Abstract
The Na+-translocating F-ATPase of the thermoalkaliphilic bacterium Clostridium paradoxum harbors an oligomeric ring of c subunits that resists dissociation by sodium dodecyl sulfate. The c ring has been isolated and crystallized in two dimensions. From electron microscopy of these c-ring crystals, a projection map was calculated to 7 Å resolution. In the projection map, each c ring consists of two concentric, slightly staggered, packed rings, each composed of 11 densities representing the α-helices. On the basis of these results, it was determined that the F-ATPase from C. paradoxum contains an undecameric c ring.
The greatest challenge to alkaliphilic bacteria living at extreme pH values (i.e., >pH 9) is the maintenance of their intracellular pH near neutral in order to prevent cytoplasmic alkalinization. These bacteria maintain a ΔpH of 1.5 to 2.5 units (acid inside), which opposes the bioenergetics of ATP synthesis using a H+-coupled F1Fo-ATP synthase. Due to the inverted pH gradient, the total proton motive force (ΔμH+) may drop to values well below −100 mV, which is not sufficient for ATP synthesis by conventional mechanisms. Clostridium paradoxum is an anaerobic thermoalkaliphile that ferments glucose rapidly to acetate, ethanol, and CO2 and produces ATP by substrate phosphorylation (3, 10). C. paradoxum has recently been reported to contain a Na+-translocating F1Fo-ATPase, and several of the enzyme's properties suggest that its primary operation is that of an ATP-driven Na+ ion pump rather than that of an ATP synthase (7). The net result of the pumping is the formation of an electrochemical gradient of sodium ions (ΔμNa+), which is purportedly used to drive bioenergetic processes (e.g., transport and motility).
The Fo subcomplex of F1Fo-ATP synthases is involved in trafficking ions across the membrane. Either the electrochemical ion gradient across the membrane produces torque and drives ATP synthesis or ATP hydrolysis drives the active transport of the ions. An assembly of c subunits in the membrane forms an hourglass-shaped rotary cylinder, known as the c ring. As has been documented for the Na+-dependent ATP synthases of other bacteria (1, 9, 15), the ATP synthase of C. paradoxum contains an oligomeric c ring that is stable on sodium dodecyl sulfate (SDS)-polyacrylamide gels (7) and is therefore well-suited for structural analysis (12). Structural data on c-ring stoichiometries have been determined only for Na+-ATP synthases that operate under physiological conditions (e.g., during succinate or tartrate fermentation) in the ATP synthesis direction (6) and for a Na+-translocating V-ATPase that operates in the ATP hydrolysis direction (13). In this communication, we present the structural analysis of the c ring isolated from the ATP-hydrolyzing Na+-ATPase of C. paradoxum and show that the Fo rotor is assembled as an undecameric c ring.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
Clostridium paradoxum DSM 7308 was maintained as a spore stock preparation in sterile distilled water at 4°C. Spores were germinated by heat shocking at 75°C for 2 min, followed by rapid cooling on ice for 45 s. C. paradoxum was grown anaerobically at 56°C in YTG medium, which contained, per liter, 5.3 g of Na2CO3, 0.36 g of Na2HPO4 · 2H2O, 0.075 g of KCl, 5 g of yeast extract, 10 g of tryptone, 0.2 g of cysteine HCl, 0.2 g of Na2S · 9H2O, and 0.3% (wt/vol) glucose (7). The pH of the medium was adjusted to 10.5 at 25°C with 5 M NaOH. Bacterial growth was routinely monitored by following the optical density of the culture at 600 nm. For large-scale growth, 200 liters of C. paradoxum cells was grown anaerobically in a 300-liter fermentor. The fermentor was inoculated with 5 liters of a preculture, and after 6 h at 55°C, the bacteria were harvested at a final optical density at 600 nm of 0.4 by continuous centrifugation. The cells were frozen in liquid nitrogen and stored at −80°C.
Purification of the c ring from C. paradoxum.
The F1Fo-ATPase of C. paradoxum was purified as previously described (7). The ATPase was mixed with 1% N-lauroylsarcosine and incubated for 10 min at 65°C. After the mixture was cooled to room temperature, ammonium sulfate was added to yield 67.5% saturation. After 20 min of incubation at 25°C, precipitated protein was removed by centrifugation (23,000 × g for 20 min). The supernatant, which contained the c ring, was filtered through a 0.22-μm polyvinylidene fluoride filter unit (Millipore) and dialyzed against 10 mM Tris-HCl buffer (pH 8.0) overnight at 25°C. The c ring was then adsorbed to 1/50 volume of hydroxyapatite in an Eppendorf tube (Bio-Gel HT; Bio-Rad, Richmond, CA), and after two washes with 10 ml of dialysis buffer containing 0.15% (wt/vol) Zwittergent 3-12, the c ring was eluted with 1 M potassium phosphate buffer (pH 8.0) containing 0.15% Zwittergent 3-12. To further concentrate the c ring and remove excess salt, the sample was loaded on an Amicon Ultra-4 tube (50,000-Da molecular mass cutoff). The sample was then washed three times with dialysis buffer containing 0.15% Zwittergent 3-12. The final concentration of the purified c ring was 4.7 mg/ml.
2D crystallization of the c ring.
For crystallization in two dimensions (2D), Zwittergent 3-12 (0.4% [wt/vol])-solubilized c ring (1 mg/ml) was mixed with 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine (POPC; Avanti Polar Lipids Inc., Alabaster, AL) at lipid-to-protein ratios of 0.5, 1.0, and 1.5 (wt/wt). The mixture was dialyzed in 25-μl buttons (Hampton Research, Aliso Viejo, CA) against 250 ml of 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 3 mM NaN3 buffer for 24 h at 25°C and another 24 h at 37°C. The two-dimensional crystals were stored at 4°C until further analysis.
Electron microscopy and image processing.
Two-dimensional crystal samples were prepared in 4.5% (wt/vol) trehalose on molybdenum grids (Pacific Grid-Tech, San Diego, CA) (19) by the back-injection method (8). Grids were examined in a JEOL 3000 SFF helium-cooled electron microscope at 4 K at an accelerating voltage of 300 kV. Images were recorded by a spot-scanning procedure, with 24 spots by 30 spots per image on Kodak SO-163 film at a magnification of ×70,000 and with an electron dose of 20 to 30 electrons/Å2. The films were developed for 12 min in full-strength Kodak D-19 developer. Images selected by optical diffraction were digitized on a Zeiss SCAI scanner using a pixel size of 7 μm, corresponding to 1 Å on the specimen. Images were processed using the MRC (4) and CCP4 (2) program suites. Data were merged to a resolution of 7 Å.
Other methods.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously (16), and gels were stained with silver (14). ATPase activity was determined by the coupled spectrophotometric assay as described previously (7). The protein concentration was determined according to the bicinchoninic acid method (17), with bovine serum albumin as the standard. To dissociate the c ring, c-ring samples (2 to 5 μg of protein) were treated with 5 volumes of 12% (wt/vol) trichloroacetic acid solution and incubated on ice for 5 min. The pellet was collected by centrifugation (11,000 × g for 5 min at 4°C) and dissolved in 1% (wt/vol) SDS-containing loading buffer. Prior to being loaded on an SDS-gel, the sample was neutralized by the addition of 1-μl aliquots of 0.5 M NaOH until the color of the bromphenol blue-stained loading buffer switched from yellow to blue.
RESULTS AND DISCUSSION
Purification of the native c ring from C. paradoxum F1Fo-ATPase.
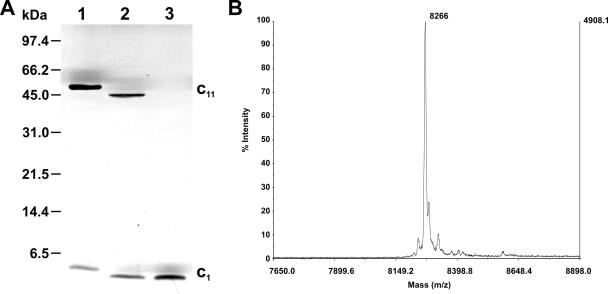
Previous investigations have shown that c-ring assemblies of Na+-translocating F1Fo-ATP synthases are of exceptional stability, even sustaining being boiled in SDS (1, 9, 15). This remarkable stability is shared by the c ring of the Na+-translocating F1Fo-ATPase from the thermoalkaliphilic bacterium C. paradoxum (7). A Na+-binding motif is also visible in the amino acid sequence of the c subunit of this bacterium (Fig. 1). As a consequence, the oligomeric c ring of C. paradoxum could be isolated by a modification of the protocol developed for the purification of the c ring from the Ilyobacter tartaricus ATP synthase (11). The results of an analysis of the purified c-ring sample of C. paradoxum by SDS-PAGE and mass spectrometry-matrix-assisted laser desorption/ionization (MS-MALDI) are shown in Fig. 2. After SDS-PAGE, the c ring migrated as a distinct band with an apparent molecular mass of 45 kDa (Fig. 2A, lane 2). Another protein band migrating with an apparent molecular mass of <6.5 kDa indicated a partial dissociation of the c ring into its monomeric units. After acidification of the sample with trichloroacetic acid, the c ring was completely dissociated into monomeric c subunits (Fig. 2A, lane 3). The mobility of the c ring from C. paradoxum was slightly faster than that of I. tartaricus, and a corresponding difference in the mobilities of the c monomers was also observed. The calculated molecular mass of subunit c from C. paradoxum was 8,257 Da (Fig. 2B), 538 Da smaller than that of the c ring from I. tartaricus (8,795 Da), which could account for the different mobilities of the c monomers and of the c rings if these contained the same number of subunits (see below).
FIG. 1.
Sequence alignment of the atpE gene products from selected species. The sequences form an α-helical hairpin. A cytoplasmic loop (shown in black) connects both helices. The H+- or Na+-binding glutamate on the C-terminal helix is shown in red. The residues involved in Na+ binding and the hydrogen bridge-providing tyrosine are shown in yellow. At the end of the alignment, the corresponding coupling ion (C) of the corresponding ATP synthase is indicated. The amino acids which are in the area of the 2D crystal contacts are shown in dark green (cytoplasmic side) and blue (periplasmic side). The numbering of the amino acids is according to that of the C. paradoxum sequence (7). The selected species are Clostridium paradoxum (CP), Ilyobacter tartaricus (IT), Acetobacterium woodii c2/3 (AW), Bacillus strain PS3 (PS3), Spinacia oleracea (SO), and Escherichia coli (EC.
FIG. 2.
(A) SDS-PAGE of the c ring isolated from the F-ATP synthase of Clostridium paradoxum cells. Lane 1, c ring (c11) from Ilyobacter tartaricus; lane 2, c ring isolated from C. paradoxum; lane 3, subunit c monomer (c1) from C. paradoxum, derived from the oligomer by acidification with trichloroacetic acid. The gel was stained with silver and a standard gel marker is indicated on the left side. (B) MS-MALDI analysis of isolated c-ring preparations. The identified mass (Da) is indicated. The samples were prepared as described in Materials and Methods.
Structural investigations of the C. paradoxum c ring.
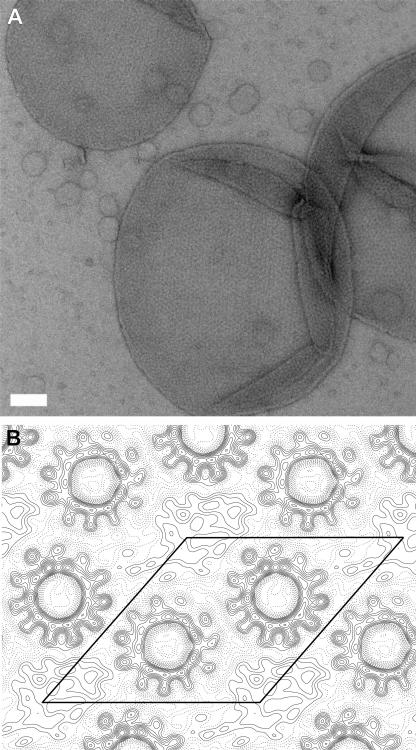
Two-dimensional crystals of the C. paradoxum c ring were obtained at a lipid-to-protein ratio of 0.5 to 1.0 with the lipid POPC. Figure 3A shows an overview of a two-dimensional crystalline C. paradoxum c-ring sample which consists of vesicles containing crystalline areas with dimensions up to 0.5 μm. The 2D crystals are of plane group p1 (Table 1). A unit cell has the dimensions 99.2 ± 0.9 Å for a, 98.5 ± 0.4 Å for b, and 131.1 ± 0.3° for β and contains two c rings, each with a stoichiometry of 11. Each ring is in contact with three neighboring c rings. Such a packing was also observed in the 2D crystals of the I. tartaricus (18, 20) and Propionigenium modestum (11) c11 rings. The projection map, calculated to 7 Å resolution (Fig. 3B), shows that each c ring contains two concentric, slightly staggered, packed rings, each composed of 11 densities. Whereas the inner ring of densities is tightly packed, the outer one is more loosely arranged, and, by analogy with the I. tartaricus c ring, the densities correspond to the N- and C-terminal α-helices of the c ring, respectively. The inner and outer diameters of the rings, measured at the density borders, are the same as in the I. tartaricus c rings, approximately 17 Å and 50 Å.
FIG. 3.
Electron microscopy of 2D crystals from C. paradoxum c rings. (A) Overview of 2D crystals, negatively stained with uranyl acetate. The white scale bar represents 100 nm. (B) Projection map of six averaged images at a resolution of 7 Å. A unit cell containing two c rings is indicated.
TABLE 1.
Statistics of the data seta
| Resolution range (Å) | No. of unique reflections | Phase residual (°)b |
|---|---|---|
| ∞-22.4 | 23 | 22.2 |
| 22.4-15.8 | 24 | 25.0 |
| 15.8-12.8 | 20 | 15.5 |
| 12.8-11.1 | 21 | 7.3 |
| 11.1-9.9 | 25 | 13.8 |
| 9.9-9.1 | 18 | 32.1 |
| 9.1-8.4 | 25 | 24.1 |
| 8.4-7.8 | 26 | 19.2 |
| 7.8-7.4 | 22 | 16.3 |
| 7.4-7.0 | 19 | 27.3 |
| ∞-7.0 | 223 | 21.0 |
Number of images, 6; plane group, p1; cell parameters, 99.2 ± 0.9 Å (a), 98.5± 0.4 Å (b), and 131.1 ± 0.3° (β); defocus range, 3,530 to 5,450 Å.
The random phase residual was 90°.
Comparison of the crystal packing (I. tartaricus and C. paradoxum).
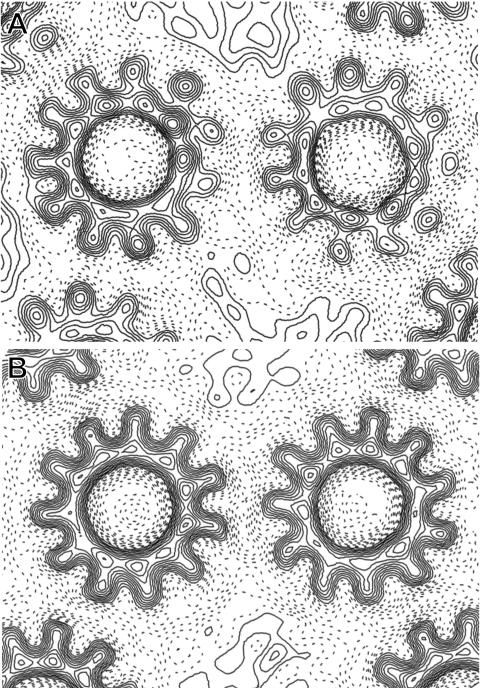
The packing of the 2D crystals of c rings from C. paradoxum and I. tartaricus ATPase appears to have the same unit cell containing two rings, with each ring having three oppositely oriented neighboring rings (Fig. 4). In both crystals, all three crystal contacts in the unit cell are identical. However, the two p1 unit cells differ significantly in their dimensions and angles and also the packing of adjacent rings is different. The crystal contacts in I. tartaricus always involve two helices from one of the rings and one helix from the other, in a staggered arrangement, whereas in C. paradoxum, two helices from each ring are involved. The shape of the c ring resembles an hourglass, and therefore, the amino acids at the ends of the helices are responsible for the contact sites. In order to determine which amino acids are involved, we have built the high-resolution X-ray structure of the I. tartaricus c ring (12) into the electron density obtained from the I. tartaricus c-ring 2D crystals (20). According to this model, the crystal contacts consist on one side of the membrane of the side chains between K50 and S55 (ring 1) and between P83 and G86 (ring 2). On the other side, the contact sites are between Y80 and L87 (ring 1) and between K50 and L59 (ring 2) (Fig. 1). Between the I. tartaricus and the C. paradoxum c subunits, four amino acids differ in the K50-L59 peptides and four amino acids differ in the Y80-L87 peptides. For clarity, we have marked in Fig. 1 the residues in the crystal contact regions with colors. In a C. paradoxum homology model that is based on the crystal structure of the I. tartaricus c ring, several side chains that differ from those in I. tartaricus would cause several steric clashes, involving K50 to Q, I54 to L, S55 to R, V58 to L, and G86 to N (I. tartaricus numbering). To avoid these clashes, the rings are therefore reoriented versus each other in the C. paradoxum 2D crystal.
FIG. 4.
Comparison of (A) I. tartaricus and (B) C. paradoxum crystal contacts on the basis of the projection maps. Both maps are calculated to 7 Å resolution.
The Na+-binding site in the C. paradoxum c ring.
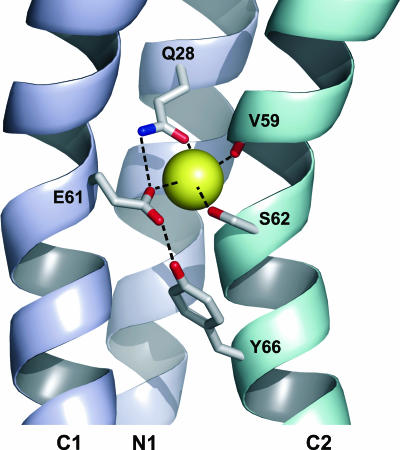
The Na+-binding site in the c ring is the most notable feature in the structure. The way a Na+ ion is coordinated between two subunits has profound consequences in the mechanism of ion translocation and, hence, torque generation. Therefore, we have looked at the binding site in the homology model (Fig. 5). Based on the I. tartaricus model, the coordination site is located in the middle of the membrane at the tightest part of the ring and the entire ring is able to bind 11 sodium ions. The Na+ ion is bound at the interface of three helices: an N-terminal helix and two C-terminal helices. A putative Na+ coordination sphere can be made up by the conserved side chains of Q28, E61, S62, and the carbonyl oxygen of V59. Moreover, one could assume that E61 acts not only as one of the four Na+-binding ligands but also as the recipient of hydrogen bonds, one hydrogen bond being provided by Y66. In addition, the homology model shows that all side chains in close proximity to the ion-binding site are conserved as well. Almost all the differences between residues from the C. paradoxum and I. tartaricus rings are located at the termini or face towards the center of the ring. Based on these observations, we propose that the sodium ion translocation mechanisms are likely to be similar between the two c rings (6).
FIG. 5.
Structure of the C. paradoxum c11 ring Na+-binding site obtained from homology modeling in ribbon representation. Two c subunits are shown in different colors. Each subunit is formed by an inner (N1) and an outer (C1 and C2) helix. The view is normal to the external surface, with the ring axis being vertical. The amino acid side chains mentioned in the text are indicated in stick representation. The Na+ coordination and selected hydrogen bonds are indicated with dashed lines. The image was created with PyMOL (5).
For an anaerobic bacterium like C. paradoxum, the consequence of having an 11-mer oligomeric c ring is that, for every three ATP molecules hydrolyzed by the ATPase during glucose fermentation, 11 Na+ ions are pumped across the membrane to generate the ΔμNa+. In the Na+ F-type ATP synthases that operate primarily in the synthesis direction during succinate or tartrate fermentation (e.g., those in P. modestum or I. tartaricus, respectively), the c ring also exists as an 11-mer. We conclude, therefore, that an undecameric stoichiometry is suitable for both operational modes in the F-ATP synthases.
Acknowledgments
We thank Peter Tittmann and Heinz Gross (EMEZ, Electron Microscopy Center of the ETH, Zürich, Switzerland) for the examination of the 2D crystals and Deryck Mills for assistance with cryoelectron microscopy. We also acknowledge Kay Diederichs (Structural Biology, University of Konstanz, Germany) for help in the creation of the homology model, Christoph von Ballmoos for MS-MALDI measurement, and Stefanie Keis for critically reading the manuscript.
G.M.C. was supported by a Marsden grant from the Royal Society of New Zealand. S.A.F. was supported by an Otago postgraduate scholarship from the University of Otago.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Aufurth, S., H. Schägger, and V. Müller. 2000. Identification of subunits a, b, and c1 from Acetobacterium woodii Na+-F1F0-ATPase. Subunits c1, c2, and c3 constitute a mixed c-oligomer. J. Biol. Chem. 275:33297-33301. [DOI] [PubMed] [Google Scholar]
- 2.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Cook, G. M., J. B. Russell, A. Reichert, and J. Wiegel. 1996. The intracellular pH of Clostridium paradoxum, an anaerobic, alkaliphilic, and thermophilic bacterium. Appl. Environ. Microbiol. 62:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 5.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, Calif. http://pymol.sourceforge.net/overview/index.htm.
- 6.Dimroth, P., C. von Ballmoos, and T. Meier. 2006. Catalytic and mechanical cycles in F-ATP synthases. EMBO Rep. 7:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson, S. A., S. Keis, and G. M. Cook. 2006. Biochemical and molecular characterization of a Na+-translocating F1Fo-ATPase from the thermoalkaliphilic bacterium Clostridium paradoxum. J. Bacteriol. 188:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaeser, R. M., and K. H. Downing. 1990. The “specimen flatness” problem in high-resolution electron crystallography of biological macromolecules, p. 98-99. In L. D. Peachy and D. B. Williams (ed.), Proceedings of the XIIth International Congress for Electron Microscopy, vol. 1. San Francisco Press, Inc., San Francisco, Calif. [Google Scholar]
- 9.Laubinger, W., and P. Dimroth. 1987. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1Fo type. Eur. J. Biochem. 168:475-482. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., L. Mandelco, and J. Wiegel. 1993. Isolation and characterization of a moderately thermophilic anaerobic alkaliphile, Clostridium paradoxum sp. nov. Int. J. Syst. Bacteriol. 43:450-460. [Google Scholar]
- 11.Meier, T., U. Matthey, C. von Ballmoos, J. Vonck, T. Krug von Nidda, W. Kühlbrandt, and P. Dimroth. 2003. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J. Mol. Biol. 325:389-397. [DOI] [PubMed] [Google Scholar]
- 12.Meier, T., P. Polzer, K. Diederichs, W. Welte, and P. Dimroth. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659-662. [DOI] [PubMed] [Google Scholar]
- 13.Murata, T., I. Yamato, Y. Kakinuma, A. G. Leslie, and J. E. Walker. 2005. Structure of the rotor of the V-type Na+-ATPase from Enterococcus hirae. Science 308:654-659. [DOI] [PubMed] [Google Scholar]
- 14.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, S., U. Matthey, G. Kaim, and P. Dimroth. 1998. Purification and properties of the F1Fo ATPase of Ilyobacter tartaricus, a sodium ion pump. J. Bacteriol. 180:3312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schägger, H., and G. Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 17.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. (Erratum, 163:279, 1987.) [DOI] [PubMed] [Google Scholar]
- 18.Stahlberg, H., D. J. Müller, K. Suda, D. Fotiadis, A. Engel, T. Meier, U. Matthey, and P. Dimroth. 2001. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vonck, J. 2000. Parameters affecting specimen flatness of two-dimensional crystals for electron crystallography. Ultramicroscopy 85:123-129. [DOI] [PubMed] [Google Scholar]
- 20.Vonck, J., T. Krug von Nidda, T. Meier, U. Matthey, D. J. Mills, W. Kühlbrandt, and P. Dimroth. 2002. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 321:307-316. [DOI] [PubMed] [Google Scholar]