Abstract
Members of a family of serine/threonine protein kinases (STPKs), unique to gram-positive bacteria, comprise an intracellular kinase domain and reiterated extracellular PASTA (for “penicillin-binding protein and serine/threonine kinase associated”) domains. PASTA domains exhibit low affinity for β-lactam antibiotics that are structurally similar to their likely normal ligands: stem peptides of unlinked peptidoglycan. The PASTA-domain STPKs are found in the actinobacteria and firmicutes and, as exemplified by PknB of Mycobacterium tuberculosis, they are functionally implicated in aspects of growth, cell division, and development. Whereas the kinase domains are well conserved, there is a wide divergence in the sequences of the multiple PASTA domains. Closer inspection reveals position-dependent evolution of individual PASTA domains: a domain at one position within a gene has a close phylogenetic relationship with a domain at a similar position in an orthologous gene, whereas neighboring domains have clearly diverged one from one another. A similar position-dependent relationship is demonstrated in the second family of proteins with multiple PASTA domains: the high-molecular-weight type II penicillin-binding protein (PBP2x) family. These transpeptidases are recruited to the division site by a localized pool of unlinked peptidoglycan. We infer that protein localization is guided by low-affinity interactions between structurally different unlinked peptidoglycan stem peptides and individual PASTA domains. The STPKs possess a greater multiplicity and diversity of PASTA domains, allowing interactions with a wider range of stem-peptide ligands. These interactions are believed to activate the intracellular kinase domain, allowing an STPK to coordinate peptidoglycan remodeling and reproduction of a complex cell wall structure.
Of all infectious diseases, tuberculosis is the biggest killer worldwide. The causative agent, the gram-positive bacterium Mycobacterium tuberculosis, can lie quiescent in unidentified sites in the human host for years without producing overt disease and then revive to cause lesions and, in many cases, progressive tuberculosis. To improve the outlook for combating this pathogen, it is particularly important to understand the genes that govern the cell cycle and latency state of infection. Serine/threonine protein kinases (STPKs) play an important part in bacterial signaling pathways, particularly in more complex prokaryotes such as Mycobacterium and Streptomyces, which possess multiple STPK genes (reviewed in reference 1). One family of these STPKs, unique to gram-positive bacteria, consists of transmembrane proteins, with the N-terminal kinase domain inside the cell and a C-terminal sensory component outside the cell (Fig. 1). The extracellular component is made up of three or four reiterated PASTA (for “penicillin-binding protein and serine/threonine kinase associated”) domains (30). These domains were first identified associated with a high-molecular-weight type II penicillin-binding protein, PBP2x, in another medically important gram-positive bacterium Streptococcus pneumoniae. Indeed, PBP2x, which has two C-terminal PASTA domains, is the only protein for which the PASTA domain structure has been determined by X-ray crystallography (25). Each domain consists of an alpha helix and three beta strands, with a loop region of variable length between the first and second strands. A further crystal structure was determined in the presence of cefuroxime, a β-lactam antibiotic (13). Surprisingly, two molecules of the antibiotic were bound: one as a covalent complex with the active-site serine residue, the second associated by Van de Waal's interactions between the β-lactam ring and the first PASTA domain. This part of the antibiotic structurally resembles an unlinked peptidogylcan stem peptide, the likely normal ligand of a PASTA domain.
FIG. 1.
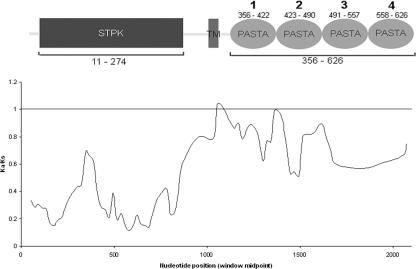
Sliding-window analysis of 14 actinobacterial pknB orthologs, using Ka/Ks as a measure of selective pressure (window = 72, step = 21 nucleotides). Values are averages of pairwise comparisons between all individual sequences in the data set. Shown above the graph is the domain structure for PknB of M. tuberculosis aligned to the same scale as the x axis. Small numbers refer to amino acids constituting each domain. The numbering system for the four individual PASTA domains is also indicated. TM, transmembrane region.
The multiple PASTA domain STPKs are found in the firmicutes and actinobacteria. PknB of M. tuberculosis is representative of one of the two different PASTA domain STPKs commonly found in the latter and has been the most extensively studied. A pknB (Rv0014c) ortholog is found in all sequenced actinomycete genomes in a highly conserved gene context and location, close to the chromosomal origin of replication, oriC. The gene is linked with others involved in signaling: its cognate phosphatase gene and two genes coding for fork-head-associated (FHA) proteins, mediators of serine and threonine phosphorylation (9, 24). In addition, the conserved gene cluster contains critical morphogenetic genes: pbpA, encoding a penicillin-binding protein important for growth and cell division (5); a rodA gene; and an ortholog of crgA, a gene implicated in the control of cell division in Streptomyces (6, 7). PknB undergoes autophosphorylation and is dephosphorylated by its cognate phosphatase in vitro (3). It can in turn phosphorylate FHA domains, including that of the neighboring gene Rv0020c in vitro (14), and also two threonine residues of PbpA (5). Overexpressing PknB in mycobacterial cells alters cell division and cell wall synthesis to produce swollen cells with incomplete division septa that also show occasional branching, whereas partial depletion results in long, thin cells (16). pknB and the neighboring rodA are both essential genes in M. tuberculosis (27).
The firmicutes generally contain a single multiple-PASTA-domain STPK. A null mutation of the Bacillus subtilis kinase gene prkC results in a decrease in stationary-phase cell density and of loss of efficiency in the formation of spores and biofilms (11, 19). In Streptococcus pneumoniae, the orthologous kinase (StkP) is implicated in virulence (10). A likely target for phosphorylation by StkP is phosphoglucosamine mutase, GlmM, involved in cell wall synthesis (23).
We have undertaken the first study of how these STPKs have evolved. Our analysis reveals an unexpected diversity among the PASTA domains and allows us to predict how they may function.
MATERIALS AND METHODS
Sequence selection.
Bacterial species containing multiple PASTA domain proteins were identified by using the Conserved Domain Architecture Retrieval Tool (CDART [http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd = rps]). The protein sequence of the PASTA domains from S. coelicolor was used as a query sequence. The same sequence was used to perform BLAST searches against the individual genomes. A representative sample of genomes was selected by using the genomes list at the GOLD database (http://www.genomesonline.org/) (2). Mycobacterium smegmatis sequences were obtained by using BLAST searches at TIGR (http://www.tigr.org). Sequences giving an e-value of ≤0.0001 were selected, and the domain architecture was checked by using the SMART tool (18). Only proteins showing N-terminal kinase, transmembrane and C-terminal PASTA domains were selected, as well as PBP2x orthologs. Representative samples of firmicute and actinomycete multiple PASTA domain proteins were selected for further detailed analysis (Table 1) . Transmembrane domains were confirmed by using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Sequences of individual domains were retrieved from the SMART alignment. The DNA sequences encoding these domains were obtained by aligning the reverse-translated protein sequences with the actual gene sequence.
TABLE 1.
Organisms and genes discussed in this studya
| Gene type and organism | STPK gene | Ortholog(s)
|
|
|---|---|---|---|
| pknB | pbp2x | ||
| Actinomycetes genes | Rv0014c | ||
| Mycobacterium tuberculosis H37Rv | ML0016 | ||
| Mycobacterium leprae TN | MAP0016c | ||
| Mycobacterium avium subsp. paratuberculosis | Mb0016c | ||
| Mycobacterium bovis AF2122/97 | MSMEGPKNB | ||
| Mycobacterium smegmatis MC2 155 | SAV4338 | ||
| Streptomyces avermitilis MA-4680 | SCO3848 | ||
| Streptomyces coelicolor A3(2) | CE0033 | ||
| Corynebacterium efficiens YS-314 | DIP0053 | ||
| Corynebacterium diphtheriae NCTC 13129 | CG0057 | ||
| Corynebacterium glutamicum ATCC 13032 | BL0589 | ||
| Bifidobacterium longum NCC2705 | NFA800 | ||
| Nocardia farcinica IFM 10152 | LXX00210 | ||
| Leifsonia xyli subsp. xyli | PPA0184 | ||
| Propionibacterium acnes KPA171202 | |||
| Bacillus genes | |||
| Bacillus clausii KSM-K16 | ABC2315 | ABC2362 | |
| Bacillus halodurans C-125 | BH2504 | BH2573 | |
| Bacillus licheniformis ATCC 14580 | BL02302 | − | |
| Bacillus anthracis strain Ames | BA4000 | BA4055 | |
| Bacillus cereus ATCC 14579 | BC3860 | BC2662, BC3916 | |
| Bacillus thuringiensis serovar Konkukian strain 97-27 | BT9727_3603 | BT9727_2431, BT9727_3568 | |
| Bacillus subtilis subsp. subtilis strain 168 | BSU15770 | BSU15160 | |
| Other firmicute pbp2x orthologs | |||
| Clostridium acetobutylicum ATCC 824 | CAC2130 | ||
| Clostridium perfringens strain 13 | CPE0564, CPE1863, CPE1880 | ||
| Clostridium tetani E88 | CTC01633 | ||
| Enterococcus faecalis V583 | EF0991 | ||
| Lactobacillus johnsonii NCC 533 | LJ0969 | ||
| Lactobacillus plantarum WCFS1 | LP_2200 | ||
| Lactococcus lactis subsp. lactis Il1403 | L89079 | ||
| Listeria innocua Clip11262 | LIN2145 | ||
| Listeria monocytogenes EGD-e | LMO2039 | ||
| Oceanobacillus iheyensis HTE831 | OB1464 | ||
| Staphylococcus aureus subsp. aureus MRSA252 | SAR1157 | ||
| Streptococcus agalactiae 2603V/R | SAG0287 | ||
| Streptococcus mutans UA159 | SMU.455 | ||
| Streptococcus pneumoniae R6 | SPR0304 | ||
| Streptococcus pyogenes SSI-1 | SPS0461 | ||
| Symbiobacterium thermophilum | STH1205, STH2392 | ||
| Thermoanaerobacter tengcongensis MB4 | TTE1651 | ||
Actinobacterial STPK genes are known either by the identifiers shown or as “pknB”. The STPK gene of bacilli was originally characterized in B. subtilis as “prkC” (formerly “yloP”) and the pbp2X ortholog was characterised as “ftsI”. Other firmicute pbp2X ortholog gene names include pbpA (SAR1157), pbpB (LIN2145), pbpC (EF0991), pbp2B (LJ0969 and some bacilli), spoVD (all clostridia), and pbp2B2 (LP_2200). −, Not present.
Data analysis.
Alignments were obtained by using MEGA3 software (17) for analysis in DNASP (26); these alignments were checked with the version of CLUSTALX current at the time. Maximum-likelihood trees were obtained by using PHYML (15) and visualized by using MEGA3. Sliding window analysis was achieved by using DNASP, comparing each sequence against all others in the data set in a pairwise manner. Ka/Ks values were calculated by using the Nei-Gojobori method (22), using the Jukes-Cantor correction for substitutions at multiple sites. Average Ka/Ks values were calculated from individual values of all pairwise comparisons between all members of each data set. Some comparisons were made impossible due to a high (>75%) proportion of synonymous substitutions, thus preventing the use of the Jukes-Cantor correction. These comparisons were excluded from the analysis. For pknB orthologs, the differences between Ka/Ks values for kinase and extracellular domains were tested for significance by using a paired t test, while the Ka/Ks values for the PASTA domains of pbp2x orthologs and pknB orthologs were compared by using a Mann-Whitney U test. These statistical analyses were done by using SPSS.
Structural analysis.
Structures of PASTA domains were determined by using the NPS server (4), using the MLRC, DSC, and PHD programs to give a consensus secondary structure prediction.
RESULTS AND DISCUSSION
Evolutionary comparison of the intracellular and extracellular components of actinomycete pknB orthologs.
A comparison of the amino acid sequences of the cytoplasmic kinase domains of four actinomycete (S. coelicolor, S. avermitilis, M. tuberculosis, and Corynebacterium diphtheriae) PknB orthologs revealed 55% conservation of identical residues and 11.8% conservation of similar residues, with an overall similarity of 66.8%. In contrast, the predicted extracellular portion of these proteins, consisting of four reiterated PASTA domains, has only 24.5% overall similarity, with 10.7% conserved residues and 13.8% similar residues. This suggests that different selection pressures are acting on either side of the plasma membrane. To quantify these differences, we examined the ratio of nonsynonymous (Ka) and synonymous (Ks) nucleotide changes in 14 orthologs of pknB (Table 1). Figure 1 illustrates the average values for Ka/Ks ratios using a window size of 72 and a step size of 21 nucleotides. When the sliding-window Ka/Ks ratios are mapped against the domain structure of the protein, clear differences in the values either side of the membrane are evident. Overall, the intracellular STPK domain is under much tighter selective constraint than the juxtamembrane, transmembrane, or extracellular (PASTA) domains. Indeed, the average Ka/Ks value for pknB kinase domains is 0.266, whereas for the extracellular reiterated PASTA domains the value is 0.787. When the two sets of pairwise comparisons for PASTA and kinase domains were examined, the values were found to be significantly different at the 95% confidence level using the paired t test (df = 90, t = −16.5).
Position-dependent evolution of individual PASTA domains.
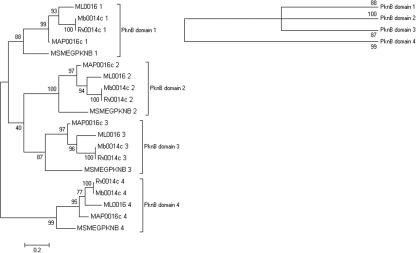
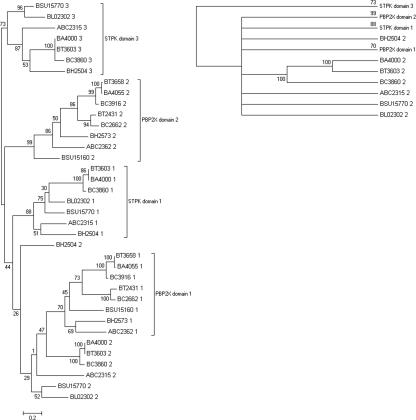
Phylogenetic comparison of the individual PASTA domains from actinobacterial STPKs reveals that they cluster according to their position in orthologous genes, rather than with neighboring domains in the same gene. This is clearly shown in the maximum-likelihood tree drawn for five mycobacterial pknB orthologs (Fig. 2), and trees drawn for other genera give a similar outcome. A large subset of the actinobacteria contains a second transmembrane kinase gene that lies within 20 genes of the division cell wall gene cluster. In these organisms each PASTA domain is more related to its orthologous domain than its homologous domain (results not shown). The positional clustering of PASTA domains is also true for pbp2x orthologs. Comparison of the nucleotide sequences of PBP2X- and STPK-associated PASTA domains of seven bacilli species (Table 1) reveals orthologous clustering of PASTA domains (Fig. 3), with PBP2X-associated domains forming their own unique clusters. However, the nucleotide diversity for the STPK-associated PASTA domain 2 is too great to allow grouping together with any degree of confidence. The nucleotide diversity is also too great to allow comparisons between PASTA domains from the same protein family from different genera.
FIG. 2.
Phylogenetic tree drawn for mycobacterial PASTA domains associated with pknB orthologs drawn using maximum likelihood. PASTA domains are numbered according to the scheme shown in Fig. 1. Percent bootstrap values from 500 replicates are shown on branches from the main nodes. Also shown (top right) is the topology tree obtained when bootstrap values of <70% are excluded.
FIG. 3.
Phylogenetic tree for PASTA domains associated with Bacilli STPKs and pbp2X orthologs. The scheme for numbering individual PASTA domains is sequential, starting with the most N terminal, similar to the scheme used for PknB in Fig. 1. The percent bootstrap values from 500 replicates are shown on branches from the main nodes. Also shown (top right) is the topology tree obtained when bootstrap values of <70% are excluded.
The significantly higher average Ka/Ks value for multiple STPK-associated PASTA domains compared to that for the kinase domains could be explained by either positive selection or relaxation of purifying selection. However, the phylogenetic analysis indicating position-dependent evolution of individual PASTA domains could only be consistent with positive selection favoring PASTA domain diversity. We hypothesize that each domain has evolved a binding affinity toward a specific stem peptide ligand. The complement of PASTA domains associated with a particular protein is specific to each genus, and this is likely to reflect differences in peptidoglycan composition existing between families of bacteria (see below). Moreover, the complement of PASTA domains is specific to each family of gene orthologs within a genus. For example, the actinomycete pknB orthologs have a distinct complement of PASTA domains compared to the division cell wall-linked STPKs from the same bacteria. This is consistent with a particular complement of PASTA domains recognizing a specific set of the stem peptide ligands. In this way, the diversity of gram-positive peptidoglycan composition may influence the function of the various PASTA domain proteins in different ways.
Identification of variable regions within PASTA domains.
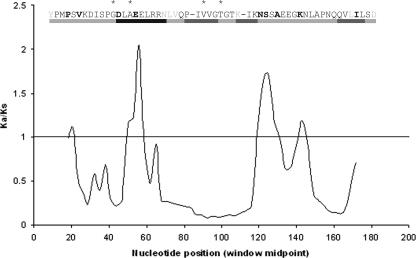
The variability of peptidoglycan structure in gram-positive bacteria is likely to provide a selection pressure for the rapid evolution of PASTA domains. Indeed, the pbp2x-associated PASTA domains of penicillin-resistant S. pneumoniae isolates are mutational hotspots (8, 30). More than 70% of the stem peptides in these isolates can consist of abnormal branched structures (12), and PASTA domain mutations presumably allow these structures to be recognized. Uniquely for the first PASTA domain of this pbp2x, there is experimental data on amino acid residues that contact the β-lactam cefuroxime (13, 30) and also the positions of amino acid substitutions present in variants that recognize abnormal stem peptides in penicillin-resistant strains (8). To identify regions of variability in this domain that could be evolving to recognize different ligands, a sliding-window analysis of Ka/Ks ratios was performed for the first PASTA domain sequence of S. pneumoniae pbp2x compared to the same domain of 27 other firmicute pbp2x orthologs (Table 1). Using a window size of 12 and a step size of three nucleotides, codon-by-codon analysis of variability could be mapped against the corresponding domain structure, with attention to amino acid residues implicated in Van der Waal's interactions with cefuroxime and the positions of amino acid substitutions present in pbp2x genes from penicillin-resistant isolates (Fig. 4). Of particular note is a correlation between the highly variable regions of this domain (Ka/Ks > 1) with the locations of amino acid substitutions. Moreover, the most divergent sequence aligns to the first half of the α-helix containing two amino acids involved in interactions with the β-lactam antibiotic. The analysis is consistent with the evolution of PASTA domain diversity to recognize different peptidoglycan stem peptides.
FIG. 4.
Sliding-window analysis of S. pneumoniae pbp2x PASTA domain 1 compared to the same domain of 27 other firmicute pbp2x orthologs. The corresponding protein structure (black bars, α-helix; gray bars, β-sheet) drawn to scale is shown above the graph, together with the S. pneumoniae PBP2x amino acid sequence alignment. Amino acids residues implicated in interactions with cefuroxime are denoted by asterisks, and residues shown in boldface are mutated in penicillin-resistant isolates. Amino acids shown in gray are not included in the sliding window analysis since they correspond to gaps in the alignment. The sliding-window analysis starts at position 18 due to “overhanging” amino acids from other sequences in the alignment.
The overall average Ka/Ks values for the first and second PASTA domains of 28 firmicute pbp2x orthologs belonging to 11 genera are 0.659 and 0.524, respectively. In contrast, the overall average Ka/Ks values for the 14 actinomycete pknB-associated PASTA domains from 7 genera are 0.895 (domain 1), 0.722 (domain 2), 0.779 (domain 3), and 0.859 (domain 4). With the exception of the comparison between the values for pknB-associated PASTA domain 2 and pbp2x-associated PASTA domain 1, in all other comparisons the diversity of the pknB-associated PASTA domains is significantly greater than that of the pbp2x-associated PASTA domains (at the 95% confidence level using a Mann-Whitney U test). A sliding-window codon-by-codon analysis of each pknB-associated domain revealed many more regions within each domain with Ka/Ks values greater than 1 than were present in either pbp2x-associated PASTA domain (results not shown). When mapped against domain structure, no consistent patterns of variability could be identified, although in general regions with Ka/Ks values of greater than 1 mapped to linker regions. Hence, regions of high variability appear to be largely constrained by protein tertiary structure.
The complexity of peptidoglycan in gram-positive bacteria: a clue to PASTA domain multiplicity, diversity, and function.
Peptidoglycan is a highly complex and essential macromolecule of bacterial cell walls (except mycoplasma, which coincidentally lack PASTA domain proteins) that contributes to cell shape and confers rigidity and resistance to osmotic pressure. It is constantly remodeled to allow cell growth and division. Whereas gram-negative bacteria have a single or few peptidoglycan layers composed of a uniform primary structure, gram-positive bacteria have up to 40 layers that can exhibit a great variation in composition and structural arrangement (28). Species-to-species differences are attributed to variation in the composition of the stem peptide linked to the glycan backbone. For example, the diamino acid component at position 3 of the peptide that is critical for cross-linking peptidoglycan chains can be any one of six different amino acids, or it can be unsubstituted, requiring that cross-linking is via a trifunctional amino acid at position 2. Moreover, a further increase in structural complexity is provided by frequent cross-bridges that link two chemically distinct stem-peptides. A corollary to this is that, in contrast to gram-negative peptidoglycan, the composition of murein in a gram-positive species can be nonuniform. In addition, the composition can be modified in response to environmental pressures. For example, up to 22 novel stem peptides are observed after the induction of vancomycin resistance in S. aureus (29), and heritable changes to peptidoglycan structure are a basis for resistance to β-lactam antibiotics (8).
Given the chemical diversity of gram-positive peptidoglycan, how is the cross-linked polymer assembled reproducibly? Mechanisms are required during cell growth and division to ensure that peptidoglycan is remodeled in a manner to reproduce a specific template. De novo peptidoglycan synthesis is preceded by localized murein hydrolysis, involving the breakage of existing cross-bridges, to allow the insertion of newly synthesized polymers that can then be cross-linked with “old” strands. For growth (elongation), the extent of incorporation of newly synthesized peptidoglycan is limited, but occurs at many sites, whereas multiple new strands are inserted at the two poles of a cell during septation. Genetic and cytological evidence suggests that these two processes are mediated by separate enzyme machinery. It is vital that appropriate peptidoglycan-synthesizing machinery is produced at the requisite time in the cell cycle and correctly positioned to allow for either growth or division.
The PASTA domains of PBP2x are likely to have a critical role in the positioning of this FtsI ortholog at the division site in S. pneumoniae. In this organism, deletion of the small carboxypeptidase, PBP3, thought to be responsible for generating a tripeptide stem transpeptidation substrate of PBP2x, leads to frequent incorrect positioning of the transpeptidase (20). Normally, PBP2x colocalizes with FtsZ and FtsW, and this positioning is due to recruitment by the localized pool of substrate stem peptides. Low-affinity binding to the substrate by a PASTA domain can thereby guide the localization of PBP2x to the division site. A second PASTA domain, recognizing an alternative stem peptide ligand, can provide greater specificity for PBP2x guidance, contributing to the reproduction of a nonuniform peptidoglycan template.
In comparison to the PBP2x orthologs, the STPKs possess a greater number of PASTA domains, and these exhibit even greater diversity (higher overall Ka/Ks ratios), suggesting evolution to recognize a wider spectrum of ligands. In actinobacteria, PknB is likely to function by sensing unlinked peptidoglycan in order to direct synthesis and/or localization of the machinery required for cell wall modification to permit growth. Likely ligands for the four PASTA domains of the STPK are unlinked stem peptides of different composition resulting either from the action of modifying endopeptidases on cross-linked peptidoglycan or from the synthesis of new peptidoglycan. Another potential source of ligands is the release of muropeptides. Recent research on resuscitation-promoting factors of actinobacteria has revealed that their biological activity in stimulating the revival of quiescent bacteria correlates with their murolytic activity (21). Extremely low concentrations of these cytokines are required for their biological activity, indicating that a limited release of muropeptides is likely to trigger a signaling cascade that amplifies the response. PknB is a good candidate as a kinase that can trigger such a phosphorelay and thus may be crucial in signaling the switch between latency and the onset of progressive tuberculosis.
In addition to growth and cell division, various developmental processes in prokaryotes also demand peptidoglycan remodeling. The PASTA domain STPKs of firmicutes appear to be dedicated to these changes, with evidence for a coordinating role in developing competence, biofilm formation, and sporulation. We postulate that different unlinked stem peptides may participate in the activation of these STPKs, leading to remodeling of the peptidoglycan appropriate to a specific developmental pathway.
Concluding remarks.
Even for the uniform and relatively simple cell wall structure of E. coli, our understanding of peptidoglycan remodeling associated with growth and division is limited. The increased chemical complexity and variety of the gram-positive cell wall would appear to demand a higher-order coordination of peptidoglycan remodeling. The evolution of proteins with multiple diverse PASTA domains that can recognize different unlinked stem peptides is likely to have been driven by the complexity and variety of gram-positive murein. The multiple PASTA domains of the STPKs can contribute to the reproduction of complex peptidoglycan by interaction with diverse stem-peptide ligands. As a consequence, signaling pathways can be triggered that coordinate the remodeling machinery. Since these interactions are important in pathogenic organisms such as M. tuberculosis, this underlines the importance of investigating the nature of the ligands that individual PASTA domains recognize.
Acknowledgments
We thank David Skibinski for help and encouragement.
G.J. was supported by a BBSRC studentship.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 2.Bernal, A., U. Ear, and N. Kyrpides. 2001. Genomes OnLine Database (GOLD): a monitor of genome projects worldwide. Nucleic Acids Res. 29:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boitel, B., M. Ortiz-Lombardia, R. Duran, F. Pompeo, S. T. Cole, C. Cervenansky, and P. M. Alzari. 2003. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 49:1493-1508. [DOI] [PubMed] [Google Scholar]
- 4.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta, A., P. Datta, M. Kundu, and J. Basu. 2006. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152:493-504. [DOI] [PubMed] [Google Scholar]
- 6.Del Sol, R., J. G. Mullins, N. Grantcharova, K. Flardh, and P. Dyson. 2006. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor. J. Bacteriol. 188:1540-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Sol, R., A. Pitman, P. Herron, and P. Dyson. 2003. The product of a developmental gene, crgA, that coordinates reproductive growth in Streptomyces belongs to a novel family of small actinomycete-specific proteins. J. Bacteriol. 185:6678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dessen, A., N. Mouz, E. Gordon, J. Hopkins, and O. Dideberg. 2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate. J. Biol. Chem. 276:45106-45112. [DOI] [PubMed] [Google Scholar]
- 9.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 10.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M.-C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 72:2434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaidenko, T. A., T. J. Kim, and C. W. Price. 2002. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J. Bacteriol. 184:6109-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Bustos, J., and A. Tomasz. 1990. A biological price of antibiotic resistance: major changes in the peptidoglycan structure of penicillin-resistant pneumococci. Proc. Natl. Acad. Sci. USA 87:5415-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, E., N. Mouz, E. Duee, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 14.Grundner, C., L. M. Gay, and T. Alber. 2005. Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 14:1918-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 16.Kang, C.-M., D. W. Abbott, S. T. Park, C. C. Dascher, L. C. Cantley, and R. N. Husson. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 18.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: toward genomic data integration. Nucleic Acids Res. 32(Database Issue):D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Seror. 2002. Characterization of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis, implicated in developmental processes. Mol. Microbiol. 46:571-586. [DOI] [PubMed] [Google Scholar]
- 20.Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The d,d-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641-1648. [DOI] [PubMed] [Google Scholar]
- 21.Mukamolova, G. V., A. G. Murzin, E. G. Salina, G. R. Demina, D. B. Kell, A. S. Kaprelyants, and M. Young. 2006. Muralytic activity of Micrococcus luteus Rpf and its relationship to physiological activity in promoting bacterial growth and resuscitation. Mol. Microbiol. 59:84-98. [DOI] [PubMed] [Google Scholar]
- 22.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 23.Novakova, L., L. Saskova, P. Pallova, J. Janecek, J. Novotna, A. Ulrych, J. Echenique, M.-C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 272:1243-1254. [DOI] [PubMed] [Google Scholar]
- 24.Pallen, M., R. Chaudhuri, and A. Khan. 2002. Bacterial FHA domains: neglected players in the phospho-threonine signaling game? Trends Microbiol. 10:556-563. [DOI] [PubMed] [Google Scholar]
- 25.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 26.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 27.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high-density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Severin, A., K. Tabei, F. Tenover, M. Chung, N. Clarke, and A. Tomasz. 2004. High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279:3398-3407. [DOI] [PubMed] [Google Scholar]
- 30.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27:438. [DOI] [PubMed] [Google Scholar]