Abstract
Bacillus subtilis has developed an intricate signal transduction cascade to respond to the imposition of a variety of stresses on the cell. Reversible protein phosphorylation and the formation of alternative protein-protein complexes modulate the activity of σB, the RNA polymerase sigma factor subunit responsible for the transcription of the general stress response genes. Some of the regulators of σB, such as RsbR and RsbS, are known to associate in a 25S complex, called the stressosome, that can bind RsbT until RsbT phosphorylates target residues in RsbR and RsbS. To date, the RsbR-RsbS complex appears to be the most upstream component of the σB regulatory pathway. This large structure is thought to play an important role in sensing and/or integrating signals from different physical stresses. The roles of the paralogues of RsbR that are found in B. subtilis remain unclear. We describe here how the RsbR paralogues copurify with RsbR from B. subtilis cell lysates, and we demonstrate in vitro that the paralogues form large complexes either with RsbS or with a prepurified RsbR-RsbS binary complex. We conclude from these biochemical studies that stressosomes in B. subtilis cells contain minimally RsbS and all of the RsbT-phosphorylatable RsbR paralogues.
Changes to the growth conditions of Bacillus subtilis, such as fluctuations in the temperature, pH, or ionic strength, can trigger the expression of two distinct sets of genes. One set constitutes a response specific to the imposed stress; the second set corresponds to a more general response to stress (11, 19). The expression of the general stress regulon, comprising more than 150 genes, is under the control of σB. This sigma factor provides multiple stress resistances to the cell (18, 20, 22). The activity of σB is regulated by at least a dozen proteins that participate in protein-protein interactions and/or reversible serine/threonine phosphorylation. In unstressed cells σB is prevented from interacting with core RNA polymerase (2-4, 9, 10, 23) by RsbW, an anti-sigma factor. RsbW also acts as a kinase toward the anti anti-sigma factor, RsbV, to produce phosphorylated RsbV (RsbV-P). Under stressful conditions the σB-RsbW complex is disrupted by unphosphorylated RsbV, inducing the release of σB to bind to RNA polymerase and to direct transcription of the σB regulon.
Two serine/threonine protein phosphatases, RsbP and RsbU, catalyze the dephosphorylation of RsbV-P, and they define two pathways by which stress signals converge on RsbV-P (21, 24). Energy stress signals lead to the activation of RsbP (5, 21, 26), whereas environmental stress signals activate RsbU. The phosphatase activity of RsbU is stimulated greatly by interacting with the kinase RsbT (8, 12, 25). Before stress, RsbT is believed to be trapped by RsbR and RsbS and is unavailable for interacting with RsbU in a high-molecular-weight complex of 25 S called the stressosome (6, 17), recently confirmed to be distinct from ribosomes (15). At the onset of stress, RsbT phosphorylates RsbS and consequently is liberated from the RsbR-RsbS complex. RsbT then interacts with its alternative binding partner, RsbU (6), stimulating its phosphatase activity toward RsbV-P and hence activating σB. In the prestressed cell the kinase activity of RsbT toward RsbS is counterbalanced by the phosphatase activity of RsbX toward RsbS-P (13), and overall RsbS is maintained in the unphosphorylated state through the sequestration of RsbT by stressosomes.
In addition to trapping RsbT prior to physical stress, the stressosome complex may have some other, but not necessarily mutually exclusive, roles. For instance, the complex could act as a receptor of the environmental stress signal itself. The observation of the globin fold of the N-terminal domain of RsbR (17) is evocative of the role that globins play in sensing gaseous diatoms. Alternatively, the complex could act as an attenuator or amplifier of the stress signal by controlling the number of RsbT molecules that are released from stressosomes as a function of the intensity of the imposed stress. The presence of three RsbT-phosphorylatable paralogues of RsbR (1) in the B. subtilis genome indicates that cellular stressosomes may be more complicated than simply being made of RsbR and RsbS. We show here that the RsbR paralogues YkoB, YqhA, and YojH also have the ability to form high-molecular-weight complexes with RsbS in vitro. The gross structure of the YkoB-RsbS complex is indistinguishable by electron microscopy from that of the RsbR-RsbS complex, suggesting a role for the STAS domains in RsbR paralogues in stressosome formation. Finally, we present evidence that the paralogs of RsbR are found in a complex with RsbR and RsbS both in vitro and in vivo.
MATERIALS AND METHODS
Cloning and protein purification.
The overexpression and purification of wild-type RsbR and RsbS has already been described (6). Wild-type ykoB, yqhA, and yojH were amplified by PCR and cloned into the pET11a (Novagen) expression vector. After overexpression of these proteins in Escherichia coli (BL21) cells were disrupted by sonication into 20 mM Tris.HCl (pH 8), 1 mM dithiothreitol, 1 mM EDTA, and 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride] and centrifuged at 29,000 × g for 45 min at 4°C. Supernatants were loaded on a Q-Sepharose column (GE Healthcare) and eluted with a linear gradient of the same buffer supplemented with 1 M NaCl. Fractions containing the proteins of interest were pooled and concentrated by ultracentrifugation and further purified with a Sephadex 200 gel filtration column (GE Healthcare). To prepare the binary YkoB-RsbS, YqhA-RsbS, and YojH-RsbS complexes, the purified proteins were mixed with a molar excess of RsbS and incubated at 4°C overnight before loading them onto a Sephadex 200 gel filtration column. The minimum mass expected to be excluded in the void volume of this column is ∼600 kDa.
RsbT kinase activity.
The RsbT-dependent phosphorylation of RsbS, RsbR, and YkoB was measured as described previously (6).
Purification of the RsbR-RsbS complex from B. subtilis cell extracts.
To purify the RsbR-RsbS complex for the subsequent identification of associated proteins, B. subtilis 168 was grown in liquid LB medium to stationary phase. Cells were harvested by centrifugation, and a cell pellet corresponding to 1 liter of cell culture was resuspended in 20 ml of phosphate-buffered saline (PBS; pH 7.4). Cells were disrupted by sonication and clarified by centrifugation. The supernatant was loaded onto an anti-N-RsbR antibody affinity column (6) and washed successively with 10 ml of PBS (20 column volumes), 2 ml of PBS supplemented with 250 mM NaCl, and then 2 ml of PBS. The column resin was resuspended by gentle pipetting in the presence of 200 μg of N-RsbR and, after 30 min of incubation at room temperature, the bound proteins were eluted with 8 ml of PBS. The eluate was concentrated by ultrafiltration to 0.5 ml before being loaded onto an analytical Superdex S200 HR 16/30 gel filtration column (GE Healthcare) to separate the complex and any associated proteins from the ∼30-kDa dimeric N-terminal domain of RsbR. The fractions that eluted around the void volume of the column were collected and precipitated with 20% trichloroacetic acid overnight at 4°C. The precipitated proteins were collected by centrifugation, and the pellet was washed with acetone and collected again by centrifugation. The acetone-free pellet was resuspended in 20 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by SDS-12% PAGE. Bands on the gel were visualized by silver staining and were excised manually from the gel for subsequent identification by peptide mass fingerprinting. Trypsin was used to generate peptide fragments, accurate masses for which were obtained using an ABI Voyager DE STR matrix-assisted laser desorption ionization-time of flight mass spectrometer. Peptide sequences were identified from the Mass Spectrometry Protein Sequence Database, which at the time of analysis contained 1.4 million protein sequences, using the MASCOT search engine (Matrix Science) and a probability cutoff of 0.05.
Electron microscopy.
For the analysis of the YkoB-RsbS complex, negative-stain grids were prepared as previously described (6) using a stain of 1% phosphotungstic acid (pH 6.9). Negative-stain electron micrographs of YkoB-RsbS were taken on a Philips CM200FEG operating at 200 kV at a magnification of 66,000 and scanned at a resolution of 1.3 Å per pixel.
RESULTS
RsbR and its paralogues.
The B. subtilis genome encodes four clear paralogues of RsbR: YkoB, YojH, YqhA, and YtvA and also a split paralogue composed of a pair of adjacent gene products, YetI-YezB. All of the paralogues, including the YetI-YezB couple, show significant sequence homology only in their C-terminal domains (Fig. 1) and clearly belong to the STAS domain family of anti-sigma factor antagonists such as RsbV and SpoIIAA. Among the RsbR paralogues only YkoB, YojH, and YqhA are phosphorylatable by RsbT (1), presumably on the conserved threonines present in their C-terminal, STAS domains. The equivalent domain in YtvA does not contain the conserved threonines; instead, it codes for glutamate at these positions, which may mimic phosphoserine/threonine, and therefore YtvA is probably not regulated by phosphorylation by RsbT. Indeed, the N-terminal domain of YtvA belongs to the PAS/LOV domain family and is light sensitive (16). Furthermore, individually, YetI and YezB are not substrates for RsbT, and a deletion in yetI-yezB has no effect on σB activity in response to salt, ethanol, or energy stress (1). In the present study, we did not investigate the properties of YtvA and YetI-YezB since they have not yet been demonstrated to be substrates for RsbT, and thus their roles in environmental stress signaling remains obscure.
FIG. 1.
Sequence alignment of RsbR and paralogues from B. subtilis. The amino acid sequences were aligned by using default settings in CLUSTAL W. The positions of the phosphorylatable residues (and their nonphosphorylatable equivalents in YtvA and RsbS) are underlined. The pairwise sequence identities and positives for the N-terminal domains of the paralogues in relation to RsbR are ∼24 and 44%, respectively. Indeed, with default settings in BLAST it was not possible to identify YtvA and YqhA with the N-terminal domain of RsbR as the query. This observation is not surprising as N-YtvA adopts the LOV domain (16), a variation on the ubiquitous PAS fold and is structurally unrelated to the globin fold of N-RsbR (17). It remains to be seen what folds are taken by the N-terminal domains of the phosphorylatable RsbR paralogues; they may also be globin-like. In contrast, the sequence identities and positives for the C-terminal, STAS domains are on the whole, much higher, ranging, respectively, from 29 and 57% for RsbS and C-YtvA to 40 to 52% and 60 to 72% for the other paralogues.
The N-terminal domains of RsbR, YkoB, YojH, and YqhA, all of which are comparable in size (∼140 amino acids), contain only four residues that are completely conserved (Fig. 1). Two (Lys47 and Glu108) of the four invariant amino acids are surface exposed on opposite faces of the structure of N-RsbR (17) approximately 30 Å apart. Gln140 is not visible in the structure but could not approach within 30 Å of the other two exposed amino acids in any event for steric reasons. The fourth conserved residue, Trp22, is completely buried. Given this lack of identity, even if the overall fold—a globin (17)—of these domains is maintained, the surface physicochemical properties will be quite different. In contrast, 38 residues in the C-terminal domains (∼120 amino acids) of these four proteins are conserved (Fig. 1). We wondered whether the differences among the N-terminal domains of the RsbR paralogues could affect the formation of high-molecular-weight complexes of the paralogues with RsbS, the binding of RsbT to these complexes, or the kinetics of phosphorylation of these complexes by RsbT.
YkoB, YqhA, and YojH each form a high-molecular-weight complex with RsbS.
We have previously demonstrated (6) that RsbR associates with RsbS to form high-molecular-weight complexes both in vitro and in vivo. We tested whether the three RsbR paralogues, YkoB, YqhA, and YojH, share this property. The purification of each of the three RsbR paralogues was performed in the same way, making use of a size exclusion column for the last purification step. A large proportion of each of the paralogues eluted in the void volume of the gel filtration column, suggesting an apparent mass far in excess of 200 kDa. About 40% of the total RsbR eluted from the gel filtration column as a dimer of ∼60 kDa, with the rest eluting in the void volume. Under the same overexpression conditions YkoB, YojH, and YqhA mostly remain as high-molecular-weight species. The tendency of YkoB, YojH, and YqhA to self-assemble is therefore higher than for RsbR.
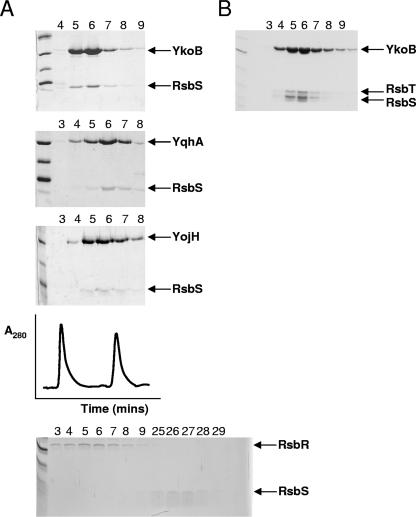
For each of the proteins YkoB, YojH, and YqhA the fractions containing the high-molecular-weight forms were mixed at 4°C overnight with an excess of RsbS, a dimer of ∼26 kDa. Overnight incubation of RsbS alone at 4°C does not alter its elution profile, it still elutes as though it were a dimer. The following day the mixture was applied to a Superdex 200 gel filtration column, and the fractions corresponding to the void volume of the column were analyzed by SDS-PAGE. In each case RsbS was present in the void volume of the column, indicating that each of the three RsbR paralogues was able to incorporate RsbS into a high-molecular-weight complex (Fig. 2A). A significant difference in behavior between RsbR on the one hand and the paralogues on the other is that YkoB, YqhA, and YojH can integrate with RsbS even when they have already formed high-molecular-weight species, whereas RsbR can assimilate RsbS only when RsbR is dimeric; once RsbR has aggregated, it cannot associate with RsbS (Fig. 2A).
FIG. 2.
The RsbR paralogues form high-molecular-weight complexes with RsbS and RsbT. (A) YkoB, YqhA, and YojH were incubated with a molar excess of RsbS at 4°C overnight and subsequently subjected to S200 size exclusion chromatography before analyzing the fractions across the void volume of the gel filtration column by SDS-PAGE. Bands corresponding to the paralogues and RsbS are highlighted. Also shown in this panel is a representative gel filtration trace of the reconstitution of the YkoB-RsbS complex; the two peaks in the chromatogram correspond to fractions 6 ± 2, where the binary complexes eluted, and fractions 27 ± 2, where excess RsbS eluted. Shown at the bottom is the result of mixing oligomeric RsbR with RsbS—the two proteins do not form a complex and elute separately. The absence of a defined peak for oligomeric RsbR may illustrate the nonspecific nature of its aggregation in the absence of RsbS. (B) The prepurified YkoB-RsbS complex was mixed with RsbT, and the mixture was subsequently loaded onto the same S200 size exclusion column as in panel A. The SDS-PAGE analysis of the fractions collected across the void volume reveal that the YkoB-RsbS complex is capable of recruiting RsbT under the same experimental conditions as for RsbT and the RsbR-RsbS complex. As an example, we only show the data for YkoB, but the other paralogues behaved identically. In both panels, the lane numbering refers to fraction number across the void volume of the column; these fraction numbers correspond to the numbering shown in Fig. 4.
In previous experiments, we have estimated that the stoichiometry of the RsbR-RsbS complex is 3:1 (6). Analysis by SDS-PAGE of the complexes formed between RsbS and each of the RsbR paralogues suggested that the same stoichiometry held also for these complexes (Fig. 2A). The ratios of the intensities of the Coomassie-stained bands between RsbS and the RsbR paralogues are ∼7-fold different. Taking into account the ∼2.4-fold difference in mass between RsbS and the RsbR paralogues, the stoichiometry, in molar terms, can be estimated at ∼3:1. It seems likely, therefore, that the mode of interaction between the proteins is maintained. Given that only the C-terminal (STAS) domain of RsbR and its paralogues display significant sequence identity (on average, ∼45%), we presume that this domain of RsbR and its paralogues mediate interactions with RsbS in the complex assemblies.
RsbT binding to and kinase activity in YkoB-RsbS and RsbR-RsbS complexes.
We have previously demonstrated that the RsbR-RsbS complex binds RsbT (6, 7) and that the RsbR-RsbS-RsbT complex cannot activate RsbU (6). We thought it possible that, in the prestressed cell, RsbT would be bound by the complexes that RsbS forms with the RsbR paralogues. RsbT would then be released during stress to bind to and activate RsbU and thus induce the σB-dependent general stress response. We therefore tested the ability of the RsbR paralogue-RsbS complexes to bind to RsbT. After incubation of the purified complexes with RsbT for 20 min at room temperature, the mixtures were loaded onto an analytical gel filtration column, and fractions across the void volume of the column were analyzed by SDS-PAGE. A 20-min incubation at room temperature does not affect the gel filtration elution profile of RsbT, it still behaves as though a monomer of ∼14 kDa. The presence of a band corresponding to RsbT in the same fractions as those containing the YkoB-RsbS (Fig. 2B) and YqhA-RsbS and YojH-RsbS (data not shown) complexes in the void volume of the column reveals that the YkoB-RsbS and YojH-RsbS complexes are capable, as is RsbR-RsbS, of binding RsbT.
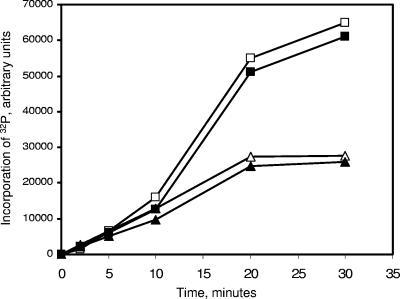
We have already demonstrated that N-RsbR is involved in the binding of RsbT (17), and we wondered whether the different N-terminal domains might affect RsbT kinase activity toward the paralogue itself and/or RsbS. To address this question, we compared the kinetics of phosphorylation of YkoB-RsbS, as an example of the RsbR paralogues, and RsbR-RsbS. An equal amount of RsbT was added to each complex in the presence of radiolabeled ATP, and the phosphoproteins were separated by SDS-PAGE and analyzed by autoradiography. As shown in Fig. 3, the kinetics of phosphorylation of RsbR and RsbS in the RsbR-RsbS complex were almost identical to that of YkoB and RsbS in the YkoB-RsbS complex. Therefore, at least under laboratory conditions, RsbT activity is not affected by the N-terminal domains of RsbR and YkoB.
FIG. 3.
Kinetics of the phosphorylation of RsbS, RsbR, and YkoB by RsbT. The phosphorylation of the substrates of RsbT was monitored by using radiolabeled [γ-32P]ATP. At various time intervals, indicated on the x axis, samples were taken, and the reaction was stopped by adding boiling 3× SDS loading buffer before SDS-PAGE. Proteins with incorporated radioactive phosphates were revealed by autoradiography of the SDS-PAGE gel. The total intensities of the bands corresponding to RsbS-P, RsbR-P, and YkoB-P were estimated by using Scion image after digitization of the autoradiograph and plotted on the y axis in arbitrary units. The kinetics of the incorporation of radioactivity in RsbS-P (▵) and RsbR-P (□) in the RsbR-RsbS complex closely resembles that found in RsbS-P (▴) and YkoB-P (▪), respectively, in the YkoB-RsbS complex. The higher total radioactivity counts in YkoB and RsbR than RsbS may reflect the stoichiometry of these proteins in their complexes.
In vivo purification of the RsbR-RsbS complex and identification of interacting components.
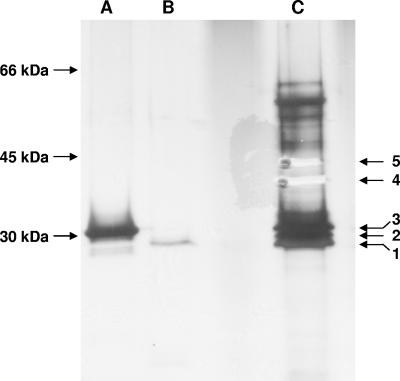
Our in vitro studies have revealed that each of the phosphorylatable paralogues of RsbR can form a high-molecular-weight complex when mixed with RsbS (Fig. 2A). However, it has not been established whether multiple, discrete complexes exist in the cell, each containing just one RsbR paralogue in complex with RsbS, or whether a single complex containing all of the RsbR paralogues with RsbS predominates. In order to discriminate between these two possibilities, we used immunoaffinity chromatography to purify RsbR and RsbR-containing complexes from B. subtilis 168 cell lysates with an anti-RsbR antibody column. The anti-RsbR antibodies were raised against recombinant N-RsbR as the immunogen and do not cross-react with YkoB, the paralogue with the greatest similarity in the N-terminal domain (24% identity) to RsbR. After disruption by sonication and clarification by centrifugation, B. subtilis cell lysate was loaded onto the anti-RsbR immunoaffinity column, and the retained proteins were eluted with an excess of N-RsbR, concentrated, and loaded onto a Superdex 200 gel filtration column. The fractions corresponding to the void volume of the column, in which the RsbR-RsbS complex prepared in vitro elutes, were precipitated with trichloroacetic acid and analyzed by SDS-PAGE and silver staining. We have previously shown that RsbS copurifies from B. subtilis cell lysate with RsbR using the same experimental procedures (6) and hence in the present study we concentrated on the identification of the RsbR paralogues. In the experiment presented in Fig. 4, five bands were excised from the gel for subsequent identification of interacting partners. In repeat experiments, a band corresponding to RsbR was always present, as well as other bands in the gel that ran in the immediate vicinity of RsbR. Faint bands corresponding to proteins of more than 40 kDa were also observed, but less consistently. The identities of the proteins with masses similar to RsbR (band 1) were obtained by peptide mass fingerprinting, which revealed that the paralogues YojH (band 2) and YkoB (band 3) copurified with RsbR. For RsbR, the peptide mass fingerprinting with the MASCOT search engine successfully identified 6 peptides with sequence coverage of 31% and an E-value of 0.0065. For YkoB, eight peptides were matched, covering 44% of the sequence, and an E-value of 0.0014, whereas for YojH 9 peptides were found corresponding to 40% of the sequence with an E-value of 0.0055. The identification of YkoB and YojH after immunopurification of RsbR and subsequent size exclusion chromatography suggests that, in the cell, RsbR, YkoB, and YojH all associate together with RsbS in a single complex. It was not possible using these experimental procedures to detect YqhA, YtvA, or the split RsbR paralogues, YetI-YezB. Given that YetI and YezB have masses quite different from that of RsbR, our procedures are unlikely to have identified them. We may have failed to identify YqhA, which can form a complex with RsbS (Fig. 2A), because of the limitations of these experimental procedures. Alternatively, YqhA may not be particularly abundant in the cell.
FIG. 4.
Copurification of RsbR paralogues with RsbS in vivo. The RsbR-RsbS complex and other, bound proteins were purified from B. subtilis cell lysate by immunoaffinity chromatography using an α-N-RsbR antibody column. Retained proteins were subsequently loaded onto a size exclusion column. Proteins from fractions around the void volume of the column were precipitated with 20% trichloroacetic acid, separated by SDS-PAGE, and visualized by silver staining (lane C). Recombinant YkoB (A) and RsbR (B) were used as markers, in addition to a commercial ladder (D). Bands from the gel were excised and identified by peptide mass fingerprinting of a tryptic digest of protein recovered from each gel slice. The B. subtilis proteins identified were RsbR (band 1), YojH (band 2), YkoB (band 3), Hag (band 4), and PdhA (band 5), the bands for which have already been excised in this figure. Hag migrates aberrantly in SDS-PAGE, appearing as a band of Mr ∼40 kDa, not its expected 32 kDa. The pattern of protein bands with an Mr of >45 kDa varied between repeat experiments and were not analyzed further.
Two other proteins were identified by the peptide mass fingerprinting procedure, Hag (band 4), a flagellin protein, and PdhA (band 5), the E1α subunit of pyruvate dehydrogenase. Both of these proteins are involved in the formation of high-molecular-mass complexes; Hag polymers form the rigid flagellum, and PdhA is a component of the pyruvate dehydrogenase complex with an expected mass in excess of 106 Da. It is therefore highly likely that these two abundant proteins are contaminants that the size exclusion column could not separate from the immunoaffinity-purified complexes containing RsbR.
The multiprotein complexes are dynamic.
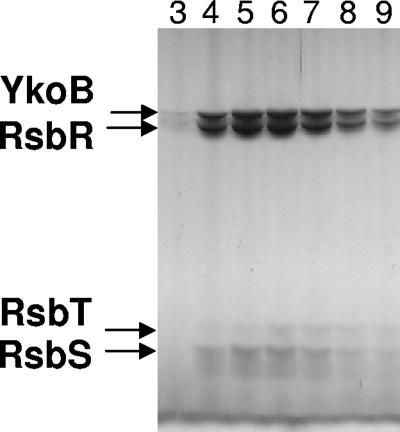
In the present study, we have demonstrated that the RsbR paralogues YkoB, YojH, and YqhA can each form a high-molecular-weight complex with RsbS in vitro. Furthermore, peptide mass fingerprinting of the complex purified from B. subtilis cell lysate revealed the presence within the same complex of three of the four RsbT-phosphorylatable paralogues of RsbR. Although rsbR and rsbS are adjacent on the genome within an operon, the genes encoding the paralogues are scattered across the chromosome and therefore are not cotranscribed with rsbR and rsbS. Furthermore, whereas YqhA is probably σB dependent (20), nothing is known of the regulation of expression of the other paralogues. The entire rsb operon is σA dependent, and the rsbV-rsbW-sigB-rsbX genes are also under the control of an internal σB promoter, which is activated during stress (9, 19). Can the formation of the multiple-paralogue complex be reproduced in vitro? To address this question, a YkoB-RsbS-RsbT complex was prepared in vitro, purified, mixed with the dimeric form of RsbR at room temperature, and incubated overnight. The mixture was then applied to an anti-RsbR immunoaffinity column and washed with 0.5 M NaCl, and the bound proteins were eluted with a glycine buffer at pH 3 into fraction tubes containing sufficient Tris buffer to neutralize the pH. The retained proteins were then subjected to size exclusion chromatography, and the fractions corresponding to the void volume of the gel filtration column were analyzed by SDS-PAGE. All four proteins were found in the fractions corresponding to the highest-molecular-weight species, with a ratio between YkoB and RsbR of ∼1:1 (Fig. 5). The stringent washing procedure probably excludes the possibility that RsbR molecules have simply bound peripherally to the YkoB-RsbS-RsbT complex. RsbR can integrate into the YkoB-RsbS-RsbT complex, which means that either the quaternary YkoB-RsbR-RsbS-RsbT complex has increased in size, in comparison to YkoBR-RsbS-RsbT, to accommodate RsbR or that RsbR has displaced molar equivalents of YkoB molecules to maintain the same [RsbR+YkoB]:RsbS ratio as that of YkoB-RsbS.
FIG. 5.
The ternary YkoB-RsbS-RsbT complex is dynamic. Prepurified YkoB-RsbS-RsbT complex was mixed with RsbR at room temperature overnight. The mixture was then applied to the α-N-RsbR immunoaffinity column, and the retained proteins were loaded onto an S200 gel filtration column. The fractions around the void volume of the column were analyzed by SDS-PAGE, and proteins were visualized by silver staining. The coelution of RsbR, YkoB, RsbS, and RsbT by size exclusion chromatography indicates that RsbR has integrated into the YkoB-RsbS-RsbT complex, presumably by displacing a molar equivalent of YkoB.
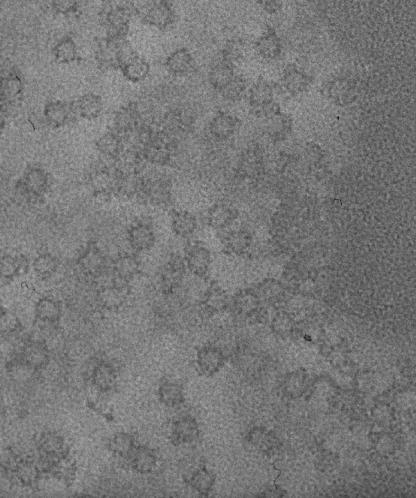
Structural analysis by electron microscopy.
Electron microscopy studies of negatively stained samples of the RsbR-RsbS complex revealed that these proteins form hollow spheres or flat discs of homogeneous size of 20 nm in diameter (6). Similar studies of the aggregates of RsbR alone were barely distinguishable from the RsbR-RsbS complexes (6). The YkoB-RsbS and RsbR-RsbS complexes both elute in the void volume of size exclusion chromatography. Negative stain electron micrographs of the YkoB-RsbS complex show the same structure we have described previously for RsbR-RsbS (Fig. 6). With the electron micrographs from this and our previous study (6), it is not possible to discriminate between the possibilities regarding the gross structure of these complexes. The particles are ring-shaped in projection, which is compatible with cyclic, dihedral, or icosahedral point group symmetries. However, most of the particles appear to lie in the same orientation in these micrographs, perhaps indicating that the complexes are not flat disks, but spherical. Moreover, the predicted mass of RsbR-RsbS from its sedimentation coefficient of 26 S (6) is ∼1.5 MDa, which is consistent with a hollow icosahedron containing a total of 60 molecules of RsbR and RsbS. The dimensions of the particles are also ∼20 nm in diameter, and at this level of detail the YkoB-RsbS and RsbR-RsbS complexes appear identical, suggesting that the complexes form similarly.
FIG. 6.
Negative-stain electron microscopy of the YkoB-RsbS complex. A region showing well-defined individual particles selected from negatively stained micrographs of the YkoB-RsbS complex.
DISCUSSION
At the onset of physical stress RsbT, a positive regulator of σB activity, is thought to switch from one partner, the stressosome complex, to its alternative partner, RsbU. In forming the RsbT-RsbU complex, the phosphatase activity of RsbU for its specific substrate, RsbV-P, is activated. Although this model of the sequestering and release of RsbT from stressosomes has received support from in vitro studies (6) using the RsbR-RsbS complex as a model, no definitive demonstration in vivo has been published. Furthermore, RsbR acts as a co-antagonist with RsbS; neither protein alone can sequester RsbT (6, 14). In the present study, we have analyzed some of the biochemical properties of the RsbR paralogues.
In a quadruple rsbR-yojH-ykoB-yqhA mutant, σB activity is constitutive (14), and this B. subtilis strain has a severe growth defect (14). This observation would indicate that the regulation of σB activity is dependent upon the formation of the 25S stressosome complex: without the complex σB activity is unregulated. The complex would appear to act as an antagonist of σB activity by trapping RsbT before stress. The in vivo and in vitro results presented in the present study confirm the results obtained previously by others (14). Among the three paralogues of RsbR, two of them, YkoB and YojH, have been identified by mass spectrometry in the in vivo purification of the native complex by immunoaffinity chromatography (Fig. 4). The identification of YkoB and YojH that copurify in a complex with wild-type RsbR is consistent with a previous study where YkoB was found to copurify with a His6-myc-tagged version of RsbR (14). The third paralogue, YqhA, which also forms a high-molecular-weight complex with RsbS, was not detected by our peptide mass fingerprinting procedures. This may reflect the limitations of this technique or may indicate that YqhA is present in the cell at a low concentration compared to the other RsbR paralogues. Nonetheless, our data confirm that in the cell, the stressosomes are not bivalent but are composed of RsbS and the paralogues of RsbR, YkoB, YojH and, probably, YqhA.
In triple-mutant backgrounds, in which any three genes of the four rsbR paralogues are deleted, the sole paralogue that remains can display co-antagonist function (14). This is because the remaining paralogue can still form a complex with RsbS and is effective in the prestress sequestration of RsbT. RsbR and YkoB are the most effective co-antagonists and YojH and YqhA are less potent (14), perhaps reflecting the relative levels of the paralogues in the cell. This is consistent with our preparations of the RsbR-containing complexes from B. subtilis cell lysates, where bands that we subsequently identified as RsbR and YkoB were always present on silver-stained SDS-PAGE gels. The band corresponding to YojH was not always visible and, by these procedures, YqhA was undetectable (Fig. 4). Much of our attention has focused on YkoB since this paralogue is as effective as RsbR in controlling σB activity, whereas YojH and YqhA are both much less effectual (14).
Although the RsbR paralogues are expressed from different loci, they are still capable of forming part of the stressosome complex (Fig. 2). Our data suggest that, at least in vitro, this might be achieved by the simple replacement of one paralogue by another (Fig. 5). The observation that a complex made of YkoB and RsbS alone can integrate dimeric RsbR implies that the stressosome is a rather dynamic and flexible structure. This view is also supported by our finding that RsbS can still integrate with the paralogues of RsbR even after the paralogues had already formed the particles that we have observed by electron microscopy (Fig. 6). Therefore, a degree of flexibility exists in this structure that allows the integration and, perhaps, replacement of RsbR paralogues even after complex formation.
The specific functions of each RsbR paralogue remain, however, to be determined. Almost all of the variability in the primary sequence between the paralogues is restricted to the N-terminal domain. Our recent structural and biochemical analysis of N-RsbR revealed two functions for this domain: (i) it is directly involved in the binding of RsbT to the stressosome and (ii) the globin fold adopted by the N-RsbR evokes a role in stress sensing (17). If all of the paralogues sense the same signal, perhaps they are responsive to different levels of stress. Other components acting upstream of the stressosome are likely to be discovered, and it is tempting to propose that the N-terminal domains of RsbR and of its paralogues would be entry points for the stress signal into the stress signaling pathway that controls σB activity.
Acknowledgments
We thank Helen Prescott for cloning yojH and yqhA, Joe Gray for proteomics support, and Rishi Matadeen and Steven Fuller for help with electron microscopy.
This study was funded by the BBSRC, the Wellcome Trust, and the University of Newcastle.
Footnotes
Published ahead of print on 8 September 2006.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor sigma(B) of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 3.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis sigmaB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of sigmaB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigmaB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. C., R. J. Lewis, J. R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signaling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. C., M. D. Yudkin, and O. Delumeau. 2004. Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J. Bacteriol. 186:6830-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delumeau, O., S. Dutta, M. Brigulla, G. Kuhnke, S. W. Hardwick, U. Völker, M. D. Yudkin, and R. J. Lewis. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279:40927-40937. [DOI] [PubMed] [Google Scholar]
- 9.Delumeau, O., Lewis, R. J., and Yudkin, M. D. 2002. Protein-protein interactions that regulate the energy stress activation of sigmaB in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecker, M., and U. Völker. 1998. Nonspecific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 12.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 13.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 186:6124-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, S., S. Zhang, R. L. Woodbury, and W. G. Haldenwang. 2004. Associations between Bacillus subtilis sigmaB regulators in cell extracts. Microbiology 150:4125-4136. [DOI] [PubMed] [Google Scholar]
- 16.Losi, A., E. Polverini, B. Quest, and W. Gärtner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray, J. W., O. Delumeau, and R. J. Lewis. 2005. Structure of a nonheme globin in environmental stress signaling. Proc. Natl. Acad. Sci. USA 102:17320-17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 20.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 21.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 22.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the sigmaB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Völker, U., A. Völker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-sigmaB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Völker, U., A. Völker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate sigmaB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, S., and W. G. Haldenwang. 2003. RelA is a component of the nutritional stress activation pathway of the Bacillus subtilis transcription factor sigmaB. J. Bacteriol. 185:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]