UV irradiation experiments, serial dilutions of bacterial cultures were spotted on LB medium plates and irradiated with several doses of UV light (254 nm) at a dose of 0.5 J/m2/s. The plates were incubated at 37°C for 24 to 48 h before the survivors were counted. In γ irradiation experiments, bacteria were pelleted by centrifugation, resuspended in cold phosphate buffer, and irradiated on ice with a 60Co source at a dose rate of 12 Gy/s. Appropriate dilutions of irradiated cells were plated on LB agar, and colonies of survivors were scored after 24 to 48 h of incubation at 37°C.
Conjugational crosses.
Hfr crosses were performed as described by Miller (28). Inheritance of the chromosomal Pro+ marker was assayed. Donor (BW6156) and recipient strains were grown at 37°C to an OD600 of 0.3 before they were mixed at a ratio of 1:10. Mating was allowed to proceed for 25 min. proAB+ recombinants were selected on M9 plates supplemented with glucose (0.4%), vitamin B1 (1 μg/ml), and all required amino acids (100 μg/ml) except proline. Streptomycin (100 μg/ml) was also added to the plates to counterselect donors.
RESULTS
Effects of sbcB15 and ΔsbcB mutations on recombinational DNA repair in recBC (sbcC) cells.
Early work on the RecF pathway was performed with strains initially considered to be recBC sbcB mutants. These strains were obtained by heavy mutagenic treatment of the recB21 recC22 strain JC5519 (23), and one of them, JC7623 carrying the sbcB15 mutation, is the strain that has been most widely used in further genetic studies. Later work by Lloyd and Buckman (25) revealed that JC7623 and some other sbcB derivatives of JC5519 had also acquired another suppressor mutation designated sbcC. To a certain degree, this finding brought into question previous interpretations of results obtained with strains believed to be recBC sbcB strains, including the observation of Templin et al. (40) that recBC sbcB15 and recBC ΔsbcB strains display equal recombination proficiencies.
To compare the effects of sbcB15 and ΔsbcB mutations (individually or in combination with sbcC) on DNA recombination and recombinational repair, we introduced these mutations into the recBC strain JC5519 by P1 transduction (Table 1). It was previously demonstrated that recBC sbcB15 mutants grow slowly and that fast-growing variants with mutations in sbcC or sbcD tend to accumulate spontaneously (18, 25). To avoid the possibility that uncharacterized suppressor mutations would influence our results, we carefully monitored the growth rates of our recBC sbcB constructs, frequently measuring the optical densities of exponential cultures and checking their typical small-colony phenotype. In addition, the SbcCD+ phenotype of recBC sbcB mutants was confirmed by the low efficiency of plating of λpal571 phage.
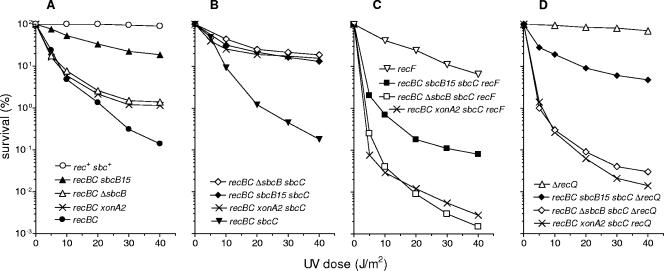
The DNA repair proficiency of the mutants constructed was first tested after exposure to UV radiation (Fig. 1). As expected, the recBC mutant showed pronounced UV sensitivity; at the highest UV dose applied (40 J/m2), its survival decreased almost 3 orders of magnitude compared to the survival of the wild-type strain (Fig. 1A). Both ΔsbcB and sbcB15 mutations considerably improved the survival of recBC cells exposed to UV light. However, while the effect of ΔsbcB was only partial, (ΔsbcB increased the survival about 10-fold at a dose of 40 J/m2), the sbcB15 mutation showed stronger suppression, resulting in a level of repair proficiency much closer to the wild-type level (Fig. 1A).
FIG. 1.
Effects of sbcB15, ΔsbcB, and xonA2 mutations on survival of different recB21 recC22 derivatives exposed to UV irradiation. The strains used were AB1157 (rec+ sbc+), JC5519 (recBC), LMM1124 (recBC ΔsbcB), LMM1128 (recBC sbcB15), and LMM1749 (recBC xonA2) (A); LMM1298 (recBC sbcC), LMM1329 (recBC ΔsbcB sbcC), LMM1330 (recBC sbcB15 sbcC), and LMM1745 (recBC xonA2 sbcC) (B); WA576 (recF), LMM1331 (recBC ΔsbcB sbcC recF), LMM1332 (recBC sbcB15 sbcC recF), and LMM1746 (recBC xonA2 sbcC recF) (C); and SWM1003 (ΔrecQ), LMM1362 (recBC ΔsbcB sbcC ΔrecQ), LMM1363 (recBC sbcB15 sbcC ΔrecQ), and LMM1747 (recBC xonA2 sbcC recQ) (D). The data for each strain are averages of results from at least three independent experiments.
In agreement with previous results (25), the sbcC mutation alone had no suppressive effect on the RecBC− phenotype (Fig. 1B). It also had no further effect on recBC UV sensitivity when it was combined with sbcB15. However, the sbcC mutation enhanced the suppressive effect of ΔsbcB so that the recBC ΔsbcB sbcC strains showed approximately the same UV resistance as recBC sbcB15 and recBC sbcB15 sbcC strains (Fig. 1A and B). As the recBC sbcB15 mutant behaved in UV repair exactly like the recBC sbcB15 sbcC mutant, we also examined whether there was a cryptic sbcC or sbcD suppressor mutation that was responsible for the high level of UV resistance of the former strain. Assuming that the presence of such a cryptic suppressor would also increase the survival of recBC ΔsbcB cells, we transduced the ΔsbcB mutation into the recBC sbcB15 strain and tested the transductants to determine their repair efficiencies. The ΔsbcB derivatives of the recBC sbcB15 strain showed decreased UV survival compared to the survival of the parental strain (not shown), just as previously observed with the recBC ΔsbcB mutant (Fig. 1A). The results described above suggest that the sbcB15 mutation alone is sufficient for maximal induction of UV repair via the RecF pathway (consistent with results of Lloyd and Buckman [25]), whereas with the sbcB deletion an additional mutation in sbcC is required to obtain the same repair efficiency.
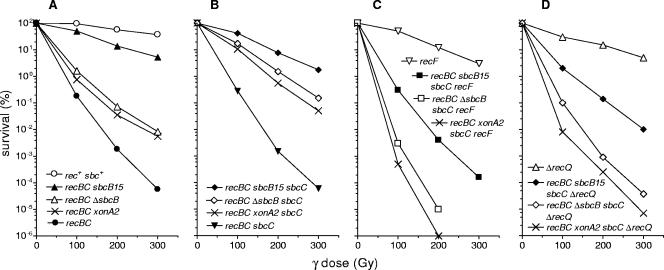
In a further analysis the strains described above were examined to determine their sensitivity to γ irradiation. Again, the ΔsbcB and sbcB15 mutations showed a marked difference in suppressing recBC sensitivity (Fig. 2A). As observed in the UV experiment, although ΔsbcB significantly increased the survival of recBC cells, the effect was moderate compared with that of the sbcB15 mutation, which almost completely restored resistance to γ irradiation.
FIG. 2.
Effects of sbcB15, ΔsbcB, and xonA2 mutations on the γ irradiation sensitivity of different recB21 recC22 derivatives. The strains used are listed in the legend to Fig. 1. The values are averages of results from at least three independent experiments.
The sbcC mutation had no significant effect on sensitivity to γ irradiation in either the recBC or recBC sbcB15 background (Fig. 2B), but it considerably increased the survival of recBC ΔsbcB cells. However, unlike the results of the UV experiment, the recBC ΔsbcB sbcC cells were still more sensitive to γ irradiation than either recBC sbcB15 or recBC sbcB15 sbcC cells (Fig. 2A and B). Therefore, we concluded that after γ irradiation, the sbcB15 mutation alone almost completely restores the repair proficiency of recBC cells, whereas the sbcB deletion cannot provide full suppression even when it is combined with sbcC.
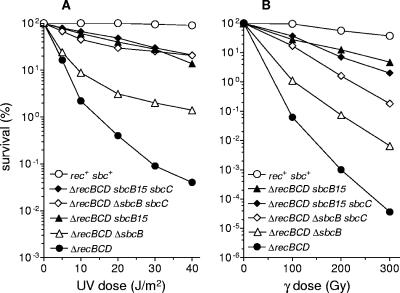
The analysis of sbcB mutations described above was performed in a recB21 recC22 background so that our results could be readily compared with previous analyses in which the workers predominantly used the same genetic background. The recB21 and recC22 mutations are classical mutations that are widely used in genetic studies, and according to all available data, either of these mutations is sufficient to abolish all known activities of the RecBCD enzyme (for reviews, see references 21 and 38). The recB21 allele contains a 1.4-kb insertion (IS186) in its coding region and is polar on recD (1, 2), whereas recC22 carries a UGA nonsense mutation (41) that presumably results in a truncated protein. To exclude the possibility that mutated RecB21 and RecC22 proteins influenced our results, the effects of sbcB mutations on recombinational repair were also examined with a strain having a complete deletion of the recBCD genes. We found that in a ΔrecBCD background, the sbcB mutations behaved essentially in the same way that they behaved in recB21 recC22 cells; i.e., sbcB15 proved to be a stronger suppressor of the RecBCD− phenotype than ΔsbcB (Fig. 3A and B).
FIG. 3.
Effects of sbcB15 and ΔsbcB mutations on survival after UV and γ irradiation in a ΔrecBCD background. The strains used were AB1157 (rec+ sbc+), LMM1724 (ΔrecBCD), LMM1725 (ΔrecBCD ΔsbcB), LMM1726 (ΔrecBCD sbcB15), LMM1732 (ΔrecBCD ΔsbcB sbcC), and LMM1733 (ΔrecBCD sbcB15 sbcC). The values are averages of results from at least three independent experiments.
sbcB15 and ΔsbcB mutations affect conjugational recombination differently in recBC (sbcC) mutants.
We further tested the recombination proficiency of different sbcB derivatives in conjugational crosses. During conjugation, ssDNA is transferred from the Hfr donor strain to the F− recipient, where it provides a template for DNA synthesis (for a review, see reference 17). When mating terminates, the transferred DNA is released as a double-stranded linear fragment that recombines with the recipient chromosome. This type of recombination can occur via either the RecBCD pathway (in wild-type cells) or the RecF pathway [in recBC sbcBC(D) mutants].
As shown in Table 2, in the recBC mutant the frequency of conjugational recombination decreased more than 100-fold, a result in accord with previous studies (16, 26). Introducing the ΔsbcB mutation into the recBC background had only a mild effect, increasing the frequency of recombination approximately twofold. The sbcB15 mutation had a much stronger effect, showing 10-fold-stronger recBC suppression than ΔsbcB. However, the recBC sbcB15 mutant still displayed significantly lower recombination than the wild-type strain.
TABLE 2.
Conjugational recombination with different sbcB recipient strains
| Recipient strain | Relevant genotype | Relative viabilitya | Relative yield of recombinantsb |
|---|---|---|---|
| AB1157 | Wild type | 1 | 1 |
| JC5519 | recB21 recC22 | 0.27 ± 0.022 | 0.008 ± 0.0014 |
| LMM1124 | recB21 recC22 ΔsbcB | 0.25 ± 0.04 | 0.015 ± 0.0016 |
| LMM1128 | recB21 recC22 sbcB15 | 0.18 ± 0.042 | 0.18 ± 0.054 |
| LMM1298 | recB21 recC22 sbcC201 | 0.33 ± 0.037 | 0.005 ± 0.001 |
| LMM1329 | recB21 recC22 ΔsbcB sbcC201 | 0.39 ± 0.088 | 0.16 ± 0.038 |
| LMM1330 | recB21 recC22 sbcB15 sbcC201 | 0.75 ± 0.125 | 1.4 ± 0.24 |
| LMM1331 | recB21 recC22 ΔsbcB sbcC201 recF400 | 0.08 ± 0.022 | 0.0022 ± 0.0013 |
| LMM1332 | recB21 recC22 sbcB15 sbcC201 recF400 | 0.12 ± 0.045 | 0.1 ± 0.02 |
| LMM1362 | recB21 recC22 ΔsbcB sbcC201 ΔrecQ | 0.36 ± 0.042 | 0.007 ± 0.002 |
| LMM1363 | recB21 recC22 sbcB15 sbcC201 ΔrecQ | 0.52 ± 0.086 | 0.087 ± 0.014 |
| LMM1749 | recB21 recC22 xonA2 | 0.21 ± 0.026 | 0.017 ± 0.004 |
| LMM1745 | recB21 recC22 xonA2 sbcC201 | 0.41 ± 0.012 | 0.14 ± 0.046 |
| LMM1746 | recB21 recC22 xonA2 sbcC201 recF400 | 0.15 ± 0.061 | 0.0017 ± 0.0005 |
| LMM1747 | recB21 recC22 xonA2 sbcC201 ΔrecQ::kan | 0.40 ± 0.056 | 0.0075 ± 0.0007 |
| LMM1724 | ΔrecBCD | 0.29 ± 0.032 | 0.0046 ± 0.0006 |
| LMM1725 | ΔrecBCD ΔsbcB | 0.23 ± 0.061 | 0.0075 ± 0.0039 |
| LMM1726 | ΔrecBCD sbcB15 | 0.19 ± 0.04 | 0.16 ± 0.026 |
| LMM1732 | ΔrecBCD ΔsbcB sbcC201 | 0.42 ± 0.11 | 0.13 ± 0.026 |
| LMM1733 | ΔrecBCD sbcB15 sbcC201 | 0.73 ± 0.035 | 1.1 ± 0.10 |
Viability was determined for cultures grown to an OD600 of 0.3 and is expressed relative to the number of CFU per milliliter in cultures of control recipient strain AB1157, which averaged 1.5 × 108 CFU/ml. The values are averages ± standard deviations of results from at least three independent experiments.
The yields of recombinants are relative to the yield of control strain AB1157 and were corrected for any deficiency in the viability of the recipient strain. The average yield for control strain AB1157 was 4.5 × 105 CFU per ml of the mating mixture. The values are averages ± standard deviations of results from at least three independent experiments.
The sbcC mutation further improved the recombination proficiency of both recBC sbcB mutants. In recBC ΔsbcB cells it caused a 10-fold increase in recombination, whereas in recBC sbcB15 cells it caused an 8-fold increase, resulting in a moderate hyper-rec phenotype (Table 2). Interestingly, the recBC ΔsbcB sbcC strain showed the same recombination frequency as the recBC sbcB15 strain, indicating that the combined effect of ΔsbcB and sbcC is necessary to match the level of suppression of the single sbcB15 mutation.
Taken together, the results described above suggest that the suppressive effects of the sbcB15 and ΔsbcB mutations on conjugational recombination differ significantly in both recBC and recBC sbcC backgrounds. As observed in UV and γ irradiation experiments, the sbcB15 mutation proved to be a stronger suppressor of the RecBC− phenotype than ΔsbcB was. These findings were corroborated by the experiments in which the effects of the two sbcB mutations on conjugational recombination were studied in the ΔrecBCD background, the results of which showed the same pattern of suppression that was observed in recB21 recC22 cells (Table 2).
xonA2 mutation affects recombinational repair and conjugational recombination in recBC (sbcC) cells in the same way as ΔsbcB.
In addition to two sbcB mutations, we tested the effects of the xonA2 mutation on DNA repair after UV and γ irradiation. We found that xonA2 had almost the same effect on UV and γ irradiation repair as ΔsbcB, moderately improving the survival of irradiated recBC mutants and providing greater resistance when it was combined with sbcC (Fig. 1A and B and 2A and B).
In conjugational crosses, the xonA2 mutation resulted in a negligible increase in recombination in recBC mutants, and the effect was quite similar to that of ΔsbcB (Table 2). The frequency of recombination was significantly increased after introduction of an additional sbcC mutation. The recBC xonA2 sbcC mutant recombined with the same efficiency as the recBC ΔsbcB sbcC strain, indicating again that there was a striking similarity between the xonA2- and ΔsbcB-associated phenotypes. Given the results described above, we concluded that in suppression of the RecBC− defects, the xonA2 mutation has characteristics of an sbcB null mutation.
Effects of sbcB15, ΔsbcB, and xonA2 mutations on the viability of recBC cells.
Populations of exponentially growing recB, recC, or double-mutant cells contain large proportions (70 to 80%) of nonviable cells (11) (Table 2), suggesting that there are frequent spontaneous dsDNA breaks that cannot be repaired in the absence of the RecBCD enzyme (10). In contrast to recB(C) strains, the sbcB15 sbcC derivatives are highly viable, indicating that endogenous DNA damage can be efficiently repaired by the RecF pathway (25). However, it seems that the joint effects of sbcB15 and sbcC mutations are critical for this type of repair since recB(C) sbcB15 strains are no more viable than their recB(C) parents (25) (Table 2).
To our knowledge, the effects of an sbcB deletion on the viability of E. coli recB(C) cells have not been described previously. According to our data, the viability of recBC ΔsbcB cells is as low as that of recBC and recBC sbcB15 mutants (Table 2). As observed previously with the recBC sbcB15 strain (25), an additional sbcC mutation also improved the viability of recBC ΔsbcB cells. However, in the latter case the effect was quite modest; the viability of recBC ΔsbcB sbcC cells was about one-half that of the recBC sbcB15 sbcC strain. Interestingly, the viability of the recBC ΔsbcB sbcC strain was similar to that of recBC sbcC (Table 2), suggesting that the improvement in viability of the former strain was primarily due to the sbcC mutation rather than to ΔsbcB. We therefore concluded that the ΔsbcB mutation has no effect on viability in the recBC mutant and has only a modest effect in the recBC sbcC background.
We also measured the viability of recBC xonA2 and recBC xonA2 sbcC strains. Briefly, the viability of xonA2 derivatives was almost identical as that of their ΔsbcB counterparts, showing that xonA2 and ΔsbcB have the same effect on repair of spontaneously occurring DNA damage in recBC (sbcC) cells (Table 2).
Effects of recF and recQ mutations on recombination in recBC sbcBC and recBC xonA sbcC mutants.
The results described above show that the sbcB15 mutation produces a stronger suppressive effect in the recBC (sbcC) background than ΔsbcB produces. It is possible that this effect is due to some residual activity of the SbcB15 mutant protein, which could modify the enzymology of reactions in the RecF recombination pathway. To test this hypothesis, we examined the effect of a recF null mutation on recombination processes in recBC sbcB15 sbcC and recBC ΔsbcB sbcC strains. This mutation inactivates the RecF protein, a component of the RecFOR complex known to play an important role in the initiation of recombination in the RecF pathway (i.e., in the formation of the RecA nucleoprotein filament) (30).
After exposure to UV light, the recF mutation moderately decreased the survival of wild-type cells and had more pronounced effects in both the recBC sbcB15 sbcC and recBC ΔsbcB sbcC strains (Fig. 1C). However, the recBC ΔsbcB sbcC recF mutant proved to be much more UV sensitive than its sbcB15 counterpart. The results obtained with γ-irradiated cells were quite similar to those obtained in UV experiments. Again, the recF mutation severely affected the survival of both recBC sbcBC strains, with a more severe effect in the ΔsbcB derivative (Fig. 2C). These results suggest that the sbcB15 mutation alleviates the requirement for recF function during UV and γ irradiation repair in the RecF pathway.
The difference in the effects of the recF mutation on the recombination proficiency of recBC sbcB15 sbcC and recBC ΔsbcB sbcC strains was also observed in conjugational crosses. The decreases in recombination due to the recF mutation were about 10-fold in the sbcB15 background and about 70-fold in the ΔsbcB background (Table 2). The collective results of the conjugational experiments suggest that the recF mutation completely nullifies suppression of the RecBC− phenotype by ΔsbcB sbcC mutations. In contrast, inactivation of the recF gene only partially impaired recBC suppression by sbcB15 sbcC.
The results obtained with recF mutants suggested that the SbcB15 protein influences the initial phase of the recombination process. To verify this suggestion, we tested the effect of a recQ mutation on recombination in the two recBC sbcBC backgrounds. The recQ mutation inactivates the principal DNA helicase of the RecF pathway, the RecQ protein, whose activity is thought to substitute for the DNA unwinding activity of the RecBCD enzyme (21, 27).
In UV and γ irradiation experiments, the effect of a recQ mutation was quite similar to the effect of a recF mutation; i.e., inactivation of RecQ affected the repair more strongly in recBC ΔsbcB sbcC mutants than in recBC sbcB15 sbcC strains (Fig. 1D and 2D). Also, in conjugational crosses the recQ mutation reduced recombination in recBC ΔsbcB sbcC cells to the level of a recBC mutant, whereas in a recBC sbcB15 sbcC background it had a partial effect, allowing significant residual recombination to proceed (Table 2).
Since in all recombination assays performed the xonA2 mutation showed the same phenotype as ΔsbcB, we wanted to examine whether the recBC xonA2 sbcC mutant exhibits a high requirement for RecF and RecQ proteins, like the recBC ΔsbcB sbcC strain. Indeed, we found that recF and recQ mutations severely affected both DNA repair (Fig. 1 and 2) and conjugational recombination (Table 2) in recBC xonA2 sbcC cells, similar to the effects observed with the recBC ΔsbcB sbcC mutant. These results complement our general finding that the effects of the xonA2 mutation on recombination in recBC (sbcC) cells resemble those of an sbcB null mutation.
DISCUSSION
The results of this work show that the sbcB15 and ΔsbcB mutations differ substantially in their abilities to suppress the recombinational deficiency of E. coli recBC mutants. In the majority of recombinational tests performed with recBC sbcB mutants (i.e., in assays of UV and γ irradiation repair and in conjugational crosses), sbcB15 had a much stronger suppressive effect than ΔsbcB. Also, the sbcB15 mutation suppressed the low-viability phenotype of recBC cells more strongly than ΔsbcB suppressed this phenotype, although in this case the suppressive effect of sbcB mutations was detectable only in the presence of an additional sbcC mutation. The results obtained clearly indicate that maximal sbcB(C)-dependent suppression of the RecBC− phenotype cannot be brought about by ΔsbcB (i.e., by complete abolition of ExoI). Instead, it seems that some residual activity of the mutant SbcB15 protein is favorable for recombination (at least in the experimental systems that we used) and is necessary for full suppression of the recombination defect in recBC cells.
Interestingly, a class of sbcB mutations resulting in a different phenotype than the sbcB deletion has also been isolated for Salmonella enterica serovar Typhimurium, a bacterium closely related to E. coli (8). One of these mutations, designated sbcB1, was thoroughly analyzed in several recombination tests and was demonstrated to be a stronger recB suppressor than ΔsbcB, suggesting that it could be a functional counterpart of the E. coli sbcB15 mutation. The similarity of the results previously described for S. enterica serovar Typhimurium and those that we obtained with E. coli indicates that the two organisms have essentially the same mechanisms for regulating initiation of homologous recombination and repair in the RecF pathway.
The results of our study are, however, contrary to an old report suggesting that sbcB15 and ΔsbcB have the same effect in restoring UV repair and conjugational recombination proficiency in recB mutants of E. coli (40). Using almost the same experimental conditions, we showed that the recBC sbcB15 strain is considerably more proficient in recombination than the recBC ΔsbcB strain (Fig. 1A and Table 2). The discrepancy in the results could be partially explained if it is assumed that the recB sbcB strains used in the previous study carried in addition uncharacterized mutations in the sbcC and/or sbcD genes. According to our results obtained in UV irradiation experiments, an sbcC mutation abolishes the distinction between recBC sbcB15 and recBC ΔsbcB strains, resulting in UV resistance close to that of the wild-type strain (Fig. 1). However, the same explanation cannot account for the discrepancy in conjugational crosses. Although we found that both recBC sbcB15 sbcC and recBC ΔsbcB sbcC strains recombine better than the recBC mutant, they still display a marked difference (almost 10-fold) in recombination frequency in favor of the sbcB15 derivative (Table 2). A similar difference was also observed in conjugational crosses with newly constructed ΔrecBCD sbcB15 sbcC and ΔrecBCD Δsbc sbcC derivatives of AB1157 (Table 2), as well as with the classical recBC sbcB15 sbcC strain JC7623 and its ΔsbcB derivative (not shown). Furthermore, this difference was also confirmed in transductional crosses involving the recBC sbcBC derivatives of strain MG1655 (not shown). A possible explanation for the high recombination proficiency of the recB ΔsbcB mutants used by Templin et al. could involve the way that these mutants were isolated; a deletion of the sbcB gene was constructed by P2 eduction, a method that leads to loss of 0.5 to 3 min of the E. coli chromosome (37, 40). We speculate that the part of the chromosome lost by P2 eduction might, in addition to sbcB, contain some other function interfering with recombination in the RecF pathway.
Besides the two sbcB mutations, we included a xonA mutation in our genetic analysis in an attempt to clarify the phenotypic similarities and differences previously reported to exist between the two classes of recBC suppressors (22). Although previous studies suggested that xonA mutations, like sbcB mutations, completely restore UV repair proficiency to recBC mutants, our results showed that the xonA2 mutation only partially improves DNA repair, whereas full recovery requires the presence of an additional sbcC mutation (Fig. 1A and B). The simplest explanation for this difference in results was that the recBC xonA strains used in previous studies also contained uncharacterized sbcC or sbcD mutations. Indeed, when we plated λpal571 phage on the original recBC xonA2 strain, strain JC8260, a high plating efficiency was obtained, revealing the sbcC(D) character of JC8260 (data not shown). Furthermore, our genetic analysis showed that in all experimental systems used, the xonA2 mutation had the same effect on recombination and recombinational repair as ΔsbcB had. This observation, together with the previous finding that expression of the xonA2 allele results in a truncated polypeptide completely devoid of nucleolytic activity (31), strongly suggests that xonA2 might be an sbcB null mutation. Hence, our results do not support the hypothesis of Phillips et al. (31) that xonA mutations leave some residual nonnucleolytic ExoI activity which could hinder recombination. Rather, we assume that xonA2 and other xonA mutants, like ΔsbcB cells, lack some feature that is present in sbcB15 strains and stimulates recombinational processes.
In addition to our observation that the two types of sbcB mutation have different suppressive effects on the RecBC− phenotype, we also found that they influence the enzymatic requirements of the RecF recombination pathway differently. The finding that the recF and recQ mutations more strongly affect recombination in recBC ΔsbcB sbcC and recBC xonA2 sbcC strains than in a recBC sbcB15 sbcC mutant suggests that the residual activity of the SbcB15 protein in the latter strain decreases the necessity for RecFOR and RecQ functions.
The possibility that SbcB15 might influence the enzymatic reactions in the RecF pathway was first proposed by Bidnenko et al. (9). These workers found that the ΔsbcB mutation facilitates the repair of broken replication forks in rep recBC sbcCD cells, whereas the sbcB15 mutation has no beneficial effect on this type of repair. It was suggested that the SbcB15 protein obstructs DNA repair via the RecF pathway in the absence of a functional Rep helicase. After homologous pairing and D-loop formation, Rep helicase might be required to remove SbcB15 from the 3′ end, allowing recombination-dependent replication to occur (9). Hence, our results together with those of Bidnenko et al. indicate that the SbcB15 protein might modulate recombination reactions in the RecF pathway, decreasing the requirement for some proteins and increasing the requirement for others. In addition, these results suggest that (at least) two types of RecF pathway can be distinguished in E. coli, one that is activated by the sbcB15 mutation and one that is activated by ΔsbcB (or other mutations that abolish all functions of ExoI).
What activity of the SbcB15 protein could account for stimulation of recombination via the RecF pathway? Previous genetic experiments indicated that in vivo the sbcB15 mutation inhibits nucleolytic processing of 3′ ssDNA more strongly than ΔsbcB inhibits this processing (32). This finding led to the hypothesis that the mutant SbcB15 protein is able to bind 3′ ssDNA ends, thus preventing other nucleases from digesting the same substrate. Hence, the stronger inhibition of DNA degradation in the presence of SbcB15 might be due to a joint effect of ExoI inactivation and DNA protection (32). In the context of the RecF recombination pathway, such blocking of DNA ends by SbcB15 might preserve recombinogenic DNA ends better than the complete elimination of the SbcB protein preserves these ends (thus having a stronger suppressive effect on recBC mutations). If it is assumed that SbcB15 protects DNA ends from degradation by other exonucleases, it could be expected to interfere with the action of the SbcCD protein, a nuclease known to partially inhibit recombination in the RecF pathway (25). In this case, the stimulating effect of SbcCD inactivation on recombination should be significantly less pronounced in recBC sbcB15 cells than in recBC ΔsbcB cells. Interestingly, our results obtained in UV and γ irradiation experiments fit this end protection model well, showing that the recombinational repair in recBC sbcB15 mutants is much more resistant to the SbcCD nuclease than the recombinational repair in recBC ΔsbcB cells is (Fig. 1 and 2). In fact, after exposure to UV and γ radiation, the recBC sbcB15 mutant was almost fully repair proficient so that inactivation of SbcCD nuclease by the sbcC mutation had little (if any) additional effect (Fig. 1 and 2). A quite different situation was observed with the recBC ΔsbcB strain, whose low repair proficiency was strongly improved by inactivation of sbcC.
Unlike the results of the irradiation experiments, in conjugational crosses the sbcC mutation significantly improved recombination in both recBC sbcB15 and recBC ΔsbcB mutants, and the net increases in recombination frequency due to SbcCD inactivation were about equal (approximately 10-fold) in the two backgrounds (Table 2). This finding suggests that during conjugational recombination the SbcB15 protein cannot efficiently prevent the activity of the SbcCD enzyme and that the beneficial effect of SbcB15 on this type of recombination must be attributed primarily to some other mechanism.
The cell viability measurements are also difficult to accommodate with the end protection model discussed above. These measurements show that neither the sbcB15 mutation nor the ΔsbcB mutation alone has any suppressive effect on the low-viability phenotype of exponentially growing recBC cells (Table 2). Only after additional inactivation of the sbcC gene was a strong increase in viability observed in recBC sbcB15 cells, whereas in recBC ΔsbcB cells the sbcC mutation caused only a slight improvement in viability. These results clearly indicate that SbcB15 cannot prevent the antirecombinogenic action of SbcCD during repair of endogenous DNA damage.
Although the results of conjugational crosses and cell viability measurements are not readily explained by the end protection model, they do not exclude the possibility that in the absence of SbcCD activity, SbcB15 protects DNA from other 3′-5′ exonucleases which might antagonize the recombination process. This hypothesis is suggested by the fact that recBC sbcB15 sbcC mutants are far more viable and show higher proficiency in conjugational crosses than recBC ΔsbcB sbcC cells. Theoretically, the interplay between SbcB15 and SbcCD (and possibly other nucleases) could be influenced by the shape of the DNA ends exposed (e.g., by the presence of single-stranded overhangs that are different lengths and have different polarities, by chemical modifications of terminal deoxynucleoside triphosphates, etc.) and by the affinity of different nucleases for a particular end type. Hence, the variety of suppression patterns observed in our experiments with recBC sbcB(C) strains might reflect different DNA substrates present, leaving the possibility that at least in some cases SbcB15 has DNA-protecting activity. To address this question in more detail, additional experiments involving new recBC sbcB(C) derivatives in which residual 3′-5′ Exo activities are depleted are needed.
An alternative explanation for the prorecombinogenic activity of SbcB15 (that does not necessarily exclude the model described above) may involve a more active role of the mutant protein in the recombination process. This possibility is derived from in vitro studies suggesting that ExoI physically interacts with two proteins that have an important role in recombination, the SSB and RecA proteins (7, 34). The interaction between ExoI and SSB may be functionally important since it was shown that SSB stimulates the deoxyribophosphodiesterase activity of ExoI during the repair of abasic sites in DNA (33), as well as its 3′-5′ exonuclease activity with an ssDNA substrate (29). On the other hand, the possible relevance of an ExoI-RecA interaction remains to be elucidated. If SbcB15 retains the interacting properties of the wild-type enzyme (or if these properties are modified due to the mutation), it could conceivably influence the recombination process at the level of RecA filament formation. It has been demonstrated in vitro that the SSB and RecA proteins compete for the same substrate (ssDNA) and that RecBCD or RecFOR activities are required to facilitate efficient loading of the RecA protein in the presence of SSB (4, 30). Since SbcB15 presumably favors recombination in recBC (sbcC) mutants, we speculate that it stimulates binding of RecA to ssDNA by dislodging the molecules of SSB protein. Such an activity could enhance the formation of RecA filaments, as well as the pairing of the filaments with homologous DNA, thus protecting DNA from nucleolytic degradation and increasing the overall efficiency of the recombination process. Our finding that the sbcB15 mutation alleviates the requirement for RecFOR activity in the RecF pathway is in accord with this hypothesis. On the other hand, the relaxed requirement for RecFOR could also result from passive DNA end protection by SbcB15, which might provide enough time for RecA filaments to be made even under restrictive conditions (i.e., in the absence of RecFOR-mediated loading). In ΔsbcB mutants deficient for RecFOR function, DNA ends would be degraded by nucleases before they are engaged in RecA filament formation and homologous pairing.
It was previously shown that in the absence of RecQ helicase, the residual DNA unwinding activity provided by UvrD (helicase II) and HelD (helicase IV) allows recombination in the RecF pathway to proceed, although it proceeds with lower efficiency (27). Our finding that the recQ mutation more strongly affects recombination in a recBC ΔsbcB sbcC strain than in a recBC sbcB15 sbcC background suggests that SbcB15 alleviates the requirement for DNA unwinding activity during initiation of recombination. We hypothesize that when DNA ends are protected by SbcB15, even reduced DNA unwinding is sufficient to ensure substantial recombination. However, in recBC ΔsbcB sbcC cells, in which DNA ends are not protected, vigorous DNA unwinding is necessary to overcome residual 3′-5′ exonucleolytic activity in order to generate recombinogenic ssDNA tails. Experiments to test the hypotheses described above and to further elucidate the role of SbcB15 in recombination are under way in our laboratory.
Acknowledgments
We thank M. Berlyn (Escherichia coli Genetic Stock Center), B. Michel, and S. R. Kushner for providing bacterial strains, M. Blažević for technical assistance with γ irradiation, and R. D'Ari and W. Ragland for critical reading of the manuscript.
This work was supported by grant 0098071 from the Croatian Ministry of Science, Education and Sports.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Amundsen, S. K., A. F. Taylor, A. M. Chaudhury, and G. R. Smith. 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83:5558-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundsen, S. K., A. F. Taylor, and G. R. Smith. 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. USA 97:7399-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and S. C. Kowalczykowski. 1997. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11:571-581. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in chi-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, D. A., and S. C. Kowalczykowski. 2000. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J. Biol. Chem. 275:12261-12265. [DOI] [PubMed] [Google Scholar]
- 6.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 7.Bedale, W. A., R. B. Inman, and M. M. Cox. 1993. A reverse DNA strand exchange mediated by RecA protein and exonuclease I. J. Biol. Chem. 268:15004-15016. [PubMed] [Google Scholar]
- 8.Benson, N. R., and J. Roth. 1994. Suppressors of recB mutations in Salmonella typhimurium. Genetics 138:11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidnenko, V., M. Seigneur, M. Penel-Colin, M.-F. Bouton, S. D. Ehrlich, and B. Michel. 1999. sbcB sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol. Microbiol. 33:846-857. [DOI] [PubMed] [Google Scholar]
- 10.Capaldo, F. N., and S. D. Barbour. 1975. DNA content, synthesis and integrity in dividing and non-dividing cells of rec− strains of Escherichia coli K-12. J. Mol. Biol. 91:53-66. [DOI] [PubMed] [Google Scholar]
- 11.Capaldo, F. N., G. Ramsey, and S. D. Barbour. 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, A. J. 1973. Recombination deficient mutants of E. coli and other bacteria. Annu. Rev. Genet. 7:67-86. [DOI] [PubMed] [Google Scholar]
- 13.Clark, A. J. 1991. rec genes and homologous recombination proteins in Escherichia coli. Biochimie 73:523-532. [DOI] [PubMed] [Google Scholar]
- 14.Connelly, J. C., E. S. de Leau, and D. R. F. Leach. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27:1039-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly, J. C., L. A. Kirkham, and D. R. F. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmerson, P. T. 1968. Recombination deficient mutants of Escherichia coli K12 that map between thyA and argA. Genetics 60:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Gibson, F. P., D. R. F. Leach, and R. G. Lloyd. 1992. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J. Bacteriol. 174:1222-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horii, Z. I., and A. J. Clark. 1973. Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K-12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 20.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 21.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kushner, S. R., H. Nagaishi, and A. J. Clark. 1972. Indirect suppression of recB and recC mutations by exonuclease I deficiency. Proc. Natl. Acad. Sci. USA 69:1366-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd, R. G., and C. Buckman. 1995. Conjugal recombination in Escherichia coli: genetic analysis in recombinant formation in Hfr × F− crosses. Genetics 139:1123-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendonca, V. M., H. D. Klepin, and S. W. Matson. 1995. DNA helicases in recombination and repair: construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J. Bacteriol. 177:1326-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics. CSHL Press, Cold Spring Harbor, N.Y.
- 29.Molineux, I. J., and M. L. Gefter. 1975. Properties of the Escherichia coli DNA-binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J. Mol. Biol. 98:811-825. [DOI] [PubMed] [Google Scholar]
- 30.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, G. J., D. C. Prasher, and S. R. Kushner. 1988. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J. Bacteriol. 170:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razavy, H., S. K. Szigety, and S. M. Rosenberg. 1996. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics 142:333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandigursky, M., and W. A. Franklin. 1994. Escherichia coli single-stranded DNA binding protein stimulates the DNA deoxyribophosphodiesterase activity of exonuclease I. Nucleic Acids Res. 22:247-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandigursky, M., F. Mendez, R. E. Bases, T. Matsumoto, and W. A. Franklin. 1996. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat. Res. 145:619-623. [PubMed] [Google Scholar]
- 35.Seigneur, M., S. D. Ehrlich, and B. Michel. 1999. recD sbcB sbcD mutants are deficient in recombinational repair of UV lesions by RecBC. J. Bacteriol. 181:6220-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, G. R. 1991. Conjugational recombination in E. coli: myths and mechanisms. Cell 64:19-27. [DOI] [PubMed] [Google Scholar]
- 37.Sunshine, M. G., and B. Kelly. 1971. Extent of host deletions associated with bacteriophage P2-mediated eduction. J. Bacteriol. 108:695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, A. F. 1988. RecBCD enzyme of Escherichia coli, p. 231-263. In R. Kucherlapati and G. R. Smith (ed.), Genetic recombination. ASM Press, Washington, D.C.
- 39.Taylor, A. F., and G. R. Smith. 1985. Substrate specificity of the DNA unwinding activity of the RecBCD enzyme of Escherichia coli. J. Mol. Biol. 185:431-443. [DOI] [PubMed] [Google Scholar]
- 40.Templin, A., S. R. Kushner, and A. J. Clark. 1972. Genetic analysis of mutations indirectly suppressing recB and recC mutations. Genetics 72:205-215. [PMC free article] [PubMed] [Google Scholar]
- 41.Templin, A., L. Margossian, and A. J. Clark. 1978. Suppressibility of recA, recB, and recC mutations by nonsense suppressors. J. Bacteriol. 134:590-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 43.Zahradka, D., K. Zahradka, M. Petranović, D. Đermić, and K. Brčić-Kostić. 2002. The RuvABC resolvase is indispensable for recombinational repair in sbcB15 mutants of Escherichia coli. J. Bacteriol. 184:4141-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]