Abstract
We recently reported the isolation and initial characterization of a transposon-generated mutation that resulted in defects in both morphogenesis and antibiotic production in Streptomyces coelicolor. The insertion identified the SCO7168 open reading frame whose predicted product is a GntR family transcriptional regulator. Here, we show that this gene acts to repress transcription of itself as well as a series of genes immediately adjacent to it on the S. coelicolor chromosome that likely encode an ATP-binding cassette (ABC)-type transporter for carbohydrate uptake. Transcription of this transporter is strongly induced by growth on relatively poor carbon sources such as trehalose and melibiose and weakly induced by lactose and glycerol but not glucose, and induction is not repressed by the presence of glucose. Constructed deletions of the ABC transporter itself resulted in the suppression of the original transposon mutation, suggesting that inappropriate expression of the ABC transporter is responsible, at least in part, for the mutant phenotype. Because this transporter responds to the presence of α-glucosides and has similarity to two other carbohydrate transporters of this class, we have named the genes of the transporter agl3E, agl3F, and agl3G and the GntR-like protein that regulates transcription of the transporter agl3R in accordance with established nomenclature. We suggest that agl3R is one of a number of homologous proteins in Streptomyces (there are 57 putative GntR family regulators in the S. coelicolor genome) that respond to nutritional and/or environmental signals to control genes that affect morphogenesis and antibiotic production.
The mechanisms by which cells sense environmental signals and transmit that information to changes in gene expression often involve the participation of transcription factors that respond either directly or indirectly to small molecules or substrates. One such family of proteins, the GntR family of transcriptional regulators, includes more than 1,300 members (31) that are distributed among a diverse group of bacteria and are involved in the regulation of a variety of different biological processes. These regulators have been shown to act as environmental sensors for controlling genes involved in responding to external stimuli. DasR, for example, is a GntR family protein that regulates cell-cell communication in Streptomyces griseus (40) independent of the A factor, a member of the γ-butyrolactone family of microbial hormone morphogens. Of special interest in this context is the GntR-like whiH gene of Streptomyces coelicolor, which plays a crucial role in the septation of aerial hyphae, and mutants fail to produce the gray spore pigment associated with mature spores. WhiH is required for the transcription of a number of genes that constitute a regulatory cascade for the differentiation of S. coelicolor (32, 33).
Streptomyces spp. undergo an elaborate program of cellular development in response to environmental changes. When nutrients are plentiful, the organism grows as a branching “substrate” mycelium that penetrates and solubilizes organic material in the soil. As food sources are exhausted, development begins with the erection of aerial hyphae that ultimately septate to form uninucleoid spores. Many of the same signals that lead to the initiation of development trigger the production of a large number of medically important natural-product antibiotics. Most of what is known about the regulation of pathways that contribute to morphogenesis and antibiotic production comes from the study of mutants defective in one or both of these processes (8, 13). Mutants blocked in the production of aerial hyphae are called bald (bld) mutants, many of which are also defective in antibiotic production. Mutants that are able to initiate morphogenesis but that are defective in the ability to form mature spores are called white (whi) mutants. The genes identified by these bld and whi mutants are diverse in function, and while a clear picture of how they participate is still emerging, it is clear that they hold the key to understanding both morphogenesis and antibiotic production in these complex bacteria.
At times and under conditions when the wild type has produced abundant amounts of spores, the agl3R mutant produces no spores and relatively few aerial mycelia. It also fails to produce the blue pigment associated with actinorhodin production (41). On relatively poor media such as minimal medium (MM)-glucose, MM-mannitol, and MM-maltose, the mutant grows more slowly than the wild type and fails to produce pigment. Both the morphological and antibiotic production defects of the agl3R mutant are restored by complementation with the wild-type allele (41). The agl3R open reading frame is located directly adjacent to a cluster of genes that show amino acid sequence similarity to ATP-binding cassette (ABC)-type membrane transporters (35, 41). ABC transporters have a wide variety of functions in solute transport, including sugars, peptides, and amino acids (39), as well as drug efflux (43). Perhaps the most relevant and interesting aspect of these transporters in this context is the fact that they are capable of transporting more than one type of molecule. The bldK gene cluster of S. coelicolor, for example, encodes an ABC transporter that transports a small-molecule morphogen that promotes sporulation in S. coelicolor on rich but not minimal medium (28). It also transports the drug bialaphos, and mutants defective in the transporter fail to respond to the morphogen and are bialaphos resistant (28). While 45 of the 81 predicted ABC transporters in the S. coelicolor genome are predicted to be involved in carbohydrate transport (2), the agl3EFG gene cluster described here and the dasABC gene cluster of S. griseus have also been implicated in development. DasABC encodes a probable sugar transporter, but overexpression of this cluster results in ectopic sporulation in substrate mycelia in response to glucose (40).
The S. coelicolor genome contains 57 putative GntR-like proteins (see “GntR” in the ScoDB [http://streptomyces.org.uk/]), but only whiH (32) and the agl3R open reading frame have been identified by mutation. The product of the whiH gene is likely to be a transcriptional regulator, but the direct target of its activity is not known, nor is anything known about the environmental conditions to which whiH responds. Here, we show that agl3R, also a GntR-like regulator, acts to repress transcription of itself as well as a cluster of genes directly adjacent to it that likely encode an ABC-type carbohydrate transporter. Reporter gene fusions to the promoter of the transporter showed that expression is strongly induced by growth on the α-glucosides melibiose and trehalose as sole carbon sources and that induction is not repressed by glucose. Gel mobility shift experiments using a His-tagged version of agl3R showed that it binds the intergenic region between agl3R and agl3E in vitro. Transcript analysis of the effect of the agl3R mutation on other whi genes suggests that it does not fit into the previously proposed whiG-dependent regulatory cascade for morphogenesis in S. coelicolor. We speculate that this GntR-like regulator plays a role in the regulation of morphogenesis and antibiotic production in response to the presence of complex carbohydrates as carbon sources.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
General techniques for bacterial growth were performed as described previously by Sambrook and Russell (34) and Kieser et al. (23) for Escherichia coli and S. coelicolor, respectively. S. coelicolor strains were grown in yeast extract-malt extract medium for genomic DNA isolation and mannitol-soya flour (MS) agar medium with the addition of 10 mmol MgCl2 for mating experiments. Antibiotic selections were applied by overlay with soft nutrient agar. Streptomyces RNA was isolated from cells grown on cellophane discs placed on top of maltose-yeast extract-malt extract (MYM) agar. E. coli strains and growth conditions for the preparation of cosmids for marker replacement in Streptomyces were described previously (14). Streptomyces strains used in this work are listed in Table 1 with the indicated sources. Primers used to amplify fragments for S1 mapping, construction of transcriptional fusions to the xylE gene (19), and construction of mutants are listed in Table 2.
TABLE 1.
S. coelicolor strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| M145 | SCP1− SCP2− | 23 |
| agl3R | M145 SCO7168::Tn5::apr | 41 |
| BH5 | agl3R containing the agl3R wild-type allele | 41 |
| BH6 | Deletion of SCO7167-SCO7165 | This work |
| BH7 | Deletion of SCO7167-SCO7162 | This work |
| BH8 | Deletion of SCO7167-SCO7165 and ΔSCO7168 | This work |
| BH9 | Deletion of SCO7167-SCO7162 and ΔSCO7168 | This work |
| BH10 | M145 containing the aglE-xylE fusion | This work |
| BH11 | agl3R containing the aglE-xylE fusion | This work |
| J2408 | whiH::ermE | 13 |
| J2400 | whiG::hyg | 13 |
| YU105 | pro arg red act::ermE whiE::hyg | 47 |
TABLE 2.
Primers used in this study
| Primer and purpose | Sequencea |
|---|---|
| To construct probes for S1 nuclease mapping | |
| agl3E/agl3R Forward | CGCTGCCGCCGCACGCGGTGGTCA |
| agl3E/agl3R Reverse | TGAGGTCCCCGTTGACGATGAGACC |
| HrdB Forward | CGGCCGCAAGGTACGAGTTGATGA |
| HrdB Reverse | CCATGACAGAGACGGACTCGGCG |
| To amplify the promoter fragment for XylE fusions | |
| agl3E/agl3R Forward | TCGGCGTCCCGGAAGCTTCCGATGTCGCCG (wild-type sequence, AAGCGT) |
| agl3E/agl3R Reverse | GAGATAGGGGAGAAGCTTGTCGTTGTGCGA (wt sequence, GACCTT) |
| To construct deletion mutations | |
| SCO7167 Upstream | ATCGCCTATCGATAGAACATAATCGATAGGGAGGGCGACATTCCGGGGATCCGTCGACC |
| SCO7165 Downstream | TTCGCCCCGGTGGCGGGGATGTCGTGTCGTGCGCTCATGTGTAGGCTGGAGCTGCTTC |
| SCO7162 Downstream | GCCCTGGTCCTGTGTCCACCTGTGGCCCGGCGCCCGCTGTGTAGGCTGGAGCTGCTTC |
| SCO7168 Upstream | CCGCGTACACGCTCCGCTCAGCCGTGGCCGTGATCGTGAATTCCGGGGATCCGTCGACC |
| To confirm deletion mutations | |
| agl3R Forward | CGACCGGCTCACGACCGGCCCGCGA |
| SCO7165 Reverse | CGCCGGACGGCGGAGTCCCGACGGA |
| SCO7162 Reverse | AGGGCCAGGGCCAGGGCCAGGACCG |
| P1 | ATTCCGGGGATCCGTCGACCTGCA |
| P2 | TGTAGGCTGGAGCTGCTTCGAAGT |
| To construct His6-tagged agl3R protein | |
| His6-Agl3R Forward | TTGTTGTAGAAGACGAGGGA |
| His6-Agl3R Reverse | GGGGCGGCTCAGATCTCTT |
| For RT-PCR | |
| WhiB Forward | GTCGACGACGCGGACGAGGAA |
| WhiB Reverse | AGATGCCGAAGCGCTCGTCGT |
| WhiG Forward | TGTGGCGGTCGTACAAGACGA |
| WhiG Reverse | ATCGCGTACGTCTCGAACTTG |
| WhiH Forward | AGCTGGGCCAGATGATCGTCT |
| WhiH Reverse | AAGGCACGCCATTCGATGATG |
| WhiE Forward | TCTTCATGTCCGGCAACCGGA |
| WhiE Reverse | TAGAGCAGCCGCAGCCGTTCC |
| SigF Forward | GGAGGTGCTGTCCTGCATCGA |
| SigF Reverse | GGAAACTACGTGCCAGTAGCC |
| Agl3R Forward | ACTGCTGTCCGACCAGGTGTA |
| Alg3R Reverse | CCGCGAACTCCTTCGATGATG |
| Agl3E Forward | AACATCACACCGAACCTGACC |
| Agl3E Reverse | AGGGGAGGACCTTGTCGTTGT |
| HrdB Forward | GCCTTCGAAGCTGACCAGATT |
| HrdB Reverse | CGGTCGCCTTCCTGCTGGTCA |
Underlining indicates the HindIII restriction site; boldface indicates altered bases.
Microscopic analysis of aerial mycelium development.
The procedure used for the microscopic analysis of aerial mycelium development was performed as previously described (32). Samples from colonies grown for 5 days on MYM agar medium were mounted onto an aluminum stub with optimal cutting temperature compound, submerged in liquid nitrogen slush at approximately −210°C, and transferred to a Gatan Alto 2500 cryostage and cryoprep chamber (Gatan UK, Oxford, United Kingdom) attached to a LEO 982 field emission scanning electron microscope (LEO Electron Microsopy, Inc., Thornwood, NY). The sample was sublimated to remove surface frost at −95°C for 3 min, coated with platinum, placed onto the cryostage in the main chamber of the microscope at approximately −140°C, and viewed at 5.0 kV.
RNA isolation and S1 nuclease mapping.
S1 nuclease mapping was carried out as described previously (25). RNA was isolated from cells after 16, 24, 36, 48, or 60 h of growth on MYM agar plates overlaid with cellophane discs, and 40 μg was hybridized to approximately 0.05 pmol of a 32P-5′-end-labeled DNA probe and incubated with 100 U of S1 nuclease (Roche). Probes were generated by PCR with Taq DNA polymerase (QIAGEN) using S. coelicolor genomic DNA as a template. The same oligonucleotides, S1 Forward and S1 Reverse, were used to generate a 305-bp probe for mapping the agl3R transcript (S1 Reverse labeled) and the agl3E transcript (S1 Forward labeled). Primers were labeled using [γ-32P]ATP and OptiKinase according to the manufacturer's instructions (USB) prior to the PCR. The hrdB promoter probe was described previously (22). The same labeled oligonucleotides were used in the fmol cycle DNA sequencing system (Promega) to generate the G, A, T, and C sequencing ladders. DNA fragments were separated on a 6% denaturating polyacrylamide gel, and bands were visualized by autoradiography.
Construction of transcriptional fusions to the xylE reporter gene.
Primers complementary to the DNA sequence 542 bp upstream of the agl3E translational start site (agl3E Forward) and 97 bp downstream of the agl3E ATG (S1 Forward) were used to generate a 639-bp fragment containing the agl3E promoter region. The agl3E Forward primer contained base changes that created a HindIII restriction site for subsequent cloning (Table 2). The PCR product was purified, digested with HindIII, and ligated to pBluescript II(+) that had been cut with HindIII and SphI (13, 33). An aliquot of the ligation mixture was transformed into competent E. coli XL10 Gold cells, and transformants were selected with ampicillin. Plasmid DNA was isolated, using a QIAGEN Plasmid Mini column, from transformants that had been grown overnight in liquid LB broth containing 100 μg/ml ampicillin. Plasmid DNA was then digested with HindIII and BamHI, and the promoter-containing fragments were ligated to HindIII/BamHI-cut pXE4, placing the fragments upstream of the xylE gene and in the correct orientation for transcription to generate pXE7167 and pXE7168. Plasmid constructions were verified by restriction digestion. Each plasmid was digested with HindIII and EcoRI to generate a fragment that contained the promoter-xylE fusion, which was then blunted using the Klenow fragment of polymerase I and ligated to EcoRV-digested pHygoriT (41) containing a hygromycin resistance gene. The ligated vector was transformed into chemically competent XL10 Gold E. coli cells, and transformants were selected by using hygromycin on LB agar plates. The constructions were confirmed by using XbaI and EcoRI digestion. Plasmids were transformed into electrocompetent E. coli Et12567 cells containing the nontransmissible helper plasmid pUZ8002 as previously described (14). Transformants were selected using hygromycin on LB agar plates, and single colonies were used for conjugation with S. coelicolor M145 and agl3R (14). Exconjugants were selected on MS agar plates overlaid with 50 μg/ml hygromycin after 16 h.
XylE assays.
Assays were preformed on whole cells as previously described (4), with the following modifications. S. coelicolor strains BH10 and BH11 were grown for 36 h on SMMS-maltose agar medium (23) overlaid with cellophane disks. After 36 h, cells were scraped from the cellophane, vortexed briefly with glass beads to disperse the cell mass, and diluted with liquid minimal medium (23) containing the indicated sugar. Cultures were grown for 36 h at 30°C. Cells were harvested by centrifugation for 10 min at 3,000 rpm, washed once with sterile 50 mM K2HPO4, pH 7.4, and resuspended in sterile 50 mM K2HPO4 to an optical density at 600 nm (OD600) of 0.5. Two hundred microliters of the cell suspension was added to 400 μl of sterile 50 mM K2HPO4 containing 10 mM catechol. Absorbance was measured at OD375 at 10-s intervals for 90 s.
Construction and confirmation of deletion mutants.
The deletion mutants shown in Fig. 1 were constructed by using a PCR targeting method described previously by Gust et al. (14). The Δagl3EFG and Δagl3EFGR mutants were made by replacing the chromosomal region of the deletion with the apramycin resistance cassette [acc(3)IV]. The extent and location of the deletions in each strain were confirmed by PCR. The presence of the apramycin resistance cassette was detected using specific primers, P1 and P2. The location of the cassette was determined using primers located outside the deleted sequence, SCO7168Out, SCO7165Out, and SCO7162Out. The primers within the deleted sequence, S1 Forward and S1 Reverse, were used as negative controls.
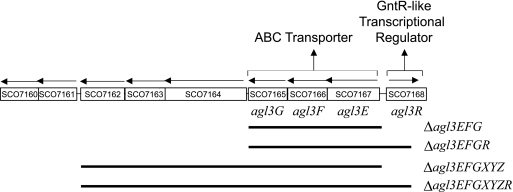
FIG. 1.
Organization of the S. coelicolor genome showing the agl3R open reading frame identified by transposon insertion and adjacent genes. Gene notations are based on the Sanger Centre Genome Sequencing Projects (http://www.sanger.ac.uk/Projects/S_coelicolor). Below the gene map is a diagram showing the limits of the DNA deleted (black lines) in constructed mutants.
Gel retardation assays.
To construct a His-tagged version of the Agl3R protein, the coding region was amplified by PCR from S. coelicolor genomic DNA using primers Agl3Rhis Forward and Agl3Rhis Reverse (Table 2). The fragment was digested with XmnI (Promega) and DdeI (Promega), treated with the Klenow fragment of DNA polymerase (Promega), and cloned into the NdeI site of pET28a that had also been blunted. DNA sequencing confirmed the correct orientation and fusion to the His tag. The resulting construction was transformed into E. coli BL21(DE3) cells, and cells were grown in 2× YT (17) at 37°C to an OD600 of 0.6, induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), grown an additional 18 h at 18°C, harvested by centrifugation, and resuspended in phosphate buffer (20 mM phosphate, 500 ml NaCl, 5% glycerol, 2 mM dithiothreitol, pH 8.0). Cells were lysed by sonication and centrifuged at 50,000 × g for 20 min. The supernatant was passed over an Ni column, and protein was eluted using a 10 mM to 300 mM imidazole gradient. Purified His6-Agl3R protein (5, 10, or 20 μg) or crude cell extract (20 μg total protein) was mixed with the same 305-bp PCR-generated DNA fragment containing the intergenic region between agl3E and agl3R used for S1 nuclease mapping experiments. The fragment was 5′-end labeled with [γ-32P]ATP (MP Biomedicals) using T4 polynucleotide kinase (Promega) and purified on a 1% agarose gel. The labeled DNA fragment (1 ng; 6,000 cpm) was incubated with cell extracts or purified His-tagged Agl3R protein for 20 min at 30°C in 20 μl (total volume) of binding buffer (20 mM Tris, 10 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, pH 8.0) containing 1 μg sonicated salmon sperm DNA and 3 μg bovine serum albumin. For competitive inhibition of the binding reaction, 100 ng of unlabeled fragment or 100 μg of sonicated salmon sperm was added to 5 μg purified His6-Agl3R protein. Reactions were displayed on a nondenaturing 6% acrylamide-Tris-borate-EDTA gel and visualized by autoradiography.
RT-PCR.
RNA was isolated from S. coelicolor M145 and the aglR mutant after 24, 36, 48, and 60 h of growth on MYM agar medium overlaid with cellophane discs as was done for S1 nuclease mapping. The One-Step PCR kit (QIAGEN) was used with primers specific for each gene. Forward and reverse primers for whiG, whiB, whiH, whiE, sigF, alg3A, agl3R, and hrdB are shown in Table 2. Reaction mixtures contained 10 pmol of each primer and 150 ng of RNA in a total volume of 20 μl. Each primer was first tested using chromosomal DNA as a template and without a reverse transcription (RT) cycle to test for DNA contamination in the RNA. HrdB was used as a control for RNA concentration. Products were displayed on a 1% agarose gel and visualized by staining with ethidium bromide.
RESULTS
An insertion into the agl3R open reading frame results in a white mutant phenotype.
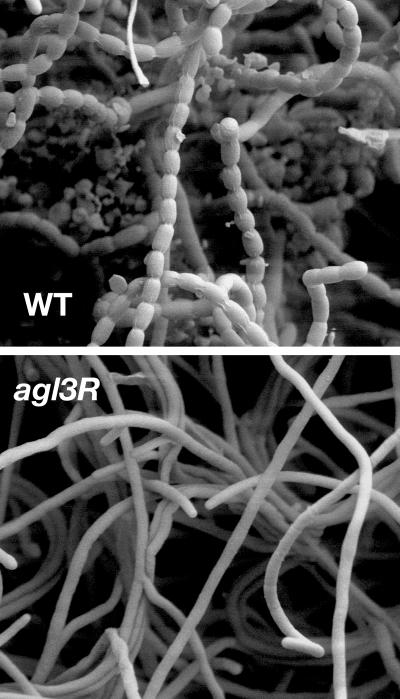
A diagram showing the location of SCO7168 (SC9A4.30), the site of the transposon insertion, and the annotated region of the chromosome at this address are shown in Fig. 1. As previously reported (41), the SCO7168/SE69 mutant (referred to hereafter as agl3R) was defective in sporulation and the production of blue pigment associated with antibiotic production in S. coelicolor. The predicted 224-amino-acid protein encoded by this open reading frame shows sequence similarity to the GntR family of transcriptional regulators. A DNA fragment containing the single SCO7168 open reading frame complemented both the morphological and antibiotic production phenotypes of the mutant, suggesting that both aspects of the mutant phenotype were the result of a single mutation. To more fully characterize the morphological phenotype of this mutant, cells were grown on the sporulation medium MYM and visualized by scanning electron microscopy. After 5 days of growth on agar plates, visual inspection of colonies showed that the morphologically wild-type strain M145 produced a dense covering of aerial mycelium and gray-pigmented spores, while the mutant remained white and failed to produce the gray pigment associated with mature spores. Such mutants in Streptomyces species are called whi mutants. As shown in Fig. 2, scanning electron microscopy revealed that the aerial hyphae made by this mutant did not coil or septate, and no spores were visible. At the same stage of growth in the wild type, coiled, fully septated aerial hyphae as well as mature spores are clearly present.
FIG. 2.
Scanning electron micrographs of aerial hyphae from wild-type S. coelicolor (M145) (WT) and the agl3R mutant at ×5,000 magnification. Both strains were grown for 5 days at 30°C on MYM medium.
The original collection of whi mutants isolated by Chater (8) were not significantly reduced in vegetative growth and made aerial mycelia at the same time as the wild type while failing to produce spores upon prolonged incubation. Compared to other well-characterized whi mutants, the morphological defects of agl3R are similar to those of whiH but perhaps most closely resemble those of whiG (13). whiG mutants make relatively straight aerial hyphae that completely lack sporulation septa. whiH mutants make loosely coiled aerial hyphae with infrequent but obvious sporulation septa. Interestingly, like agl3R, the whiH gene encodes a GntR-like transcriptional regulator, but the agl3R mutant has a more severe morphological phenotype than whiH, and whiH mutants are not defective in antibiotic production. Neither the agl3R mutant nor whiH mutants produce the gray pigment associated with mature spores. whiA or whiB mutants have abnormally long and coiled aerial hyphae that are almost devoid of septation. The products of the whiG, whiA, and whiB genes apparently act early in aerial hypha formation and may participate in the decision to enter sporulation (9). Mutations in whiG are, in fact, epistatic to those in whiH (7, 33). Analysis of the phenotypes of the agl3R and whiH mutants suggests that the agl3R gene product participates in the maturation of aerial hyphae rather than in the earliest stages of the decision to enter sporulation.
The agl3R gene product acts to repress transcription of an adjacent gene cluster that encodes a putative ABC-type transporter.
A gene cluster directly adjacent to agl3R and transcribed in the opposite direction contains a number of open reading frames (Fig. 1), three of which show similarity to ABC-type transporters (41). In accordance with established nomenclature, these open reading frames are designated agl3E, agl3F, and agl3G.
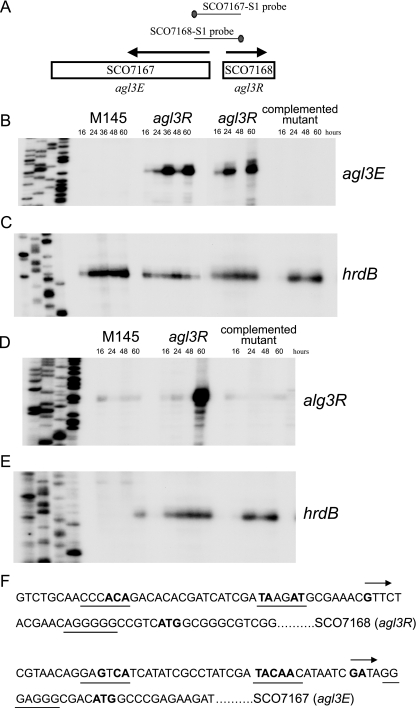
To test whether a mutation in agl3R affected transcription of this cluster, high-resolution S1 nuclease mapping experiments were performed on RNA isolated from cells at different stages of development using a PCR-generated probe overlapping the beginning of the agl3E open reading frame (Fig. 3A). RNA was isolated from morphologically wild-type strain M145 and agl3R mutant cells grown on MYM sporulation agar and harvested at different stages of development. This solid medium supports morphological development of the wild type, and the stages of development are clearly detectable. Cells harvested after 16 h were growing vegetatively (no aerial mycelia were present). Aerial mycelia began to appear between 24 and 36 h and were abundant between 36 and 48 h. The gray pigment associated with mature spores was evident at 60 h. As shown in Fig. 3B, no transcript was detected from agl3E in wild-type cells, suggesting that it was not expressed under the conditions and on the medium from which the cells were harvested. In the agl3R mutant strain, transcription of agl3E was readily detected, and two peaks of apparent transcriptional activity were observed, one at 36 h and another at 60 h. This pattern of expression was detected in three independent experiments, and the absence of transcript at 48 h was evident in every case. In fact, two independent experiments are shown in Fig. 3B. S1 nuclease mapping was also performed on RNA isolated from a strain in which the agl3R mutation had been complemented in trans with a wild-type copy of the agl3R open reading frame. The presence of a wild-type copy of agl3R restored the repression of the abundant agl3E transcripts appearing at 36 and 60 h of growth. The same RNA preparation used for the analysis of agl3E was used for the detection of the hrdB transcript, which served as a control for the level of RNA (Fig. 3C). These data suggest that the agl3R gene product acts to repress, either directly or indirectly, transcription of agl3E and perhaps some genes in the downstream gene cluster that are likely to be translationally coupled to agl3E. It is also of note that transcription of agl3E is apparently temporally regulated and is detected at 24 to 36 h and again at 60 h, with a lower level at 48 h. This observation suggests that simple repression by the agl3R gene product may not fully explain the transcriptional regulation of agl3E. If, in fact, transcription of agl3E is constitutive in the mutant, differences in message stability might account for the differences in the levels of RNA observed. If the appearance of RNA at the onset of aerial hypha production represents new initiation, either another repressor or an activator of transcription, in addition to the agl3R gene product, may be involved.
FIG. 3.
S1 nuclease mapping of transcripts originating upstream of agl3R and agl3E. RNA was isolated from the wild type (M145), the agl3R mutant, and the agl3R mutant containing a wild-type copy of the agl3R gene. Panel A shows the position of the probe used for the detection of agl3R and agl3E transcripts. Panel B shows the protected RNA of a transcript originating upstream of agl3R isolated from the wild type, the agl3R mutant (two different experiments), and the agl3R mutant containing the agl3R wild-type allele. Panel D shows the protected RNA of a transcript originating upstream of agl3E isolated from the wild type, the agl3R mutant, and the agl3R mutant containing the agl3R wild-type allele. Panels C and E show transcripts originating upstream of the hrdB gene as a control for RNA. In all cases, the sequencing reactions were loaded A, G, C, and T. (F) DNA sequence upstream of the apparent transcription start sites (indicated by boldface type and an arrow) for agl3R and agl3E. The potential −35 and −10 region RNA polymerase recognition sequences and bases with consensus to TTGACA and TACAAT, prototypical of vegetative promoters of Streptomyces, are in boldface type and underlined. Putative ribosome binding sites are underlined, and the annotated translation start sites (ATG for each) are also in boldface type.
Inappropriate expression of the gene cluster beginning with agl3E in the agl3R mutant is responsible, at least in part, for the mutant phenotype.
To establish a direct connection between the gene cluster beginning with agl3E and the defects in morphogenesis and antibiotic production observed in the agl3R mutant, various deletions of the gene cluster beginning with agl3E were constructed in both the wild type and an agl3R mutant (Fig. 1). Because each mutant was generated by marker replacement with a constructed deletion, it was not possible to construct a deletion of the ABC transporter in the original agl3R transposon mutant. Four deletions were constructed: two different deletions of the downstream gene cluster and two double mutants that deleted both the ABC transporter and the agl3R open reading frame. The genes downstream of agl3EFG do not show homology with ABC transporters or sugar transport proteins but are included in this analysis because of their proximity to the transporter and the fact that there is the possibility that they are translationally coupled to the transporter encoded by agl3EFG. All of the mutants were compared to the wild type after growth on maltose as the carbon source (the same conditions used to isolate RNA for the S1 nuclease mapping experiments).
As there was no detectable expression of the transporter in the wild-type strain, a deletion of the gene cluster in a wild-type strain should have no phenotype. In fact, mutants deleted for agl3EFG and/or the downstream gene cluster were indistinguishable from the wild type. If inappropriate expression of the gene cluster beginning with agl3E in the agl3R mutant was responsible for the mutant phenotype, the deletion of this cluster in an agl3R mutant should suppress the agl3R mutant phenotype resulting in a wild type phenotype. Deletions of the downstream gene cluster were, in fact, indistinguishable from each other, the single mutants, or the wild-type strain. We conclude from this analysis that inappropriate expression of the transporter in the agl3R mutant is responsible, at least in part, for the mutant phenotype.
The agl3R gene product represses transcription of its own synthesis.
Some GntR-like regulators (notably whiH) serve to repress their own synthesis (33). To test whether a mutation in agl3R affected transcription of itself, high-resolution S1 nuclease mapping experiments were preformed using a PCR-generated probe overlapping the beginning of the agl3R open reading frame (Fig. 3A). RNA was isolated from morphologically wild-type strain M145 and agl3R mutant cells grown on MYM sporulation agar and harvested at the same stages of development as those used to analyze agl3R transcription. As shown in Fig. 3D, in wild-type cells, a transcript originating upstream of agl3R was detected at approximately the same low level throughout growth and development. In the agl3R mutant, transcription of agl3E was approximately the same as that of the wild type until 60 h of growth, when abundant RNA was detected. While 60 h of growth is well after the onset of aerial mycelium production, the agl3R mutant does not produce spores, so it is difficult to correlate this stage of development with the wild type. At 60 h, however, under the same conditions, M145 was clearly beginning to produce the gray pigment associated with mature spores. S1 nuclease mapping was also performed on RNA isolated from a strain in which the agl3R mutation had been complemented in trans with a wild-type copy of the agl3R open reading frame. As shown in Fig. 3, the presence of a wild-type copy of agl3R restored the repression of the agl3R transcript. The same RNA preparation used for the analysis of agl3R was used for the detection of the hrdB transcript that served as a control for the level of RNA (Fig. 3E). These data suggest that the agl3R gene product acts to repress its own transcription either directly or indirectly. Interestingly, expression of agl3R in the agl3R mutant is apparently temporally regulated, with a peak of transcriptional activity appearing as the cells enter late stages of sporulation.
The Agl3R protein binds the agl3R/agl3E promoter region in vitro.
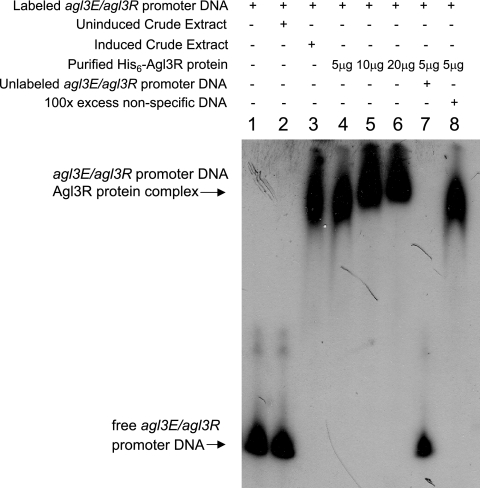
To address whether the regulatory effect of the agl3R gene product involved a direct interaction with the agl3R/agl3E promoter region, a His6-Agl3R protein was constructed and expressed in E. coli and used in gel mobility shift assays with a DNA fragment containing the intergenic region between agl3R and agl3E. As shown in Fig. 4, crude extracts from E. coli cells expressing the His6-Agl3R protein retarded the promoter-containing fragment, whereas extracts from uninduced cells did not. Furthermore, the amount of probe shifted in the retardation assay was reduced in the presence of unlabeled probe but not nonspecific DNA, suggesting that the binding of Agl3R to the promoter-containing fragment was sequence or motif specific.
FIG. 4.
Gel mobility shift assays using a DNA fragment containing the intergenic region between agl3E and agl3R and the His6-Agl3R protein. Lane 1, labeled fragment; lane 2, labeled fragment with crude extract from uninduced cells containing the His6-Agl3R construction; lane 3, labeled fragment with crude extract from induced cells containing the His6-Agl3R construction; lanes 4 to 6, labeled fragment with 5 μg, 10 μg, or 20 μg of purified His6-Agl3R protein; lane 7, labeled fragment with 100 ng unlabeled promoter-containing fragment and 5 μg purified His6-Agl3R protein; lane 9, labeled fragment with 100 μg sonicated salmon sperm DNA and 5 μg purified His6-Agl3R protein.
DNA sequences located upstream of the apparent transcription start sites for agl3E and agl3R are shown in Fig. 3F. The transcription start site of agl3R was mapped to a guanine residue 23 nucleotides upstream of the annotated translational start site. Sequences centered around −10 and −35 bp upstream of the start site show little homology to any known prokaryotic RNA polymerase binding site, suggesting that either this promoter is recognized by a novel sigma factor or transcriptional regulators facilitate RNA polymerase binding. The transcription start site of agl3E was mapped to a purine residue only 13 nucleotides upstream of the annotated translational start site. The apparent leader regions of these mRNAs are somewhat shorter than those for typical messages, especially for the transcript beginning with agl3E. There are, however, purine-rich sequences that are likely to serve as ribosome binding sites in both transcripts located between −10 and −6 with respect to the translational start site (Fig. 3). The distance between the putative ATG translational start sites of agl3E and agl3R is 135 bp, with 18 bp between the apparent −35 regions.
Transcription of agl3E is induced by growth on relatively poor carbon sources.
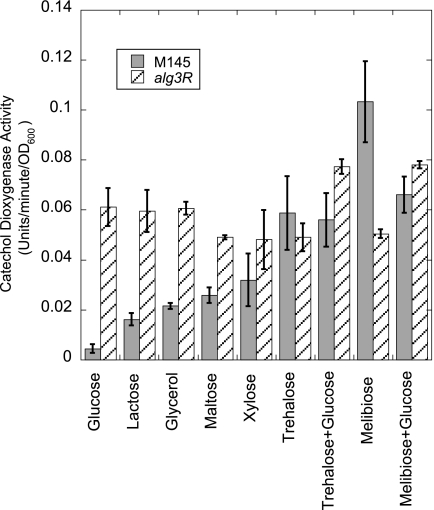
To identify conditions that might lead to the induction of the transporter, a transcriptional fusion between the promoter of agl3E and the xylE reporter gene was constructed. The xylE gene encodes a catechol dioxygenase that converts colorless catechol to a bright yellow oxidation product. The agl3E promoter-xylE fusion was constructed on a nonreplicating vector containing a φC31 attachment site and introduced into the wild-type and agl3R mutant strains, allowing the insertion of the fusion into the S. coelicolor chromosome. Cells were then grown in liquid medium containing various carbohydrate carbon sources and monitored for catechol dioxygenase activity (4, 19). As shown in Fig. 5, transcription from the agl3E-xylE fusion was not detected from cells grown on glucose or lactose but was readily detected from cells grown on melibiose or trehalose. Transcription was constitutive in an agl3R mutant, supporting the notion that the agl3R gene product serves to repress transcription of the transporter. Expression of the agl3E-xylE fusion during growth on melibiose or trehalose was not affected by the presence of glucose. While no transcription from the agl3E promoter was detected by S1 nuclease mapping from cells grown on maltose as a carbon source on plates, some transcription was detected from cells grown on maltose in liquid medium. We conclude from these data that transcription of the transporter is induced by growth on relatively poor carbon sources and that induction is not repressed by the presence of glucose. These data also suggest that these carbon sources relieve repression by the agl3R gene product either by direct interaction with the protein itself or by allowing the expression of a factor that leads to its ability to function.
FIG. 5.
Histogram showing the results of quantitative catechol dioxygenase assays from cells grown on various carbon sources. The darkly shaded bars are data from wild-type (M145) cells, and hatched bars are data from assays of the agl3R mutant, each containing a transcriptional fusion between the xylE reporter gene and the promoter region of agl3E.
The agl3R mutant has no apparent effect on the expression of whiE, whiG, whiB, whiH, or sigF, nor is its expression dependent on whiG, whiH, or whiE.
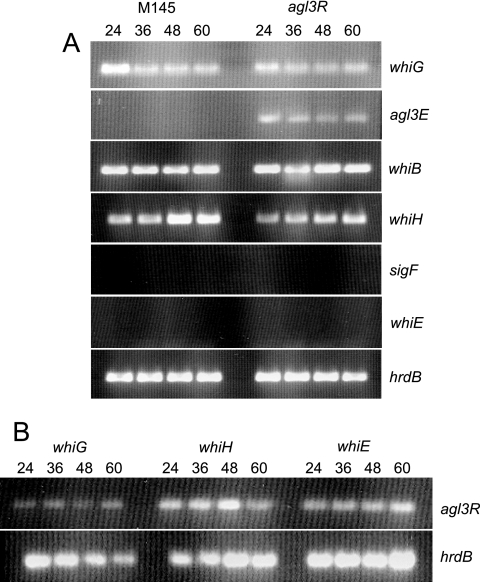
A model for how known whi genes interact has been proposed (33). To investigate whether agl3R fits into this cascade, RT-PCR was performed to determine whether a mutation in agl3R affected the expression of other genes known to be involved in morphogenesis and antibiotic production and whether its expression was dependent on the activity of other whi genes. RNA was isolated from S. coelicolor M145 (Fig. 6A, left) and agl3R (Fig. 6A, right) at 24, 36, 48, and 60 h and used as a template for the detection of whiG, whiB, whiH, sigF, and whiE orf1 transcription. The constitutively expressed hrdB transcript was used as a control for the presence of RNA. We emphasize that this method is not quantitative but that it does allow the detection of the presence or absence of transcripts. This analysis confirms the observations from S1 nuclease mapping showing that agl3R transcription is repressed in the wild type and is constitutive in the agl3R mutant. There is no detectable difference in the expression of any of the whi genes tested in the agl3R mutant (Fig. 6A). While sigF and whiE transcripts were not detected in this analysis (these genes are transcribed late in development, and their transcripts would not be present at the times examined here), the fact that no transcript from these genes was detected in the agl3R mutant suggests that it is unlikely to be a repressor of their transcription. While whiH transcription has been shown to be dependent on the activity of whiG (33), transcription of agl3R is not, suggesting that it is not part of the proposed whiG-dependent regulatory cascade.
FIG. 6.
RT-PCR analysis of transcripts from wild-type S. coelicolor and various whi mutants. (A) Transcripts from whiG, agl3E, whiB, whiH, sigF, and whiE in wild-type strain M145 and the agl3R mutant. (B) Transcripts from agl3R in whiG, whiH, and whiE mutants. hrdB transcript was used as a control for RNA.
DISCUSSION
agl3R controls transcription of itself and a group of genes that are likely involved in carbohydrate transport.
S1 nuclease mapping of transcripts originating upstream of the GntR-like protein, agl3R, and the first open reading frame of the ABC-type transporter, agl3E, suggested that the transcription of these genes is repressed by the agl3R protein, and gel mobility shift experiments using His6-Agl3R protein and the agl3R-agl3E intergenic region showed that the agl3R protein binds this region directly in vitro. Analysis of a reporter gene fusion to the agl3E promoter indicated that expression of the transporter is strongly induced by growth on relatively poor carbon sources such as trehalose and melibiose and is induced weakly by lactose and glycerol. Induction was not repressed by the presence of glucose.
Most sugar transport in Streptomyces relies on either ABC-type transporters/permeases or phosphotransferases. A recent in silico study of the S. coelicolor genome revealed 53 possible carbohydrate transport systems (3), most of which were ABC permeases. Carbohydrate uptake by ABC transporters has been described for cellobiose, cellotriose (37), and trehalose in Streptomyces reticuli (36); cellobiose and xylobiose in Streptomyces lividans (18); and maltose and maltodextrans in S. coelicolor (45). In addition, ABC transporters for maltodextran, cellobiose, cellotriose, β-xylosides, α-glucosides, xylose, chitobiose, lactose, sugar alcohols, ribose, maltose, maltodextrans, arabinose, cyclodextrans, trehalose, sorbitol, palatinose, and mannitol were identified based on similarity to previously characterized ABC clusters known to transport these sugars (3). The agl3EFG gene cluster was noted in that study as a likely sugar transporter but was not assigned to a specific carbohydrate group, and no transporters showing similarity to specific melibiose transporters were identified in that analysis. agl3E also shows similarity to the msmE, the corresponding sugar binding protein in the Streptococcus mutans msmEFG transporter, a multiple-sugar importer, one of which is melibiose (42). Here, we show that the agl3EFG transporter, which has some weak similarities to the melibiose class of transporters, responds to the presence of α-glucosides.
Most of the predicted regulatory genes for ABC transporters show similarity to the LacI/GalR family of transcriptional regulators. Transcription of the malEFG operon in S. coelicolor, for example, which encodes an ABC transporter specific for maltose and maltodextrans, is controlled by MalR, a repressor located adjacent to the malEFG operon. The malEFG operon has been shown to be induced by the presence of amylase and maltotriose and is repressed by glucose (38). There is, however, an example of an ABC transporter, dasABC in S. griseus, that is controlled by the GntR-like repressor DasR (40). Members of this family contain similar N-terminal DNA binding domains with helix-turn-helix motifs but lack significant similarity in regions involved in effector binding or oligomerization (16). Typically, oligomerization between regulatory subunits and/or conformational changes resulting from the binding or removal of inducing/repressing molecules allow correct helix-turn-helix disposition and confers DNA binding ability to the protein as a whole. While many GntR family proteins have been shown to act as repressor proteins that are responsive to carboxylate-containing intermediates in carbon metabolism, some have been shown to bind sites other than typical operator sequences, and at least one, FadR, can act as a transcriptional activator (12).
Inappropriate expression of an ABC transporter leads to defects in morphogenesis and antibiotic production.
Constructed deletions of the ABC transporter encoded by agl3EFG resulted in the suppression of the original transposon mutation, suggesting that inappropriate expression of the ABC transporter is responsible, at least in part, for the mutant phenotype. Mutations in agl3EFG alone have no growth phenotype, probably because of a redundancy in function with other α-glucoside transporters such as agl1EFG and agl2EFG and no morphological phenotype.
Interestingly, both agl3R and whiH, the two GntR-like regulators identified by mutation in S. coelicolor, encode transcriptional repressors, and while the target of whiH is unknown, presumably, the inappropriate expression of the genes that it controls is responsible, at least in part, for the mutant phenotype. The organization of the agl3EFG gene cluster is similar to that of dasABC in S. griseus, which, like agl3EFG, is controlled by a GntR-like transcriptional regulator, dasR. Constructed deletions of dasR resulted in the overexpression of dasABC and ectopic sporulation when cells were grown on glucose. Overexpression of dasA itself showed the same phenotype, suggesting that inappropriate expression of this ABC transporter affects morphogenesis in this strain as well.
The morphological phenotype of agl3R is similar to that of whi mutants of S. coelicolor in that it initiates morphogenesis, as evidenced by the emergence of aerial hyphae, but fails to develop the gray spore pigment associated with mature spores. Transcription of whiH, the other characterized GntR-like regulator in S. coelicolor, is dependent on whiG and whiH mutants that are defective in the expression of some late sporulation genes, including whiE and sigF. The whiH gene product has also been implicated in the regulation of its own synthesis (33). The whiG, whiA, and whiB genes apparently act early in aerial hypha formation and participate in the decision to enter sporulation (13). They are absolutely required for sporulation septation and for all visible signs of nucleoid condensation and partitioning as well as other changes associated with later stages of sporulation (13). In addition, whiG, whiA, and whiB are epistatic to whiH in that they prevent these structures from forming in double mutants (7, 13). These spore-like features and the synthesis of clearly detectable levels of the whiE-directed gray spore pigment were not due to any residual activity of previously studied whiH alleles since they were retained by a constructed whiH null mutant. Interestingly, there is a potential ABC transporter located directly upstream of the whiH gene, and initial BLAST searches show weak similarity to transporters of the amino acids glycine and proline (with E values of 40% [6e−16] and 46% [2e−15], respectively) and multidrug transporters (with E values of 45% [3e−16] and 53% [2e−15], respectively) but not carbohydrate transporters. In fact, of the 57 probable GntR-like proteins in the S. coelicolor genome, five are adjacent to predicted ABC-type transporters, Agl3R, WhiH, SCO0823, SCO6246, and SCO6294.
agl3R is apparently independent of the whiG regulatory pathway for morphogenesis.
Several models have been proposed for the participation of the whi genes in the development of aerial hyphae (13, 33). The sigma factor encoded by the whiG gene is pivotal in the initiation of a series of steps that involve whiH, whose expression is whiG dependent. Expression of a late-stage sigma factor encoded by sigF is dependent on whiH. Deletion of agl3R has no effect on the transcription of whiG, whiB, whiH, whiE, or sigF compared to wild-type strain M145 (Fig. 6), and transcription of agl3R was not dependent on whiG, whiH, or whiE, suggesting that it does not participate in the whiG-dependent regulatory cascade proposed previously.
What is the connection between the expression of the agl3EFG transporter, carbon utilization, and the regulation of development?
The regulation of carbon utilization is central to the most interesting and important aspects of the biology of Streptomyces. One aspect of the bld mutants, observed early in their characterization, is that growth on poor carbon sources is sufficient to partially restore the morphological and antibiotic defects of these mutants (5). With the exception of bldB (5, 6, 27, 30), when grown on minimal medium agar plates containing glucose as a carbon source, bld mutants fail to erect aerial hyphae and are also defective in antibiotic production. When grown on minimal medium containing mannitol, aerial hypha and spore production is partially restored. While growth on mannitol partially rescues the morphogenic defect of bldA mutants, the cells remain deficient in antibiotic production (5). In contrast, growth on mannitol rescues both sporulation and antibiotic production in bldH mutants (5). The most severe of the bld mutants, bldB, remains both morphologically and physiologically defective, failing to sporulate or produce antibiotics, regardless of carbon source.
While these observations clearly suggest a connection between the initiation of morphogenesis and antibiotic production and the regulation of carbon utilization, the fact that the phenotype of some morphological mutants is carbon source dependent has been difficult to interpret. Are there several independent pathways for the initiation of morphogenesis, some of which are repressed by glucose? Does relief of carbon catabolite repression in bld mutants result in the expression of genes that affect morphogen transport? Does the carbon source play a more direct role in initiation by, for example, inducing morphogen synthesis or morphogen transport?
The mechanism of carbon catabolite repression in Streptomyces is fundamentally different from that in gram-negative bacteria and even different from that in low-G+C gram-positive bacteria. While Streptomyces species do produce cyclic AMP and cyclic AMP receptor protein, there is no clear link between these molecules and glucose-mediated carbon catabolite repression (11). In S. coelicolor, glucose is not transported by a phosphotransferase, and uptake depends on the activity of glucose kinase (1, 44). A number of genes have been implicated in the global regulation of carbon utilization in Streptomyces, but no clear picture of how they interact or interconnect is evident from the present studies. As in Saccharomyces cerevisiae, glucose kinase clearly plays a role in catabolite repression in Streptomyces independent of its kinase activity (26) and perhaps involving other regulators (15). While the phosphotransferase does not transport glucose, enzyme IIA-glucose plays a role in general sugar transport and inducer exclusion (21). A mutation in ccrA1 leads to relief of glucose repression of a number of genes in S. coelicolor (20), suggesting that repression rather than activation controls the expression of many genes involved in carbon utilization. Genes with similarity to ccpA have been identified, but no clear homologue exists, and efforts to detect serine 47-phosphorylated Hpr in Streptomyces have not been successful (29).
Interestingly, many studies that focused on the regulation of catabolite repression have identified genes that also play a role in morphogenesis and antibiotic production. For example, levels of cyclic AMP change during growth and development and reach a peak during the transition from vegetative growth to aerial mycelium production in S. coelicolor, and mutations in either adenylate cyclase or cyclic AMP receptor protein result in defects in spore germination and delays in sporulation and antibiotic production (10, 11).
Many ABC transporters have multiple functions, most notably bldK in S. coelicolor, which transports the drug bialaphos and a small-molecule morphogen. Drug transport is apparently irrelevant to the morphogenic defects of bldK mutants, and the fact that the same transporter serves to import both molecules may be coincidental. Like bldK, the agl3EFG transporter may serve two functions, carbohydrate transport and morphogen signaling. The two bursts of transcription of agl3E seen in the mutant correspond to the timing of two major decision points during development, one at the initiation of aerial hypha formation and the other late in development as spore maturation begins. The fact that expression of this transporter is induced by complex carbohydrates may provide a direct connection between carbon utilization, morphogenesis, and antibiotic production.
An alternative model for the effects that we observed is that morphogen transport by this permease might be an accident of its binding chemistry and that what we are detecting is a perturbation of the signaling cascade rather than a direct effect on development. Inappropriate and/or high-level expression of the transporter, as in an agl3R mutant, might lead to the transport of one or perhaps several signaling molecules, not normal substrates for this permease, literally giving the organism mixed signals as it enters stationary phase. This model would also provide an explanation for why the relief of carbon catabolite repression in bld mutants, and the resulting inappropriate expression of carbohydrate utilization genes, might lead to defects in morphogenesis. The pathways leading to morphogenesis and antibiotic production in these complex bacteria are clearly complicated, highly regulated, and interconnected. Any serious perturbation in the progression of check points that result in normal development would result in defects in one or both of these processes.
Acknowledgments
We are greatly indebted to Mark Buttner for his generosity in offering advice on every aspect of this work and to him and Keith Chater for critical review of the manuscript. We thank Michael Adams, Michael Mceachern, Larry Shimkets, and Tim Hoover for helpful discussions at various stages of the work and David Brown for help in preparation of the manuscript. We thank Ondrej Sprusansky for advice and help with several of the S1 mapping experiments and Karen Stirrett for insights into the analysis of these results as well as contributions to the writing of the manuscript.
B.H. was supported by a predoctoral training grant from the National Institute for General Medical Sciences, GM07103, to the Genetics Department of the University of Georgia, and J.W. was supported by a grant from Microbia, Inc.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Angell, S., C. G. Lewis, M. J. Buttner, and M. J. Bibb. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol. Gen. Genet. 244:135-143. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bertram, R., M. Schlicht, K. Mahr, H. Nothaft, M. H. Saier, Jr., and F. Titgemeyer. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmachary, P., M. G. Dashti, J. W. Olson, and T. R. Hoover. 2004. Helicobacter pylori FlgR is an enhancer-independent activator of σ54-RNA polymerase holoenzyme. J. Bacteriol. 186:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champness, W. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champness, W. C., and K. F. Chater. 1994. Regulation and integration of antibiotic production and morphological differentiation in Streptomyces spp., p. 61-93. In P. J. Piggot, C. P. Moran, and P. Youngman (ed.), Regulation of bacterial development. American Society for Microbiology, Washington, D.C.
- 7.Chater, K. 1975. Construction and phenotypes of double sporulation deficient mutants in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 87:312-325. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F. 1972. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 72:9-28. [DOI] [PubMed] [Google Scholar]
- 9.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 10.Derouaux, A., D. Dehareng, E. Lecocq, S. Halici, H. Nothaft, F. Giannotta, G. Moutzourelis, J. Dusart, B. Devreese, F. Titgemeyer, J. Van Beeumen, and S. Rigali. 2004. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem. Biophys. Res. Commun. 325:983-990. [DOI] [PubMed] [Google Scholar]
- 11.Derouaux, A., S. Halici, H. Nothaft, T. Neutelings, G. Moutzourelis, J. Dusart, F. Titgemeyer, and S. Rigali. 2004. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. J. Bacteriol. 186:1893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiRusso, C., A. Metzger, and T. Heimert. 1993. Regulation of transcription of genes required for fatty acid transport and unsaturated fatty acid biosynthesis in Escherichia coli by FadR. Mol. Microbiol. 7:311-322. [DOI] [PubMed] [Google Scholar]
- 13.Flardh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 14.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, S., A. Carmona, L. Escalante, I. Imriskova, R. Lopez, R. Rodriguez-Sanoja, B. Ruiz, L. Servin-Gonzalez, S. Sanchez, and E. Langley. 2005. Pleiotropic effect of the SCO2127 gene on the glucose uptake, glucose kinase activity and carbon catabolite repression in Streptomyces peucetius var. caesius. Microbiology 151:1717-1723. [DOI] [PubMed] [Google Scholar]
- 16.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual, 1st ed. The John Innes Foundation, Norwich, United Kingdom.
- 18.Hurtubise, Y., F. Sareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 19.Ingram, C., M. Brawner, P. Youngman, and J. Westpheling. 1989. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 171:6617-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingram, C., I. Delic, and J. Westpheling. 1995. ccrA1: a mutation in Streptomyces coelicolor that affects the control of catabolite repression. J. Bacteriol. 177:3579-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamionka, A., S. Parche, H. Nothaft, J. Siepelmeyer, K. Jahreis, and F. Titgemeyer. 2002. The phosphotransferase system of Streptomyces coelicolor. Eur. J. Biochem. 269:2143-2150. [DOI] [PubMed] [Google Scholar]
- 22.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. Chater, and M. Buttner. 1996. Developmental regulation of the transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 24.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kormanec, J. 2001. Analyzing the developmental expression of sigma factors with S1-nuclease mapping. Methods Mol. Biol. 160:481-494. [DOI] [PubMed] [Google Scholar]
- 26.Kwakman, J. H., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 28.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 29.Parche, S., R. Schmid, and F. Titgemeyer. 1999. The phosphotransferase system (PTS) of Streptomyces coelicolor: identification and biochemical analysis of a histidine phosphocarrier protein HPr encoded by the gene ptsH. Eur. J. Biochem. 265:308-317. [DOI] [PubMed] [Google Scholar]
- 30.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 31.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 32.Ryding, N. J., M. J. Bibb, V. Molle, K. C. Findlay, K. F. Chater, and M. J. Buttner. 1999. New sporulation loci in Streptomyces coelicolor A3(2). J. Bacteriol. 181:5419-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flardh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29:343-357. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Saurin, W., W. Koster, and E. Dassa. 1985. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12:993-1004. [DOI] [PubMed] [Google Scholar]
- 36.Schlosser, A. 2000. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 184:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Schlosser, A., J. Jantos, K. Hackmann, and H. Schrempf. 1999. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl. Environ. Microbiol. 65:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlosser, A., A. Weber, and H. Schrempf. 2001. Synthesis of the Streptomyces lividans maltodextrin ABC transporter depends on the presence of the regulator MalR. FEMS Microbiol. Lett. 196:77-83. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, L., and R. Tampe. 2002. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12:754-760. [DOI] [PubMed] [Google Scholar]
- 40.Seo, J. W., Y. Ohnishi, A. Hirata, and S. Horinouchi. 2002. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol. 184:91-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprusansky, O., L. Zhou, S. Jordan, J. White, and J. Westpheling. 2003. Identification of three new genes involved in morphogenesis and antibiotic production in Streptomyces coelicolor. J. Bacteriol. 185:6147-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutcliffe, I., L. Tao, J. Ferretti, and R. Russell. 1993. MsmE, a lipoprotein involved in sugar transport in Streptococcus mutans. J. Bacteriol. 175:1853-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Veen, H. W., A. Margolles, M. Muller, C. F. Higgins, and W. N. Konings. 2000. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J. 19:2503-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Wezel, G. P., K. Mahr, M. Konig, B. A. Traag, E. F. Pimentel-Schmitt, A. Willimek, and F. Titgemeyer. 2005. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2). Mol. Microbiol. 55:624-636. [DOI] [PubMed] [Google Scholar]
- 45.van Wezel, G. P., J. White, M. J. Bibb, and P. W. Postma. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol. Gen. Genet. 254:604-608. [DOI] [PubMed] [Google Scholar]
- 46.Willey, J. W., R. Santamaria, J. Guijarro, M. Geislich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by Streptomyces coelicolor. Cell 65:641-650. [DOI] [PubMed] [Google Scholar]
- 47.Yu, T. W., and D. A. Hopwood. 1995. Ectopic expression of the Streptomyces coelicolor whiE genes for polyketide spore pigment synthesis and their interaction with the act genes for actinorhodin biosynthesis. Microbiology 141:2779-2791. [DOI] [PubMed] [Google Scholar]