Abstract
In this study, we show that Salmonella produces an O-antigen capsule coregulated with the fimbria- and cellulose-associated extracellular matrix. Structural analysis of purified Salmonella extracellular polysaccharides yielded predominantly a repeating oligosaccharide unit similar to that of Salmonella enterica serovar Enteritidis lipopolysaccharide O antigen with some modifications. Putative carbohydrate transport and regulatory operons important for capsule assembly and translocation, designated yihU-yshA and yihVW, were identified by screening a random transposon library with immune serum generated to the capsule. The absence of capsule was confirmed by generating various isogenic Δyih mutants, where yihQ and yihO were shown to be important in capsule assembly and translocation. Luciferase-based expression studies showed that AgfD regulates the yih operons in coordination with extracellular matrix genes coding for thin aggregative fimbriae and cellulose. Although the capsule did not appear to be important for multicellular behavior, we demonstrate that it was important for survival during desiccation stress. Since the yih genes are conserved in salmonellae and the O-antigen capsule was important for environmental persistence, the formation of this surface structure may represent a conserved survival strategy.
Extracellular polysaccharides (EPS), together with proteins and nucleic acids, form an amorphous matrix encasing multicellular microbial communities and mediate direct interactions between the bacteria and the environment. Generally, EPS is considered capsular when covalently attached to lipids, whereas secreted EPS molecules or slime are easily sloughed from the cell surface with no visible means of attachment (48). Proposed functions of bacterial EPS include provision of a stable matrix resulting in environmental or host persistence, prevention of microbial desiccation via the formation of a hydrated gel, facilitation of biofilm maturation, prevention of phagocytosis, and immune system evasion and providing hydration for swarming (27, 44). There is mounting literature indicating that EPS embedded microbial populations are culpable for chronic infections (16, 58).
Escherichia coli strains produce more than 80 chemically and serologically distinct capsules that have been separated into four groups, in addition to cellulose and colanic acid EPS (48, 66). Group 4 capsules, formerly group 1, comprise structurally identical lipopolysaccharide O-polysaccharides (LPS) and are thus known as O-antigen (O-Ag) capsules (66). Similar to the heteropolymeric LPS O-Ags, the repeat units of group 1 capsules are known to be assembled on an undecaprenol phosphate carrier lipid at the cytoplasmic face of the inner membrane, transferred across the inner membrane by a “flippase,” Wzx, and polymerized by the O-Ag polymerase, Wxy, resulting in high-molecular-weight (HMW) capsular polysaccharides that are translocated across the outer membrane for final assembly at the cell surface (66). Recently, a seven-gene operon (ymcDCBA, yccZ, etp, etk) involved in the surface translocation of the group 4 O-Ag capsule from enteropathogenic E. coli (EPEC) was identified (45). The operon is present in some E. coli strains but not in Salmonella. Each gene is required for surface translocation in EPEC; mutation of any single gene results in the loss of capsule expression.
Salmonella enterica serovar Enteritidis is a persistent food-borne enteric human pathogen capable of forming biofilms on abiotic surfaces (4). This has major economic and health consequences (34, 43). Biofilm formation in Salmonella is associated with the multicellular and aggregative behavior variously described as rdar (50), rugose (3), or lacy (26). This multicellular behavior is highly conserved among the salmonellae (64) and is characterized by the elaboration of thin aggregative fimbriae (Tafi, curli [12]), cellulose (68), and other uncharacterized EPS (63). Together, these components form the extracellular matrix that confers resistance to acid and bleach and facilitates environmental persistence (3, 51, 53, 56, 64).
AgfD (CsgD) from the Tafi (Curli) operon positively regulates elements that promote biofilm formation: Tafi (50), cellulose (49), serine hydroxymethyltransferase (10), and a biofilm-associated protein, BapA (36), and negatively regulates factors that inhibit biofilm formation (9). Thus, AgfD is a biofilm control point (24). Activation of the agfD promoter is regulated by RpoS, OmpR, MlrA, CpxR, H-NS, and IHF (23, 32). Transcription of agfD is maximal during the stationary phase of growth in media of low osmolarity and 30°C (23). A single point mutation in the promoter region of agfD in S. enterica serovar Enteritidis 27655-3b (SE 3b), the strain studied here, renders Tafi expression RpoS and temperature independent (50).
In the present study, we have identified and characterized an O-Ag capsule associated with the Salmonella extracellular matrix. Previously, we and others had observed EPS in Salmonella that was distinct from the known EPS: colanic acid, cellulose and the Vi-Antigen (46, 56, 63). Although E. coli O-Ag capsules have been characterized (66), this is the first published report of a Salmonella O-Ag capsule. We have identified two operons, yihU-yshA and yihVW, to be important for capsule surface assembly and translocation. YihQ and YihO were found to be important for capsule assembly and translocation. We show that the yih operons are differentially regulated by AgfD, in coordination with other extracellular matrix components. The O-Ag capsule plays a fundamental role in the protection of cells against desiccation stress but does not appear to affect the formation of the extracellular network between cells. The yih genes are conserved throughout salmonellae. Thus, the Salmonella O-Ag capsule appears to represent a conserved component of the extracellular matrix that is important for environmental persistence.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
SE 3b (21) was routinely grown at 37°C for 24 h 1% tryptone (T) agar (13) unless otherwise indicated. For capsule purification experiments, SE 3b ΔbcsA (63) was grown on agar supplemented with 0.05% yeast extract, 10 mM Na2HPO4, 0.1% NH4Cl, 0.3% KH2PO4, and 1% glucose at 28°C for 5 days. For cloning, E. coli XL1-Blue (Stratagene) harboring pBCKS (Stratagene), pHAG (11), pGEM-T Easy (Promega), or pHSG415 (28) was grown at 28 or 37°C for 24 or 48 h with agitation in LB broth supplemented with 50 μg of chloramphenicol/ml, 100 μg of ampicillin/ml, 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as required. For transposon mutagenesis, SE 3b transduction mixtures and ΔbcsA Tn10dCm mutants were incubated at 37°C overnight on LB medium or broth supplemented with 50 μg of kanamycin/ml, 12 μg of chloramphenicol/ml, and 10 mM EGTA. For lux assays, overnight cultures were diluted 1 in 600 in 1% tryptone (pH 7.2) supplemented with 50 μg of kanamycin/ml to a final volume of 150 μl in 96-well clear-bottom black plates (9520 Costar; Corning, Inc.) and overlaid with 50 μl of mineral oil. Cultures were assayed for luminescence (0.1 s) and absorbance (620 nm, 0.1 s) every 30 min in a Wallac Victor (Perkin-Elmer Life Sciences, Boston, Mass.) during growth at 28°C with agitation for 48 h (three 90-s shaking periods with 10-min spacing). For whole-cell enzyme-linked immunosorbent assays (ELISAs) and desiccation experiments, strains were inoculated onto T agar and grown at 28°C for 6 days.
EPS purification.
Cells scraped off agar surfaces were resuspended in 1% phenol, mixed vigorously by vortexing, and incubated at room temperature for 30 min. Cellular debris was pelleted by centrifugation (16,000 × g, 4°C, 5 h). The aqueous phase of the supernatant was removed, and 4 volumes of ice-cold acetone were added on ice with constant stirring using a glass rod for at least 10 min. The precipitated material was spooled, washed with acetone, and air dried at room temperature overnight. The material was solubilized in distilled H2O with heat and gentle agitation, dialyzed (MW 6-8; Spectrum) overnight in distilled H2O, and lyophilized. Approximately one agar plate of cells (diameter, 140 mm; Fischer) yielded 4 mg of crudely purified EPS. The EPS (10 mg/1 ml) was separated on a Superose 6 column (Pharmacia) that had been equilibrated in phosphate-buffered saline (pH 7.4) (PBS). PBS was flowed though the column at a rate of 0.4 ml/min, and 50 1-ml fractions were collected every 2 min. Fractions with a high A220 were collected and analyzed by immunoblotting, silver staining, and protein staining.
Generation of polyclonal serum against EPS and capsule.
Crudely purified EPS was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and resolved by electrophoresis. The stacking gel containing the EPS material was excised and broken up by using a tissue homogenizer. The EPS material was conjugated to keyhole limpet hemocyanin (Pierce) and bovine serum albumin (Sigma) according to established procedures from Conlan et al. (15). The EPS-keyhole lympet hemocyanin conjugate prepared in Emulsigen adjuvant (MVP Laboratories) was used to immunize a New Zealand White rabbit. Subcutaneous and intramuscular injections of 200 μg were performed three times at 2-week intervals with a final boost of 100 μg. Three weeks after the final booster injection, serum was collected, and the titer was determined by ELISA (20), using the EPS-BSA conjugate. Capsule-specific antiserum was generated by incubating equal volumes of rabbit polyclonal serum generated against purified EPS and SE 3b Tn10::galE (63) whole-cell suspensions (A600 = 1) for 1 h at 4°C rotating.
Scanning electron microscopy.
Glass coverslips that had been overlaid with 1% T agar on 1.5% T agar plates and incubated at 37°C for 24 h were picked off the agar surface, locked in a mount, and immersed in 2.5% glutaraldehyde in 0.2 M Millonigs buffer (39) at room temperature for 24 h. Cells were dehydrated in a series of alcohols, followed by incubation on an EMS 850 critical point drier. The coverslip was mounted on a specimen block and gold coated by using an Edwards S150B sputter coater and viewed with a Hitachi S-3500N SEM at 15 kV.
Transmission electron microscopy.
Cells from colonies were resuspended in 10 mM Tris-HCl (pH 8) and placed onto 0.3% Formvar-coated 200 mesh copper grids prior to immunostaining with serum generated against purified EPS, followed by goat anti-rabbit serum conjugated to 10-nm gold (Cedarlane Laboratories) and negative staining in 2% uranyl acetate (pH 7) for 15 s. All samples were viewed with a Hitachi H7600 transmission electron microscope under HC-zoom mode at 100 kV.
Transposon mutagenesis and screening for capsule-negative mutants.
SE 3b harboring pNK (5) was infected with a P22 HT 105/1 int-201 phage (54) lysate of Salmonella serovar Typhimurium ATCC 14028 containing Tn10dCm on F′ (19). The resulting transductants were pooled (20,000 to 30,000 colonies), and a P22 SE 3b ΔbcsA Tn10dCm transducing fragment library was created by using the method of Maloy (37). The resulting stable Tn10dCm mutants were picked into 55 clear flat-bottom sterile 96-well cell culture plates (Costar, Corning, N.Y.) and stored in 5% glycerol at −70°C. Tn10dCm mutants were inoculated from glycerol stocks by using a sterile 48-pronged metal stamp into 100 μl of 1% tryptone (pH 7.2) broth in clear flat-bottom sterile 96-well cell culture plates. Growth was measured at A595 using an ELx 808 Biotek Ultra microplate reader (Biotek) after 24 h growth at 37°C. Whole cells were heat fixed at 85°C in a hybridization oven for 6 h or until all of the media had evaporated. ELISAs were performed by using the method of Engvall and Carlsson (20) in flat-bottom high-binding 96-well EIA/RIA plates (Costar) using rabbit polyclonal serum generated against purified EPS, followed by goat anti-rabbit immunoglobulin G-alkaline phosphate (Cedarlane Laboratories) and 1 mg of p-nitrophenyl phosphate (Sigma)/ml. The absorbance in each well was detected at A405, and the final A405/A595 values were calculated and compared to the relative percentage of capsule on the wild-type (wt) surface where the wt was 100%, and the background was SE 3b Tn10::galE. Mutants with less than 10% capsule on the cell surface were rescreened, streaked onto EBU plates to render the mutants phage-free, and rescreened for maintenance of the phenotype. Phenotypes were confirmed by moving mutations into a clean SE 3b background using P22 phage. Genomic DNA for the PCR was prepared according to the method of Walsh et al. (60). Sites of Tn10dCm insertion in the chromosome were amplified by arbitrary primed PCR described by Welsh and McClelland (61) using the primers cat1+arb1 and cat2+arb2 and sequenced using the primer cat2 (Table 1) .
TABLE 1.
Primers used in this study
| Primera | Primer sequence (5′-3′) |
|---|---|
| cat1 | CAGGGTCGTTAAATAGCCGC |
| arb1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
| cat2 | CCGTTGCTTCTCAAATGCC |
| arb2 | GGCCACGCGTCGACTAGTAC |
| yihO_A | AGCCCAAGCTTAAAAAACCCATCCATGA |
| yihO_B | TCTAATCATGATCCGCGTTAATTAATTAACGCCACTCACTTTACTTTACCCGTTAAAACA |
| yihO_C | AGTAAAGTGAGTGGCGTTAATTAATTAACGCGGATCATGATTAGACATAATCGCCTCTCA |
| yihO_D | TCGGCGGGCTGTTAATATGG |
| yihQE_A | ACCGTAAGCTTTTACGCCTGCGTTGCGC |
| yihQE_B | AATTCTCTACCACAACGTTAATTAATTAACCTGCAAAACGACGACCTGCACCATAACCGT |
| yihQE_C | GTCGTCGTTTTGCAGGTTAATTAATTAACGTTGTGGTAGAGAATTCATTGTAATCTCCG |
| yihQE_D | TCGGTTAGATGCCGCCAGTG |
| agfD_A | GACGAATTCGTGTGTTATGCCGCCATGGG |
| agfD_B | GGACTGCAGTAAACATGATG |
| agfD_C | GCCCTGCAGCAAACGATAATCTCAGGCGG |
| agfD_D | GCCAAGCTTTGTCCGTGACGTTGAGCTGG |
| ΔPyih_A | AGGGGAAGCTTGCTCTACTTGCTCCGCC |
| ΔPyih_B | CGAAATGTTTAAAAAAAAAAAGCTGTCAATGTATTGACCCCCCCCCCCAAATACGAAAAAAAACCCCCCCCCCCCTGCTCATTAATGAC |
| ΔPyih_C | GTCATTAATGAGCAGGGGGGGGGGGGTTTTTTTTCGTATTTGGGGGGGGGGGTCAATACATTGACAGCTTTTTTTTTTTAAACATTTCG |
| ΔPyih_D | AGCCTGACGCGCCAGCGTAA |
| yihV1 | GCCCTCGAGCCGATAAATGCTATAACTG |
| yihV2 | GCCGGATCCCAAGCAATACGAACCATG |
PCR primers were purchased from Alpha DNA or University Core DNA and Protein Services (www.dnaservices.myweb.med.ucalgary.ca; University of Calgary).
Generation of SE 3b mutants.
Isogenic ΔyihO (298-bp deletion; primers yihO_A to yihO_D), ΔyihQ (296-bp deletion; primers yihQ_A to yihQ_D), and ΔagfD (394-bp deletion; primers agfD_A to agfD_D) mutants containing six in-frame translational stop codons at the beginning of the gene and a ΔPyih mutant (primers ΔPyih_A to ΔPyihD) with the putative promoter region replaced with random sequence was generated according to the methods of White et al. (62, 62a). All primers are listed in Table 1.
ELISA.
Cells were resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0) and adjusted to A600 0.1 in the wells of flat-bottom high-binding 96-well EIA/RIA plates (Costar). Cells were treated as described above using capsule-specific serum.
Immunofluorescence microscopy.
Cells were scraped off agar surfaces and resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0). Cell suspensions were adjusted so each slide had an A600 of 0.05. Cells were incubated for 1 h at room temperature with capsule-specific serum diluted in 5% normal mouse serum in PBS. Cells were harvested by centrifugation (6,000 × g, 2 min), washed twice with 500 μl of 5% normal mouse serum in PBS, incubated in goat anti-rabbit immunoglobulin G-fluorescein isothiocyanate conjugate (Cedarlane Laboratories) for 1 h at room temperature in the dark and washed again. Cells were viewed under a fluorescence microscope (Nikon Labophot) and imaged digitally.
SDS-PAGE, protein staining, and immunoblotting.
Cells were resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0) and adjusted to an A600 of 0.1. The cells were harvested by centrifugation (6,000 × g, 10 min), resuspended in SDS-PAGE sample buffer, and boiled for 10 min prior to electrophoresis. SDS-PAGE was carried out according to the method of Laemmli (35) with a 5% stacking gel and a 12% resolving gel. The EPS was stained with Gelcode (Bio-Rad), Alcian blue (Sigma-Aldrich), or LPS silver stain (Bio-Rad). For immunoblots, EPS material was transferred to nitrocellulose by using a Mini-Trans-Blot electrophoretic transfer cell (Bio-Rad Laboratories) in buffer recommended by the manufacturer. Rabbit polyclonal immune serum generated against purified EPS or whole Tafi or specific to capsule followed by goat anti-rabbit immunoglobulin conjugated to IRDye800 (LI-COR, Ltd.) was used for detection. Immunoreactive material was visualized by using an Odyssey Biosciences scanner (LI-COR, Ltd.). Goat polyclonal immune serum generated against E. coli lipid A (Biodesign International), followed by swine anti-goat immunoglobulin G-alkaline phosphatase conjugate (Cedarlane Laboratories) was used for the detection of LPS. Immunoreactive material was visualized by incubating in BCIP (5-bromo-4-chloro-3-indolylphosphate) and 4-nitroblue tetrazolium chloride (Sigma-Aldrich).
Generation of lux reporters.
The yih intergenic region was PCR amplified from SE 3b or Salmonella serovar Typhimurium ATCC 14028 chromosomal DNA using the primers yihV1 and yihV2. PCR products were purified and sequentially digested with XhoI and BamHI (Invitrogen Canada, Inc.). After restriction digestion, promoter regions were ligated by using T4 DNA ligase (Invitrogen Canada, Inc.) into pCS26-Pac (XhoI-BamHI) or pU220 (BamHI-XhoI) reporter vectors containing the luxCDABE operon from Photorhabdus luminescens (6). DNA sequencing was performed by Macrogen (Seoul, South Korea) using primers pZE05 and pZE06 (6). The construction of agfD and agfB reporters has been described elsewhere (64).
Desiccation experiment.
Six individual colonies (diameter, ∼1 mm) from SE 3b, ΔagfD, ΔagfA, ΔbcsA, ΔagfA ΔbcsA, ΔyihO, ΔyihQ, ΔPyih, and Salmonella serovar Typhimurium ATCC 14028 strains were removed from the agar surface and frozen. Three colonies were lyophilized for 1 week. All colonies were resuspended in 500 μl of PBS for 1 h, vigorously vortexed, and homogenized by using a sterile applicator stick until even turbidity was reached. Cell mixtures were serially diluted in triplicate and plated in duplicate to determine CFU values for each individual colony. The difference between the averaged CFU values for each desiccated colony and the averaged CFU value from initial colonies was statistically analyzed by using a one-way analysis of variance with Bonferroni's post-test using GraphPad Prism version 3.00 for Windows (GraphPad Software, San Diego, CA).
RESULTS
Salmonella EPS.
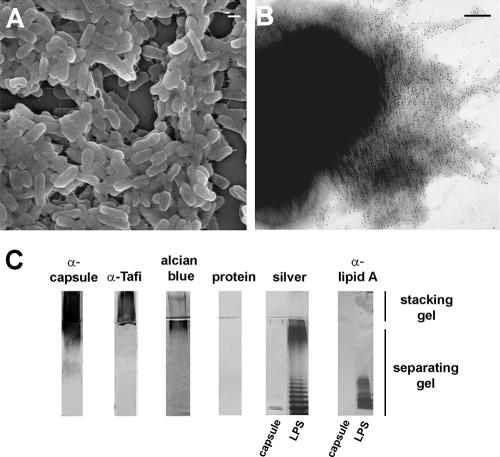
To characterize the EPS associated with the extracellular matrix (Fig. 1A), polysaccharides were purified from a cellulose-deficient background strain (ΔbcsA) of SE 3b. Serum generated against purified EPS was used to immunostain the cell surface of SE 3b (Fig. 1B) and revealed that the EPS formed an amorphous layer around the cell. To analyze the EPS biochemically, crudely purified fractions were resolved by SDS-PAGE and stained directly or transferred to nitrocellulose for immunoblotting. A diffuse band corresponding to the stacking gel did not react with protein- or LPS-specific silver stains or serum generated against LPS lipid A but was stained by the acidic polysaccharide stain, Alcian blue, (Fig. 1C), similar to other EPS (33, 59). The diffuse material was recognized by immune serum generated against whole Tafi or crudely purified EPS (Fig. 1C) in addition to serum generated against group D O-Ag (Difco; data not shown). These tests indicated that low-molecular-weight (LMW) LPS was not a significant component of the purified material, but the HMW material may be structurally related to O-Ag. Furthermore, HMW material from commercially purified LPS was stained by Alcian blue and slightly cross-reacted with serum generated against crudely purified EPS (Fig. 1C, blots on right). Thus, commercially purified LPS contained immunologically similar HMW material that was distinct from LMW LPS. These results indicated the fraction of EPS(s) contained immunoreactive, nonproteinaceous polymer(s) of HMW, some of which were acidic.
FIG. 1.
Salmonella EPS associated with the extracellular matrix. (A) Scanning electron micrograph of SE 3b cells embedded in the extracellular matrix (bar = 1 μm). (B) Transmission electron micrograph of SE 3b expressing the extracellular matrix immunostained with serum generated against the purified EPS (bar = 100 nm). (C) SDS-PAGE and immunoblot analysis of 10 μg of purified EPS from SE 3b ΔbcsA or 10 μg of LPS from Salmonella serovar Enteritidis (Sigma).
SE 3b produces an O-Ag capsule.
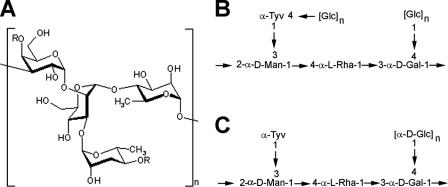
Compositional analysis of the purified EPS fraction indicated the presence of one major, predominant polysaccharide and another minor polysaccharide. Several different fatty acids were also detected, including common LPS components, 3-hydroxytetradecanoic, dodecanoic, tetradecanoic, and hexadecanoic acids, as well as octadecanoic acid and the unsaturated fatty acids hexadecenoic acid and octadecadienoic acid, which are not normally detected in the LPS of enteric bacteria (55). The observed levels of fatty acid were consistent with the possibility that they represented a lipid anchor for a glycolipid (55). The repeating oligosaccharide unit for the major polysaccharide (Fig. 2A and B) was similar to the reported LPS O-Ag of Salmonella serovar Enteritidis (29) but with some structural differences: the tyvelose residue was partially substituted with a polymeric glucose side chain at the 4 position and the galactose residue was glucosylated in a similar manner as structure II HMW O-Ag (47). Furthermore, the major EPS was composed of >2,300 repeat units that is 23 to 143 times greater than that of normal HMW LPS. In E. coli, an EPS that is identical to a given strain's O-Ag is called an O-Ag capsule (66). Furthermore, several lines of evidence indicated that the major EPS was capsular: (i) a solution of purified, nonhydrazinolyzed material was turbid, as would be expected for a lipidated molecule (55); (ii) SE 3b stained positive using Maneval's capsule stain (41), and (iii) the material did not slough off the cell surface unless the membrane was disrupted with phenol treatment (data not shown), indicating that the EPS was tightly associated with the cell. Based on this biochemical evidence and to be consistent with the E. coli nomenclature, the major EPS produced by SE 3b was termed here the O-Ag capsule. Descriptions of the minor EPS and the methodology behind structural analysis of both the major and minor polysaccharides will be reported elsewhere (D. L. Gibson and S. D. Snyder et al., unpublished data).
FIG. 2.
SE 3b capsular polysaccharide structure. (A and B) Configuration (B) and linear drawing (B) of the repeating oligosaccharide unit. R, nonstoichiometric glucose substitution (55). (C) Linear drawing of the O-Ag repeating unit from Salmonella serovar Enteritidis (29, 47). The mannose residue has also been reported in the β-configuration (47).
The yih operons are important for capsule assembly and translocation.
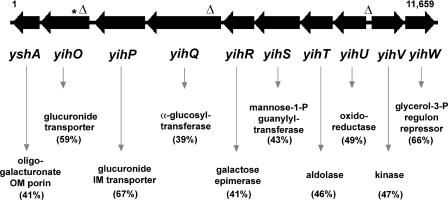
Tn10dCm mutagenesis was used to inactivate and identify prospective genes involved in capsule synthesis, regulation, or transport. The serum generated to the whole EPS fraction primarily recognized the O-Ag capsule and had slight cross-reactivity to the uncharacterized minor EPS (data not shown) but did not strongly recognize LMW and HMW LPS (Fig. 1C). This suggested that the serum recognized conformational epitopes formed by extended helices in the O-Ag capsule, similar to serum generated against other HMW polysaccharides (8). The specificity of the serum allowed us to screen Tn10dCm mutants by ELISA for loss of the capsule. Since the O-Ag capsule repeating unit contained a galactose residue, a galE::Tn10 reference strain was used as a negative control. One of the mutants that was identified had a Tn10dCm insertion in the yihO gene, the seventh gene in a putative eight-gene operon that is adjacent to a divergent putative yihVW operon (Fig. 3). The yih gene products had homologies and conserved domains to known proteins involved in carbohydrate transport, biosynthesis, and regulation (Fig. 3 and Table 2). This led us to speculate that the yih operons may be involved in capsule assembly and translocation.
FIG. 3.
Structure of the yih operons involved in assembly and translocation of the Salmonella O-Ag capsule. The yihU-OyshA and yihVW gene clusters, as represented from S. enterica serovar Typhimurium LT2 (NC003197), are shown to scale. The asterisk denotes the insertion site of the Tn10dCm element; insertion marks represent in-frame deletions of the majority of the gene coding sequence or promoter regions performed in SE 3b. Putative functions listed below each of the yih genes (NP462896-905) were delineated by comparing Yih sequences to protein sequences encoded in sequenced genomes from the NCBI database. The percentages of similar amino acids for each predicted orthologue are indicated in parentheses below the gene names: YshA; KdgM (YP275183) from Pseudomonas syringae pv. phaseolicola 1448A; YihO; YihP; YihQ, UidB (NP416133) from Bacillus licheniformis ATCC 14580, YihQ; CtsZ (BAD34980) from Arthrobacter globiformis, YihR; GalM (AF3058) from Agrobacterium tumefaciens, YihC; YihS (E95924) from Sinorhizobium meliloti 1021, YihT; LacD (Q6G7C2) from Staphylococcus aureus subsp. aureus MSSA476, YihU; MmsB (YP244045) from Xanthomonas campestris pv. campestris 8004, YihV; RbsK (YP177042) from Bacillus clausii KSM-K16, and YihW; GlpR (NP927560) from Photorhabdus luminescens subsp. laumondii TTO1.
TABLE 2.
NCBI conserved domain hits of the proteins encoded by the yih operonsa
| Protein | Conserved domain hit(s)b | Domain function | Score (bits) | E value |
|---|---|---|---|---|
| YihU | MmsB; gnl CDD 11792 | Dehydrogenase; lipid metabolism | 269 | 3e-73 |
| YihT | LacD; gnl CDD 13006 | Aldolase; carbohydrate transport and metabolism | 393 | 1e-110 |
| YihS | GlcNAc_2-epim, family; gnl CDD 12290 | Epimerase; carbohydrate transport and metabolism | 401 | 9e-113 |
| YihR | GalM; gnl CDD 11725 | Mutarotase; carbohydrate transport and metabolism | 188 | 7e-49 |
| YihQ | α-Glucosidases, family 31; gnl CDD 11215 | Hydrolase; carbohydrate transport and metabolism | 566 | 2e-162 |
| YihP | MelB, GPH family; gnl CDD 11918 | Carbohydrate transport | 430 | 2e-121 |
| YihO | MelB, GPH family; gnl CDD 11918 | Carbohydrate transport | 447 | 2e-126 |
| YshA | No hits | |||
| YihW | HTH_ARSR, ARSR family; gnl CDD 17771 | Metal-regulated transcriptional repressors | 37.6 | 0.001 |
| YihV | Ribokinase_group_B, pfkB superfamily gnl CDD 28258 | Synthesize nucleotides and histidine | 244 | 1e-65 |
According to A. Marchler-Bauer and Bryant (38).
Conserved domain hits of the highest scores for each protein are listed along with their respective GenBank designations.
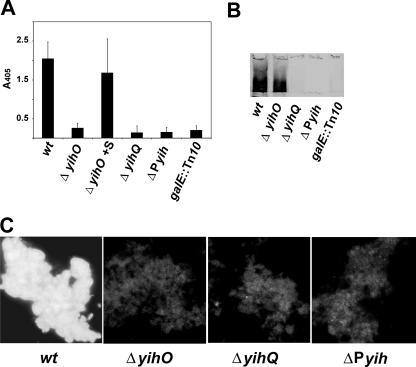
To confirm that the yih genes were important in capsule assembly and translocation, isogenic deletion mutants were generated for yihO, encoding a putative carbohydrate transporter, and yihQ, encoding a putative glucosyltransferase (Fig. 3). In addition, the putative promoter region between the divergent yih operons was replaced in-frame with a random nucleotide sequence (Fig. 3). Capsule expression in the yih mutant strains was measured by ELISA, immunoblotting, and immunofluorescence using serum that had been cross-absorbed to the galE::Tn10 strain (O-Ag capsule-specific serum). The O-Ag capsule was not detected on the cell surface of the ΔyihO mutant when analyzed by ELISA and immunofluorescence (Fig. 4A). However, when whole-cell lysates were subjected to immunoblot analysis, a diffuse band was detected in the stacking gel (Fig. 4B). This indicated that YihO was required for capsule transport but not for biosynthesis. Sonicating the ΔyihO mutant cells prior to ELISA substantiated this because, after disruption, the amount of capsule detected was similar to that of the wt (Fig. 4A). The capsule could be detected on the surface of the ΔyihO cells when they were complemented by yihO in trans (data not shown). The capsule was not detected in either ΔyihQ or the promoter mutant (ΔPyih) when analyzed by ELISA, immunoblotting, or immunofluorescence (Fig. 4). Thus, we hypothesize that YihQ is also involved in capsule assembly and translocation, and the putative promoter region controls the expression of the yih genes. Collectively, these results suggest that the yih genes are important for O-Ag capsule assembly and translocation.
FIG. 4.
Immunological analysis of O-Ag capsule expression on SE 3b yih mutants. wt and yih mutants were analyzed by using capsule specific serum by whole-cell ELISA (A), immunoblotting of whole-cell lysates (B), and immunofluorescence (C). The bars in panel A correspond to averaged values with standard deviations from triplicate samples from three independent experiments. ΔyihO cells were sonicated prior to analysis (ΔyihO +S). In panel B, only the region on the immunoblot corresponding to the stacking gel is shown. SE 3b galE::Tn10 was used as a negative control for capsule production.
The yih genes are differentially regulated via AgfD in coordination with the extracellular matrix genes.
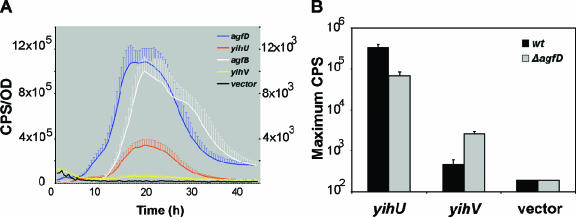
The O-Ag capsule was originally detected in association with the AgfD-regulated extracellular matrix (63). Therefore, we wanted to determine whether AgfD had a regulatory effect on yih expression. yihU and yihV promoter::lux reporter fusions were used to measure the promoter activity of each yih operon in SE 3b. During growth in 1% tryptone broth, yihU activity was high at 3.4 × 105 CPS, whereas yihV expression was similar to the inactive promoter-less vector control (Fig. 5A). Peak yihU expression coincided with peak activity of agfD and agfB promoter::lux fusions, suggesting coordinated expression with other extracellular matrix genes. However, when expression was tested in an isogenic ΔagfD mutant background, yihU activity decreased ∼10-fold, whereas yihV turned on with a peak activity of 2.6 × 103 CPS, representing an ∼10-fold increase in expression (Fig. 5B). This indicated that AgfD had a stimulatory affect on yihU expression and a repressive affect on yihV expression. Furthermore, these results show that high yihU expression corresponds with lower or inactive yihV expression in the presence of AgfD. Based on these results we speculate that AgfD positively regulates the expression of the yihU operon via repression of the yihV operon, although further experimentation is required to substantiate this. These results indicate that AgfD has a positive regulatory effect on O-Ag capsule synthesis under the specified conditions.
FIG. 5.
The yihU promoter has a similar expression profile as the extracellular matrix gene promoters and is regulated via AgfD. (A) The expression of yihU, agfB, and agfD promoter::lux fusions in SE 3b had similar expression profiles, whereas yihV was similar to the vector control near the baseline. (B) The expression of yihU and yihV in an isogenic ΔagfD strain was compared to SE 3b, and the maximum CPS is shown. AgfD positively affects yihU expression and negatively affects yihV expression. Cultures were grown in 1% tryptone at 28°C for 48 h with agitation, and readings were taken every 30 min to monitor the luminescence (in counts per second [CPS]) and cell density (optical density at 600 nm [OD]). Individual points in each expression curve represent averaged CPS/OD values calculated from triplicate samples from three independent experiments; error bars represent the standard deviations. In panel A, CPS/OD measurements for yihU, agfB, and agfD are on the left axis; measurements for yihV and vector only are on the right axis. pU220 is the promoterless vector control (6).
Conservation of yih operons in salmonellae.
The yih operons are conserved throughout all Salmonella serovars currently being sequenced (22). To determine whether the genes could be detected throughout the salmonellae, isolates from the Salmonella reference collection C (SARC [7]), representing all seven subgroups of S. enterica, along with S. bongori, were tested by a PCR specific for yihU and yihV DNA flanking the yih intergenic region. The yih intergenic region was detected in all 16 SARC isolates (data not shown), indicating that the divergent operon structure was conserved.
The O-Ag capsule promotes environmental persistence.
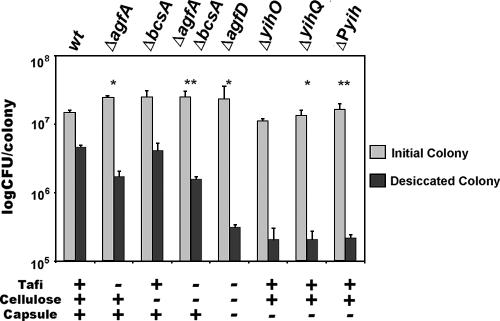
To determine whether the O-Ag capsule was involved in environmental persistence, colonies of the rdar morphotype from yih mutant strains were subjected to desiccation by lyophilization to represent the long-term storage used in our previous study (i.e., storage on plastic for nine months [64]). The wt strain capable of producing Tafi, cellulose, and capsule had the highest survival rate of 31.1% (Fig. 6). In the absence of Tafi (ΔagfA), cellulose (ΔbcsA), or Tafi and cellulose (ΔagfAbcsA), survival was decreased to 7, 16.6, and 6.3%, respectively. There was a significant loss in viability in both Tafi mutants (ΔagfA and ΔagfAbcsA) but not in the cellulose mutant (ΔbcsA) alone compared to the wt. The ΔagfD mutant was most impaired in survival (1.3%). These results closely matched the results of White et al. (64) and indicated that additional components regulated by AgfD, other than Tafi and cellulose, were responsible for survival. The O-Ag capsule mutants (ΔyihO, ΔyihQ, and ΔPyih) were reduced to levels similar to the ΔagfD mutant demonstrating that the O-Ag capsule was primarily responsible for desiccation tolerance. Surprisingly, the yih mutations did not disrupt pellicle formation, the rdar morphotype and biofilm formation similar to the other extracellular matrix components (data not shown). This indicated the capsule was not important in multicellular aggregation.
FIG. 6.
The O-Ag capsule is required for survival against desiccation stress. Survival of SE 3b and fimbrial (ΔagfA), cellulose (ΔbcsA), extracellular matrix (ΔagfD), and capsule mutants (ΔyihO, ΔyihQ, and ΔPyih) was compared after lyophilization. Colonies were grown on T agar for 6 days at 28°C, peeled off the agar surface, frozen, and lyophilized for 1 week. Bars represent the average number of CFU per colony and standard deviations corresponding to three individual colonies. The asterisks shows statistical significance compared to wt survival values. (*, P < 0.05; **, P < 0.01).
DISCUSSION
We report here that Salmonella produces an O-Ag capsule associated with the extracellular matrix. Structural analysis yielded a repeating oligosaccharide unit similar to Salmonella serovar Enteritidis LPS O-Ag with some modifications (55). The previously uncharacterized yih operons were shown to be important for capsule assembly and translocation. The yihVW operon, coding for a putative repressor of yihU transcription, was repressed in the presence of AgfD. This suggested that capsule assembly and translocation was regulated along with Tafi and cellulose as part of the extracellular matrix. The yih genes are conserved throughout salmonellae and appears to plays an important role in desiccation tolerance. Thus, the O-Ag capsule could play a role in environmental persistence facilitating Salmonella's cyclic lifestyle aiding the transmission of cells through a host, into the environment, and back into a new host (67).
The SE 3b O-Ag capsule was structurally similar to the Salmonella serovar Enteritidis LPS O-Ag; however, this is not unusual. Capsular and O-Ag polysaccharides are the same in some strains of E. coli (2, 25) and Vibrio ordalii (52) and are classified as group 4 O-Ag capsules in E. coli (66). Since the major EPS identified has some biochemical characteristics of a capsule and the repeat unit is structurally similar to the SE LPS O-Ag, we propose to classify it as a group 4 O-Ag capsule. The Salmonella O-Ag capsule does have some structural modifications, however, most notably a glucose polymer linked to the tyvelose residue. Furthermore, the O-Ag capsule has >2,300 repeat units compared to LPS, which normally has 16 to 100 repeat units (40, 65). The capsule and LPS differ in modifications, size, charge, and lipid attachment (55). These differences are adequate to confer immunological distinction to the O-Ag capsule. Other HMW polysaccharides have conformationally dependent epitopes due to extended helices formed by large polysaccharides and their modifications (8). Thus, we presume that our serum predominantly recognizes conformationally dependent epitopes on the O-Ag capsule. Recently, another report of a Salmonella capsule purified from the extracellular matrix was published (18). However, this capsule does not appear to have the same chemical composition as the O-Ag capsule since rhamnose was absent from compositional analysis (18). Furthermore, the reported capsule was important in biofilm maturation (18), whereas the O-Ag capsule did not significantly contribute to biofilm formation under similar conditions (data not shown). Finally, we found the yihU genes were inactive at 37°C in Salmonella serovar Typhimurium due to reduced AgfD levels (data not shown), but the capsule reported by de Rezende et al. (18) was detected at both 28 and 37°C. It is possible that the second, minor EPS identified here, in association with the O-Ag capsule, could represent the capsule reported by de Rezende et al. (18).
We propose that the yih operons are responsible for capsule surface assembly and translocation. From homology searches and comparisons to known gene functions, we were able to make the following putative assignments of function: YihU represents an oxidoreductase, YihT represents an aldolase, and YihR represents an epimerase, all of which have conserved domains related to carbohydrate transport and metabolism. YihS has homology to ManC (RfbM), a mannose-1-phosphate guanylytransferase, which is involved in the formation of an activated sugar nucleotide precursor for residues in cell surface polysaccharides (31). Thus, YihS may be involved in activating sugar nucleotide precursors involved in side chain modifications. YihQ has conserved domains related to carbohydrate transport, is a member of the family 31 glycosyl hydrolases, has α-glucosidase activity in E. coli (42), and has sequence identity to two members of the family 31 glycosylhydrolases that also have glucosyltransferase activity: CtsZ (25%), a 6-glucosyltransferase from Bacillus globisporus C11 (1), and YicI (24%), an α-xylosidase from E. coli (42). Furthermore, we have shown that in the absence of YihQ, the capsule is not assembled or expressed on the cell surface. Thus, YihQ could act as a glucosyltransferase that catalyzes sugar, possibly glucose transfer onto the tyvelose residue of the repeating unit and may also be required for efficient O-Ag capsule translocation. YihO is a putative membrane glucuronide transport protein; in an isogenic yihO mutant, the O-Ag capsule was not exported. We hypothesize that YihO is important in translocation and since it is predicted to be a lipoprotein (PredictProtein software [www.predictprotein.org]) containing 12 integral transmembrane spanning regions (TMHMM server [www.cbs.dtu.dk]). YihP is homologous to YihO but was not sufficient to transport the capsule in the isogenic ΔyihO mutant. For yshA, we found no predicted promoter upstream of yihO and thus assume that it is transcribed as a part of the yih operon, although currently we do not have any transcriptional evidence. YshA encodes a putative outer membrane protein (14) involved in sugar transport (17) and thus could also be involved in O-Ag capsule surface assembly or the translocation complex. From the divergent yihVW operon, yihW encodes a putative negative repressor and in the promoter-lux fusion assays described, when yihVW expression was high (i.e., in the absence of AgfD), yihU-OyshA expression was low and vice versa. It is possible that YihW negatively regulates the expression of yihU-OyshA, although other regulatory effects are also possible. Biochemical enzyme analyses will be required to conclusively assign the function of each protein in the synthesis of the O-Ag capsule. Since the yih operons contain relatively few genes coding for transferases and structural enzymes and the repeat unit is similar to LPS O-Ag, we predict that the O-Ag biosynthetic genes are utilized to synthesize the O-Ag capsule. We did not identify gene inactivations in any of the LPS O-Ag biosynthetic operons since P22 phage were used to transfer mutations. P22 phage are known to bind LPS O-Ag prior to infection and are unable to infect strains containing alterations in the O-Ag structure (57). As a result, Tn10dCm mutations causing alterations in O-Ag structure would not have been represented in the transducing lysate prepared from SE 3b. Together with biochemical, structural, and bioinformatic analyses, our results indicate that the yih operons are important for O-Ag capsule surface assembly and transport.
Capsule expression was coregulated with the extracellular matrix by AgfD from the Tafi operon. yihV expression was repressed in the presence of AgfD, and this coincided with yihU activation. AgfD controls the expression of the extracellular matrix genes for multicellular pattern formation, which is a highly conserved physiological phenomenon of salmonellae (64a). It appears the yih operons are also conserved throughout the salmonellae. This may reflect an important role the O-Ag capsule may play in the life cycle of Salmonella.
Our previous studies have shown that the extracellular matrix promotes the environmental persistence of Salmonella in the face of desiccation stress (64). We used lyophilization as a simple and efficient test of bacterial components important for long-term survival. Our results closely mirrored those of White et al. (64) performed on colonies that had been stored for 9 months on plastic. We demonstrated that the O-Ag capsule is primarily responsible for desiccation tolerance in SE 3b. We hypothesize that the main role of the O-Ag capsule may be to protect the cells against desiccation stress when faced with adverse conditions.
The Salmonella O-Ag capsule plays an important role in mediating protective effects against desiccation. This may be relevant for the proposed life cycle of Salmonella spp., aiding the transmission of encapsulated cells through a host, into the environment, and back into a new host, by enhancing survival in nonhost environments (67). It is possible the O-Ag capsule is a contributing factor to the survival of Salmonella spp. on desiccated foods (30). Furthermore, other capsules (58) and the extracellular matrix (26) are important in virulence. Thus, the O-Ag capsule reported here may also have an important role during the infection process, but this remains to be tested. Nevertheless, we have demonstrated that the O-Ag capsule is an important component of the Salmonella extracellular matrix involved in environmental persistence.
Acknowledgments
We thank K. Sanderson (University of Calgary) for providing access to SARC strains, C. Whitfield (University of Guelph) for helpful suggestions, and C. Rajotte and P. Banser for technical contributions. DNA sequencing was performed as a service from the Biomedical DNA Sequencing Center (www.uvic.ca/seq) of the University of Victoria (Canada) except when indicated.
This study was supported by grants to W.W.K. from the National Sciences and Engineering Research Council (NSERC) and the Canadian Bacterial Disease Network. D.L.G. and A.P.W. were supported by postgraduate and postdoctoral fellowships from the NSERC.
REFERENCES
- 1.Aga, H., K. Maruta, T. Yamamoto, M. Kubota, S. Fukuda, M. Kurimoto, and Y. Tsujisaka. 2002. Cloning and sequencing of the genes encoding cyclic tetrasaccharide-synthesizing enzymes from Bacillus globisporus C11. Biosci. Biotechnol. Biochem. 66:1057-1068. [DOI] [PubMed] [Google Scholar]
- 2.Ali, T., F. Urbina, A. Weintraub, and G. Widmalm. 2005. Structural studies of the O-antigenic polysaccharides from the enteroaggregative Escherichia coli strain 522/C1 and the international type strain from Escherichia coli O178. Carbohydr. Res. 340:2010-2014. [DOI] [PubMed] [Google Scholar]
- 3.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, J. W., G. Sanders, W. W. Kay, and S. K. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295-301. [DOI] [PubMed] [Google Scholar]
- 5.Bender, J., and N. Kleckner. 1992. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 11:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brisson, J. R., S. Uhrinova, R. J. Woods, M. van der Zwan, H. C. Jarrell, L. C. Paoletti, D. L. Kasper, and H. J. Jennings. 1997. NMR and molecular dynamics studies of the conformational epitope of the type III group B Streptococcus capsular polysaccharide and derivatives. Biochemistry 36:3278-3292. [DOI] [PubMed] [Google Scholar]
- 9.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 10.Chirwa, N. T., and M. B. Herrington. 2003. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 149:525-535. [DOI] [PubMed] [Google Scholar]
- 11.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collinson, S. K., P. C. Doig, J. L. Doran, S. Clouthier, T. J. Trust, and W. W. Kay. 1993. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 175:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collinson, S. K., L. Emody, K. H. Muller, T. J. Trust, and W. W. Kay. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condemine, G., C. Berrier, J. Plumbridge, and A. Ghazi. 2005. Function and expression of an N-acetylneuraminic acid-inducible outer membrane channel in Escherichia coli. J. Bacteriol. 187:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conlan, J. W., A. D. Cox, R. KuoLee, A. Webb, and M. B. Perry. 1999. Parenteral immunization with a glycoconjugate vaccine containing the O157 antigen of Escherichia coli O157:H7 elicits a systemic humoral immune response in mice, but fails to prevent colonization by the pathogen. Can. J. Microbiol. 45:279-286. [PubMed] [Google Scholar]
- 16.Conway, T., and M. Diksic. 1988. Synthesis of “no-carrier-added” carbon-11 SarCNU: the sarcosinamide analog of the chemotherapeutic agent BCNU. J. Nuclear Med. 29:1957-1960. [PubMed] [Google Scholar]
- 17.Dartigalongue, C., H. Nikaido, and S. Raina. 2000. Protein folding in the periplasm in the absence of primary oxidant DsbA: modulation of redox potential in periplasmic space via OmpL porin. EMBO J. 19:5980-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Rezende, C. E., Y. Anriany, L. E. Carr, S. W. Joseph, and R. M. Weiner. 2005. Capsular polysaccharide surrounds smooth and rugose types of Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 71:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott, T., and J. R. Roth. 1988. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol. Gen. Genet. 213:332-338. [DOI] [PubMed] [Google Scholar]
- 20.Engvall, E., and H. E. Carlsson. 1976. Enzyme-linked immunosorbent assay, p. 135-147. In G. Feldman et al. (ed.), Immunoenzymatic techniques. Elsevier/North-Holland, Amsterdam, The Netherlands.
- 21.Feutrier, J., W. W. Kay, and T. J. Trust. 1986. Purification and characterization of fimbriae from Salmonella enteritidis. J. Bacteriol. 168:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florea, L., M. McClelland, C. Riemer, S. Schwartz, and W. Miller. 2003. EnteriX 2003: visualization tools for genome alignments of Enterobacteriaceae. Nucleic Acids Res. 31:3527-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstel, U., C. Park, and U. Romling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639-654. [DOI] [PubMed] [Google Scholar]
- 24.Gerstel, U., and U. Romling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154:659-667. [DOI] [PubMed] [Google Scholar]
- 25.Goldman, R. C., D. White, F. Orskov, I. Orskov, P. D. Rick, M. S. Lewis, A. K. Bhattacharjee, and L. Leive. 1982. A surface polysaccharide of Escherichia coli O111 contains O-antigen and inhibits agglutination of cells by O-antiserum. J. Bacteriol. 151:1210-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guard-Petter, J., L. H. Keller, M. M. Rahman, R. W. Carlson, and S. Silvers. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gygi, D., M. M. Rahman, H. C. Lai, R. Carlson, J. Guard-Petter, and C. Hughes. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167-1175. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto-Gotoh, T., F. C. Franklin, A. Nordheim, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene 16:227-235. [DOI] [PubMed] [Google Scholar]
- 29.Hellerqvist, C. G., B. Lindberg, S. Svensson, T. Holme, and A. A. Lindberg. 1969. Structural studies on the O-specific side chains of the cell wall lipopolysaccharides from Salmonella typhi and S. enteritidis. Acta Chem. Scand. 23:1588-1596. [DOI] [PubMed] [Google Scholar]
- 30.Hiramatsu, R., M. Matsumoto, K. Sakae, and Y. Miyazaki. 2005. Ability of Shiga toxin-producing Escherichia coli and Salmonella spp. to survive in a desiccation model system and in dry foods. Appl. Environ. Microbiol. 71:6657-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jayaratne, P., D. Bronner, P. R. MacLachlan, C. Dodgson, N. Kido, and C. Whitfield. 1994. Cloning and analysis of duplicated rfbM and rfbK genes involved in the formation of GDP-mannose in Escherichia coli O9:K30 and participation of rfb genes in the synthesis of the group I K30 capsular polysaccharide. J. Bacteriol. 176:3126-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junkins, A. D., and M. P. Doyle. 1992. Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr. Microbiol. 25:9-17. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy, M., R. Villar, D. J. Vugia, T. Rabatsky-Ehr, M. M. Farley, M. Pass, K. Smith, P. Smith, P. R. Cieslak, B. Imhoff, and P. M. Griffin. 2004. Hospitalizations and deaths due to Salmonella infections, FoodNet, 1996-1999. Clin. Infect. Dis. 38(Suppl. 3):S142-S148. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penades, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322-1339. [DOI] [PubMed] [Google Scholar]
- 37.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett Publishers, Boston, Mass.
- 38.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millonig, G. 1961. A modified procedure for lead staining of thin sections. J. Biophys. Biochem. Cytol. 11:736-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 41.Okeke, I. N., and A. Lamikanra. 1995. Bacterial capsules: a simple method for demonstration under the light microscope. Br. J. Biomed. Sci. 52:321-322. [PubMed] [Google Scholar]
- 42.Okuyama, M., H. Mori, S. Chiba, and A. Kimura. 2004. Overexpression and characterization of two unknown proteins, YicI and YihQ, originated from Escherichia coli. Protein Expr. Purif. 37:170-179. [DOI] [PubMed] [Google Scholar]
- 43.Pang, T., Z. A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:253-255. [DOI] [PubMed] [Google Scholar]
- 44.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peleg, A., Y. Shifrin, O. Ilan, C. Nadler-Yona, S. Nov, S. Koby, K. Baruch, S. Altuvia, M. Elgrably-Weiss, C. M. Abe, S. Knutton, M. A. Saper, and I. Rosenshine. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J. Bacteriol. 187:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prouty, A. M., and J. S. Gunn. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahman, M. M., J. Guard-Petter, and R. W. Carlson. 1997. A virulent isolate of Salmonella enteritidis produces a Salmonella typhi-like lipopolysaccharide. J. Bacteriol. 179:2126-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 49.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 50.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behavior of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 51.Ryu, J. H., and L. R. Beuchat. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71:247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadovskaya, I., J. R. Brisson, N. H. Khieu, L. M. Mutharia, and E. Altman. 1998. Structural characterization of the lipopolysaccharide O-antigen and capsular polysaccharide of Vibrio ordalii serotype O:2. Eur. J. Biochem. 253:319-327. [DOI] [PubMed] [Google Scholar]
- 53.Scher, K., U. Romling, and S. Yaron. 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 55.Snyder, S. D., D. L. Gibson, C. Heiss, W. W. Kay, and P. Azadi. 2006. Structure of the O-antigen Salmonella capsule. Carbohydr. Res. CAR-4027. [DOI] [PubMed]
- 56.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 57.Steinbacher, S., S. Miller, U. Baxa, A. Weintraub, and R. Seckler. 1997. Interaction of Salmonella phage P22 with its O-antigen receptor studied by X-ray crystallography. Biol. Chem. 378:337-343. [DOI] [PubMed] [Google Scholar]
- 58.Vuong, C., M. Durr, A. B. Carmody, A. Peschel, S. J. Klebanoff, and M. Otto. 2004. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol. 6:753-759. [DOI] [PubMed] [Google Scholar]
- 59.Waldor, M. K., R. Colwell, and J. J. Mekalanos. 1994. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. USA 91:11388-11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walsh, P. S., D. A. Metzger, and R. Higuchi. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10:506-513. [PubMed] [Google Scholar]
- 61.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White, A. P., S. K. Collinson, J. Burian, S. C. Clouthier, P. A. Banser, and W. W. Kay. 1999. High efficiency gene replacement in Salmonella enteritidis: chimeric fimbrins containing a T-cell epitope from Leishmania major. Vaccine 17:2150-2161. [DOI] [PubMed] [Google Scholar]
- 62a.White, A. P., E. Allen-Vercoe, B. W. Jones, R. DeVinney, W. W. Kay, and M. G. Surette. An efficient system for markerless gene replacement applicable in a wide variety of enterobacterial species. Can. J. Microbiol., in press. [DOI] [PubMed]
- 63.White, A. P., D. L. Gibson, S. K. Collinson, P. A. Banser, and W. W. Kay. 2003. Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis. J. Bacteriol. 185:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.White, A. P., and M. G. Surette. Comparative genetics of the rdar morphotype in Salmonella. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 65.Whitfield, C., P. A. Amor, and R. Koplin. 1997. Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol. Microbiol. 23:629-638. [DOI] [PubMed] [Google Scholar]
- 66.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 67.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]