Abstract
B. melitensis 16M genome analysis revealed the presence of six putative sigma factor-encoding genes: rpoD, rpoH1, rpoH2, rpoE1, rpoE2, and rpoN. We mutated all these genes except rpoD. Phenotypic analysis of the mutants reveals that a strain carrying an rpoH2 null mutation (ΔrpoH2) is impaired for growth at 21 and 42°C and shows increased sensitivity to hydrogen peroxide. Compared to the wild-type strain, the ΔrpoH2 mutant is attenuated in all virulence models tested. Three other null mutants (ΔrpoH1, ΔrpoE1, and ΔrpoE2 mutants) are also defective for survival in mice at 4 weeks postinfection. We also demonstrated that rpoH2 deletion strongly reduces the expression of two major virulence factors in B. melitensis, the type IV secretion system and the flagellum.
During their infectious cycle, pathogenic bacteria are exposed to a wide variety of environments. Associations between alternative σ factors and RNA polymerase provide one efficient mechanism for appropriately modifying the transcriptional profile of the bacterium in response to changing environments (10). Alternative σ factors contribute to bacterial resistance to environmental stress conditions, such as high temperature, oxidative stress, carbon starvation, and low pH, and therefore contribute to virulence of pathogenic bacteria. Alternative σ factors may also be involved in the regulation of more “specific” virulence genes (4, 11). Among the σ factors involved in bacterial virulence, there are stress response σ factors (σB and σS), flagellar σ factors (σ28), extracytoplasmic function σ factors (RpoE, AlgU), and σ54. Up to now, very few examples of heat shock sigma factor (σ32) being involved in bacterial virulence have been reported (13).
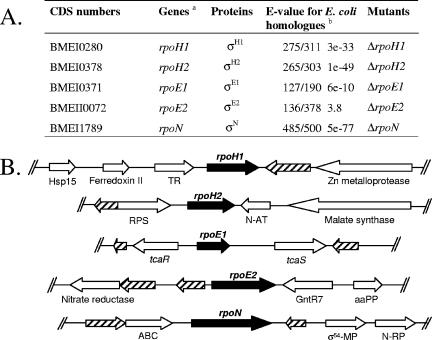
Brucellae are gram-negative, intracellular pathogenic bacteria that cause brucellosis in a variety of mammals, including humans. The availability of the complete Brucella melitensis 16M genomic sequence allowed us to identify six putative σ factors in this organism (Fig. 1A), a housekeeping σ factor (σ70, encoded by rpoD), two σ32 homologues (σH1 and σH2), two extracytoplasmic function (ECF) σ factors (σE1 and σE2), and a σ54 homologue (σN). Unlike enteric bacteria and pseudomonads, B. melitensis, as well as the other α-proteobacteria (14), does not possess an rpoS-like gene coding for the general stress σ factor, σS.
FIG. 1.
Predicted σ-factor CDS identified in the B. melitensis 16M genome. A. CDS numbers and gene, protein, and mutant names used in this study. a, in agreement with the names given to σ-coding genes in E. coli; b, numbers before the E values correspond to the fraction of the predicted protein that is aligned with the E. coli homologue, and the second number is the predicted protein length. B. Genomic organization around the B. melitensis σ CDS. Each predicted CDS is indicated by an arrow. The CDS for putative σ factors are black, and the hypothetical proteins are hatched. Abbreviations: Hsp15, heat shock protein 15; TR, transcriptional regulator; RPS, ribosomal large-subunit pseudouridine synthase; N-AT, phosphinothricin N-acetyltransferase; aaPP, leucine-, isoleucine-, valine-, threonine-, and alanine-binding protein precursor; GntR7, transcriptional regulator GntR family (8); σ54-MP, putative σ54 modulation protein; N-RP, nitrogen-regulatory protein; ABC, ATP-binding protein of the ATP-binding cassette transporter.
The genomic organization of the σ coding sequences (CDS) in B. melitensis 16M is depicted in Fig. 1B. Some interesting observations may be made. The rpoH1 CDS is found close to CDS potentially involved in adaptation to heat shock (a zinc metalloprotease homologue and a heat shock protein 15 homologue). The rpoH2 CDS is located close to the rpoE1 CDS, which is next to genes coding for proteins belonging to a two-component regulatory system, TcaR (response regulator) and TcaS (sensory transduction histidine kinase). This locus is well conserved in α-proteobacteria. TcaR contains an unusual N-terminal DNA binding domain compared to other response regulators, displaying homology to the RpoE domain (COG1595), which is usually found in σECF factors. The predicted peptidic sequence for BMEII0072 presents similarity to the Sigma_r2 domain from Pfam (E value = 1.3e−14), and it is also similar to a predicted σECF, CarQ from Myxococcus xanthus (6). These data suggest that BMEII0072 may encode a phylogenetically distant σECF homologue. The rpoN CDS is close to genes coding for a σ54 modulation protein and a nitrogen-regulatory protein, which suggests that there may be a functional relationship between RpoN and these two gene products. None of the σ CDS is predicted to be part of a transcriptional unit with other CDS.
In vitro characterization of σ mutants indicates that σH2 is involved in adaptation to heat, cold, and oxidative stress.
Each nonessential rpo gene (coding for a σ factor) was replaced by a kanamycin resistance gene, using a previously reported strategy (1) with the oligonucleotides listed in Table S1 in the supplemental material. Classical polar effects probably do not occur in these mutants, since none of these genes forms part of a predicted operon (Fig. 1). All the mutants displayed a smooth phenotype, as detected with the crystal violet colony staining method, suggesting that lipopolysaccharide O chain is present. Indeed, rough variants are frequent, and attenuation may be due to this character instead of the absence of a σ factor. The σ mutants (ΔrpoH1, ΔrpoH2, ΔrpoE1, ΔrpoE2, and ΔrpoN mutants) were characterized with regard to their sensitivity to oxidative and heat stresses.
The σ mutants were tested for survival upon oxidative stress (H2O2) using a disk sensitivity assay (3). Briefly, 100 μl of B. melitensis cultures adjusted to an optical density at 600 nm (OD600) of 0.4 were plated on 2× yeast extract-tryptone (2YT) agar, and sterile paper disks (5-mm diameter) saturated with 10 μl of H2O2 at a concentration of 5 M were layered on top prior to incubation at 37°C. The ΔrpoH2 mutant showed significantly increased sensitivity to H2O2. The assay was performed on three separate plates, and the diameter of growth inhibition for the ΔrpoH2 mutant was 5.52 cm (±0.20 cm [standard deviation]) compared with 4.18 cm (±0.04 cm) for the wild-type strain, which is a highly significant difference (Student's t test, P < 0.01). Sensitivity of the ΔrpoH2 mutant to H2O2 is reduced when this strain carries a complementation plasmid, pMR-rpoH2 (data not shown). pMR-rpoH2 is a low-copy-number plasmid (RK2-derived pMR10, made compatible for Gateway cloning to give pRH001; R. Hallez, unpublished data) carrying the rpoH2 coding sequence. Oligonucleotides used for the construction of complementation plasmids are available in Tables S2 and S3 in the supplemental material. The other σ mutants did not display a reproducible and significantly altered sensitivity to H2O2.
The growth of σ mutants was compared to that of the wild-type strain on 2YT agar plates at temperatures of 21, 37, and 42°C. Serial dilutions of bacterial cultures adjusted to an OD600 of 0.4 were spotted on 2YT plates and incubated at these temperatures. Growth at 21 and 42°C was completely abolished for the ΔrpoH2 mutant (Fig. 2A). Complementation of the ΔrpoH2 mutant with pMR-rpoH2 restored a growth comparable to the wild-type strain at 21 and 42°C (Fig. 2A). We also compared the growth curve of the wild-type strain, the ΔrpoH2 mutant, and the complemented strain at 37 and 42°C in liquid 2YT medium, starting with an OD600 of 0.1. At 37°C, the ΔrpoH2 mutant reached an optical density slightly higher than the wild-type strain in late exponential and stationary phases. Following a temperature shift to 42°C, the ΔrpoH2 mutant was unable to grow, while the wild-type strain grew, as shown in Fig. 2B. Complementation of the ΔrpoH2 mutant with pMR-rpoH2 partially restored the growth at 42°C in liquid medium (Fig. 2B). The reduced growth of the complemented strain observed at 37 and 42°C (Fig. 2B) compared to that of the wild-type strain is probably due to the lack of appropriate regulation of rpoH2, which is fused to an Escherichia coli lac promoter in the pMR10 vector. The growth curves of the ΔrpoH1, ΔrpoE1, ΔrpoE2, and ΔrpoN mutants were also monitored, but no differences was observed compared to the wild-type strain (data not shown). Measuring CFU at different culture phases indicated that rpoH2, rpoE1, rpoE2, and rpoN are not required for survival in stationary phase (data not shown).
FIG. 2.
A. Heat- and cold-sensitive growth phenotype of the B. melitensis ΔrpoH2 mutant. The wild-type strain (WT), the ΔrpoH2 mutant (rpoH2), and the strain complemented with pMR-rpoH2 (Cplt) were grown overnight in liquid 2YT medium at 37°C. After adjustment to an OD600 of 0.4, 10 μl of serial dilutions of these cultures (consecutive 1:10 dilution steps from top to bottom) was spotted onto 2YT agar plates and incubated at the indicated temperature. B. Comparison of the growth of wild-type cells (WT) with the growth of the ΔrpoH2 mutant and the complemented strain (pMR-rpoH2), as measured using OD600. Growth of the wild-type strain (triangles), ΔrpoH2 mutant (squares), and pMR-rpoH2-complemented strain (circles) in liquid medium at 37°C (open symbols) and 42°C (filled symbols) is indicated.
σH2 is required for survival of B. melitensis in several models of infection.
The pathogenicity of Brucella spp. is critically dependent on its ability to infect and to multiply within both professional and nonprofessional phagocytes (2). We tested the ability of σ mutants to invade and survive within J774 macrophages and epithelial (HeLa) cells, using a previously described protocol (9), with 105 cells/well. A ΔvjbR mutant was used as a positive control for attenuation (1). The CFU were counted after 48 h of infection. The results showed that the ΔrpoH2 mutant was strongly attenuated in both J774 and HeLa cells. The ΔrpoE1 mutant was slightly attenuated in both models of infection (Table 1).
TABLE 1.
Attenuation of σ mutants
| Mutation | LPSa | Attenuation inb:
|
||
|---|---|---|---|---|
| HeLa cells | J774 macrophages | BALB/c mice | ||
| ΔrpoH1 | S | −0.4 ± 0.2 | 0 ± 0.1 | 1.5 ± 0.1 |
| ΔrpoH2 | S | 1.4 ± 0.3 | 1.8 ± 0.2 | >3 |
| ΔrpoE1 | S | 0.7 ± 0.1 | 0.9 ± 0.1 | 2.2 ± 0.2 |
| ΔrpoE2 | S | −0.6 ± 0.2 | 0 ± 0.05 | 1.8 ± 0.2 |
| ΔrpoN | S | −0.3 ± 0.2 | −0.4 ± 0.3 | 0.1 ± 0.1 |
| ΔvjbR | NDc | 2.0 ± 0.1 | 2.6 ± 0.2 | ND |
Smooth (S) lipopolysaccharide (LPS) was detected by crystal violet colony staining.
Attenuation in cellular models (48 h postinfection) and in BALB/c mice (4 weeks postinfection) is expressed as the difference of the log CFU between the wild-type strain and the mutants ± the standard deviation, which was calculated from values obtained for each mutant in the different models of infection (n = 3 for the cellular models and n = 4 for the mouse model). Cellular infections were performed twice in triplicate, with a multiplicity of infection of 300 bacteria per cell. The ΔvjbR mutant was used as a positive control for attenuation in the cellular models (1).
ND, not determined.
The persistence of each mutant was studied in a mouse model of infection, using a previously described protocol (8). Groups of four mice were inoculated intraperitoneally with either a mutant strain or the wild-type strain. The Brucella CFU were evaluated 1 and 4 weeks postinfection in the spleens of four animals. After 1 week of infection, the ΔrpoH2 mutant was recovered from spleens at markedly lower levels (1.9 ± 0.2 log) than the isogenic parental strain (Table 1). By 4 weeks postinfection, the number of ΔrpoH2 CFU was 3 orders of magnitude lower than that of the parental strain in one mouse, while the three others had cleared the ΔrpoH2 strain from their spleens. All mutant strains except the ΔrpoN strain showed reduced spleen colonization after 4 weeks of infection. The need for four different σ factors suggests that B. melitensis faces various environments within this infection model, indicating that this model is rather complex, especially compared to the cellular models of infection tested here. In particular, the requirement for ECF σ factors is in agreement with the major role of the Brucella sp. envelope during infection (7). The involvement of four of the five nonessential σ factors predicted from the B. melitensis genome indicates that σ factors are key players in the regulation of virulence determinants required for survival in this model of infection. Moreover, our data indicate that σH2 may be required for the acute phase of the infection, while σE1, σE2, and σH1 would be required for chronicity of the infection.
σH2 is involved in the regulation of both VirB and FlgE production.
Since VirB and flagella are important virulence factors for Brucella spp. (5, 12), we investigated the abundance of these structures in the σ mutants compared to that in the wild-type strain, with the hypothesis that σ factors may contribute to the regulation of these structures.
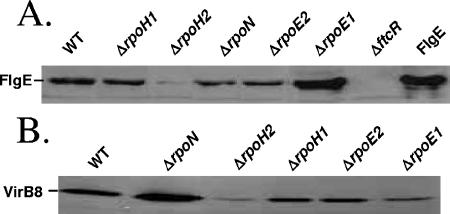
To compare VirB and FlgE production in the mutants and the wild-type strain, we carried out Western blotting analysis of whole-cell extracts by using anti-VirB8 and anti-FlgE polyclonal antisera generated in rabbits (5, 15) (Fig. 3), using a previously described protocol (1). The FlgE protein (flagellar hook monomer homolog) was used for monitoring the production of flagellar proteins. The abundance of VirB8 and FlgE proteins was tested in cells harvested at a phase of the growth curve in 2YT medium where their production is maximal (OD600 around 0.3 or 1.5 for the detection of FlgE or VirB8, respectively). The ΔftcR mutant was used as a negative control for FlgE detection, ftcR being required for fliF expression and FlgE abundance (S. Léonard, unpublished data). We observed that VirB8 is slightly less abundant in the ΔrpoE1 mutant, almost absent in the ΔrpoH2 mutant, and overproduced in the ΔrpoN mutant compared to the wild-type strain. FlgE is almost absent in the ΔrpoH2 mutant and overproduced in the ΔrpoE1 mutant. Comparable effects were observed using a fusion between the fliF promoter and lacZ in these two mutants (M. Delory, unpublished results), indicating that expression of several flagellar genes is similarly affected by the absence of σH2 or σE1.
FIG. 3.
Western blots of SDS-PAGE protein gels probed with anti-VirB8 and anti-FlgE polyclonal antisera. For blot A, early-log-phase cultures were harvested from 2YT growth medium. The ΔftcR mutant was used as a negative control for FlgE detection. Purified recombinant His6-FlgE was used as a positive control (lane FlgE). For blot B, cultures were grown in 2YT medium until late log phase. Lanes were loaded with equal cell quantities (determined by OD600 before harvesting). VirB8 and FlgE migrate at estimated molecular masses of 31.5 kDa and 41 kDa, respectively.
Recently, Delrue et al. identified and characterized VjbR, a LuxR-like transcriptional regulator essential for the expression of the virB operon and for the production of FlgE protein (1). VjbR being a possible mediator of the regulation of VirB production by a σ factor, we tested the activity of the vjbR promoter in the ΔrpoH2 strain. The activity of vjbR promoter was monitored by measuring luciferase activity using a pvjbR luxAB fusion carried by the plasmid pJD27-pvjbR. The plasmid pJD27-pvjbR was conjugatively transferred into the B. melitensis 16M Nalr strain and the mutant ΔrpoH2 strain. The wild type and the ΔrpoH2 mutant bearing the pJD27-pvjbR plasmid were grown in 2YT medium and harvested during the exponential phase. A luciferase assay was performed as described previously (1). In the ΔrpoH2 strain, the activity of the vjbR promoter was reduced more than 100-fold compared to that in the wild-type strain (1,865 ± 384 [standard deviation] relative light units for the mutant versus 231,411 ± 5,866 for the wild type; data are representative of three independent experiments), suggesting that vjbR may be an mediator of the effect of rpoH2 mutation on the abundance of VirB and FlgE proteins.
In conclusion, using a systematic targeted mutagenesis strategy, we identified a σ factor (σH2) having multiple roles in B. melitensis, since the ΔrpoH2 mutant is very sensitive to heat, cold, and oxidative stress. The essential role of σH2 in both thermotolerance and cryotolerance is unique among proteins belonging to the RpoH family. It is possible that σH2 is actually involved in generalized cytoplasmic stress response. The molecular mechanisms involving σH2 in the adaptation to low and high temperature, and to oxidative stress, in B. melitensis remain to be discovered. The function of the other σ factors should also be studied, since three of them (σH1, σE1, and σE2) are required for virulence in the mouse model of infection.
Supplementary Material
Acknowledgments
Marie Delory and Régis Hallez were supported by the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture (F.R.I.A.). This work was supported by the Fonds de la Recherche Fondamentale Collective (F.R.F.C., convention no. 2.4521.04).
We thank URPhyM (University of Namur) for DNA sequencing facilities. We thank C. Deschamps for the construction of pJD27-pvjbR.
Footnotes
Published ahead of print on 25 August 2006.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151-1161. [DOI] [PubMed] [Google Scholar]
- 2.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang, F. C., S. J. Libby, N. A. Buchmeier, P. C. Loewen, J. Switala, J. Harwood, and D. G. Guiney. 1992. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fretin, D., A. Fauconnier, S. Kohler, S. Halling, S. Leonard, C. Nijskens, J. Ferooz, P. Lestrate, R. M. Delrue, I. Danese, J. Vandenhaute, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7:687-698. [DOI] [PubMed] [Google Scholar]
- 6.Gorham, H. C., S. J. McGowan, P. R. Robson, and D. A. Hodgson. 1996. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol. Microbiol. 19:171-186. [DOI] [PubMed] [Google Scholar]
- 7.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haine, V., A. Sinon, F. Van Steen, S. Rousseau, M. Dozot, P. Lestrate, C. Lambert, J. J. Letesson, and X. De Bolle. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lestrate, P., A. Dricot, R. M. Delrue, C. Lambert, V. Martinelli, X. De Bolle, J. J. Letesson, and A. Tibor. 2003. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 71:7053-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 11.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 13.Parsot, C., and J. J. Mekalanos. 1990. Expression of ToxR, the transcriptional activator of the virulence factors in Vibrio cholerae, is modulated by the heat shock response. Proc. Natl. Acad. Sci. USA 87:9898-9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roop, R. M., J. M. Gee, G. T. Robertson, J. M. Richardson, W. L. Ng, and M. E. Winkler. 2003. Brucella stationary-phase gene expression and virulence. Annu. Rev. Microbiol. 57:57-76. [DOI] [PubMed] [Google Scholar]
- 15.Rouot, B., M. T. Alvarez-Martinez, C. Marius, P. Menanteau, L. Guilloteau, R. A. Boigegrain, R. Zumbihl, D. O'Callaghan, N. Domke, and C. Baron. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.