Abstract
The surface of Geobacillus stearothermophilus NRS 2004/3a cells is covered by an oblique surface layer (S-layer) composed of glycoprotein subunits. To this S-layer glycoprotein, elongated glycan chains are attached that are composed of [→2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-L-Rhap-(1→] repeating units, with a 2-O-methyl modification of the terminal trisaccharide at the nonreducing end of the glycan chain and a core saccharide as linker to the S-layer protein. On sodium dodecyl sulfate-polyacrylamide gels, four bands appear, of which three represent glycosylated S-layer proteins. In the present study, nanoelectrospray ionization time-of-flight mass spectrometry (MS) and infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry were adapted for analysis of this high-molecular-mass and water-insoluble S-layer glycoprotein to refine insights into its glycosylation pattern. This is a prerequisite for artificial fine-tuning of S-layer glycans for nanobiotechnological applications. Optimized MS techniques allowed (i) determination of the average masses of three glycoprotein species to be 101.66 kDa, 108.68 kDa, and 115.73 kDa, (ii) assignment of nanoheterogeneity to the S-layer glycans, with the most prevalent variation between 12 and 18 trisaccharide repeating units, and the possibility of extension of the already-known →3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→ core by one additional rhamnose residue, and (iii) identification of a third glycosylation site on the S-layer protein, at position threonine-590, in addition to the known sites threonine-620 and serine-794. The current interpretation of the S-layer glycoprotein banding pattern is that in the 101.66-kDa glycoprotein species only one glycosylation site is occupied, in the 108.68-kDa glycoprotein species two glycosylation sites are occupied, and in the 115.73-kDa glycoprotein species three glycosylation sites are occupied, while the 94.46-kDa band represents nonglycosylated S-layer protein.
Protein glycosylation is an emerging field in bacteriology and, particularly, the envisaged medical and biotechnological applications make prokaryotic glycoproteins an interesting system for further analysis and exploitation (for details, see references 19, 23, 30, 33, 36, 40, 41, and 43). This endeavor requires a thorough understanding of the protein glycosylation processes that occur in the prokaryotic cell.
The glycosylated surface layer (S-layer) protein SgsE from Geobacillus stearothermophilus NRS 2004/3a is a promising model for studies on prokaryotic glycosylation because of the homopolymeric nature of the glycan, which is composed only of rhamnose residues (35). In sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, the mature S-layer glycoprotein is separated into four bands. Three of them represent broad bands in the molecular mass range of approximately 119 to 170 kDa which also give a positive periodic acid-Schiff (PAS) staining reaction, indicating the presence of covalently linked glycan chains (7, 14, 35). The 93-kDa band is nonglycosylated, and the estimated molecular mass concurs with the calculated mass derived from the amino acid sequence of the mature structural protein, SgsE, after cleavage of the 30-amino-acid signal peptide from the precursor protein (GenBank accession number AF328862) (35). Dependent on the cultivation conditions of the organism (batch versus continuous culture), variations exist in the degree of glycosylation of the individual protein bands, all of which possess identical N termini indicative of identical protein portions (20, 35). Previous nuclear magnetic resonance (NMR) experiments have demonstrated that only one type of glycan chain is present on the individual glycoprotein species, however, with considerable length variations. The S-layer glycans consist of trisaccharide repeats with the structure [→2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→]n and of a short core saccharide consisting of α-1,3-linked l-Rhap residues, attached to carbon 3 of a β-d-galactose residue that serves as the linkage sugar to the S-layer polypeptide backbone (35). So far, two glycosylation sites for O-linked glycans have been determined after a papain degradation experiment of SgsE glycoprotein, namely, amino acids threonine-620 and serine-794 (with the numbers referring to the positions on the precursor protein) (35). These data, however, cannot satisfactorily explain the existence of three broad S-layer glycoprotein bands on SDS-PAGE gels.
To better understand the glycosylation pattern of the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a, we have investigated the intact, glycosylated S-layer protein SgsE and selected glycopeptides derived thereof using different mass spectrometric approaches. These data will allow interpretation of S-layer protein glycosylation in a more general way, because similar SDS-PAGE patterns of S-layer glycoproteins have also been observed with other organisms (24).
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Geobacillus stearothermophilus NRS 2004/3a (22, 26) was grown at 57°C on modified SVIII medium, either in pH-controlled continuous culture in a fermentor (35) or in batch culture in 1-liter Erlenmeyer flasks without pH control (containing 400 ml medium; 5-h cultivation with shaking at 200 rpm). Cells grown in the fermentor were separated from culture broth by continuous centrifugation (Sepatech 17 RS centrifuge; Heraeus, Vienna, Austria) at 16,000 × g and 4°C. The biomass was stored at −20°C.
General methods.
SDS-PAGE was carried out according to a standard protocol (15) using a Protean II electrophoresis apparatus (Bio-Rad Laboratories, Vienna, Austria). Protein bands were visualized with Coomassie blue R-250 staining reagent. The protein concentration was determined using the Bradford reagent (Bio-Rad) (4). PAS staining for carbohydrates was performed according to the methods of Hart and coworkers (11).
Preparation of S-layer glycoprotein and glycopeptide samples.
S-layer glycoprotein was isolated according to a standard procedure (21). The lyophilized protein was further purified on a Sephacryl S-200 column (1.6 by 60 cm; GE Healthcare, Uppsala, Sweden) using 2 M guanidinium hydrochloride (GHCl) in 50 mM Tris-HCl buffer, pH 7.5, as eluent. After extensive dialysis, first against 15 mM CaCl2 and then against distilled water, the protein was lyophilized and stored.
After degradation with papain (Sigma-Aldrich, Vienna, Austria), S-layer glycopeptides were isolated and purified as described elsewhere (35). Individual glycopeptide fractions were dried in a SpeedVac centrifuge and stored at −20°C.
β-Elimination and alkyl-aminylation of the S-layer glycoproteins and glycopeptides.
A 200-pmol aliquot of S-layer glycoprotein (corresponding to approximately 20 μg) or 500 pmol of glycopeptides was dissolved in 50 to 100 μl of 50% aqueous isopropylamine (Sigma-Aldrich) and incubated at 50°C for 18 h (10). The amine was evaporated at 50°C in a fume hood. The protein was washed twice with 10% ammonia to evaporate residual amines followed by two washes with distilled water and dried in a SpeedVac centrifuge. The dried protein was resuspended in distilled water by ultrasonication and proteolytically digested. Routinely, 200 pmol of S-layer protein was treated with 0.2 μg of trypsin or chymotrypsin (both from Roche Diagnostics, Mannheim, Germany) in a final volume of 20 μl of 10 mM NH4HCO3 solution. Digestion was carried out at 37°C for 18 to 36 h. Peptides were desalted prior to mass spectrometric analysis on ZipTip clean-up columns (Millipore, Eschborn, Germany) according to the manufacturer's instructions.
Release of the O-linked glycan.
The reductive release of glycans was performed according to a protocol of Huang and coworkers (13), with minor modifications. A 20-μg aliquot of glycoprotein was dissolved in 10 μl of a solution containing 5 mg of NaBH4 in 1 ml of 28% ammonia and incubated at 50°C for 18 h. After drying the sample in a SpeedVac centrifuge and redissolving it in distilled water, the released oligosaccharides were purified on house-made carbon columns (29).
MS analyses. (i) Orthogonal TOF-MS.
The orthogonal time-of-flight mass spectrometry (TOF-MS) apparatus is a modified prototype machine which was described recently (9). For infrared matrix-assisted laser desorption/ionization mass spectrometry (IR-MALDI-MS), an Er:YAG laser (Speser, Spektrum Laser, Berlin, Germany) emitting pulses of ∼100-ns duration at a wavelength of 2.94 μm was used. Analysis was performed in positive ion mode, and mass spectra were processed using the MoverZ3 software (version 2001.02.13; Genomic Solutions, Ann Arbor, MI). The S-layer protein (50 pmol/μl) was dissolved in 8 M urea, and 0.5 μl of the respective sample was premixed with glycerol and then spotted on a glass plate. Mixtures with different sample-to-matrix ratios (5:1, 1:1, 1:5, 1:10, and 1:50 [vol/vol]) were examined. A modified MALDI sample plate containing a 1.2-mm-thick groove was used to accommodate the glass plate.
(ii) ESI-QTOF.
Positive ion mode (+) MS of the (glyco)peptides and oligosaccharides was carried out by use of an electrospray ionization-quadrupole TOF (ESI-QTOF) instrument (Micromass, Manchester, United Kingdom). A house-made capillary puller was used for the production of nanospray glass capillaries. Capillary voltage applied on an internal wire electrode was 1,100 V; the cone voltage used for peptide analysis was 40 V, for oligosaccharides it was 60 V, and for glycopeptides it was 40 to 90 V. Peptide sequencing was conducted using argon as the collision gas. The applied collision energy was 15 to 40 V. The β-eliminated samples were dissolved in water, yielding a concentration of the stock solution of 20 pmol/μl. Four microliters of that solution was mixed with 5 μl of methanol and 1 μl of 10% formic acid. The final concentration of the β-eliminated samples was 8 pmol/μl. The final concentration of glycopeptides and glycan chains varied between 4 and 20 pmol/μl in 50% methanol containing various concentrations of formic acid (0.1 to 10%, vol/vol).
RESULTS AND DISCUSSION
Analysis of the S-layer glycoprotein of Geobacillus stearothermophilus NRS 2004/3a.
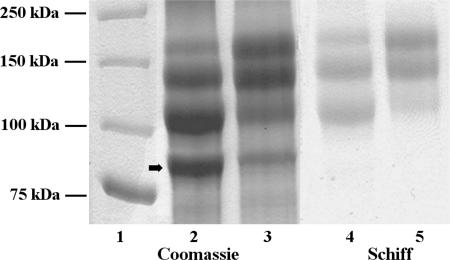
On an SDS-PA gel (Fig. 1, lanes 2 and 3), the mature S-layer glycoprotein of G. stearothermophilus NRS 2004/3a is separated into four bands with apparent molecular masses of 93, 119, 147, and 170 kDa. The three high-molecular-mass bands give a positive PAS staining reaction for carbohydrates. However, the apparent molecular masses of S-layer proteins with extended glycan chains are usually too high (16, 24, 39). The finding that the masses of nonglycosylated or chemically deglycosylated S-layer proteins correspond well with the theoretical values from protein sequencing experiments indicates that glycosylation of these proteins is responsible for the aberrant migration behavior of bacterial S-layer glycoproteins on SDS-PA gels (3, 6, 35). Upon chemical deglycosylation of the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a, a single 93-kDa band representing the nonglycosylated SgsE protein remains visible on the gel (20, 35). Notably, according to the SDS-PAGE evidence, S-layer glycoprotein preparations of cells harvested from a batch culture show an increased amount of the highest-molecular-mass glycoprotein species (Fig. 1, lanes 3 and 5, batch culture, compared to lanes 2 and 4, continuous culture).
FIG. 1.
SDS-PAGE analysis of the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a (8% gel). Lane 1, molecular mass standard (AllBlue Precision Plus Protein standard; Bio-Rad); lanes 2 and 4, S-layer glycoprotein derived from continuous culture; lanes 3 and 5, S-layer glycoprotein derived from batch culture; lanes 2 and 3, Coomassie blue staining; lanes 4 and 5, periodic acid-Schiff staining. The amounts of total protein loaded per gel lane were 10 μg in lanes 2 and 3 and 25 μg in lane 4 and 5. The arrow indicates the nonglycosylated S-layer protein protomer.
In comparison to SDS-PAGE analysis, where the migration behavior of glycoproteins on the gel is frequently influenced by the attached glycan chains, MS is a more reliable alternative for accurate mass determination of glycoproteins (12, 45). MS has also been the method of choice for the exact determination of molecular masses of various complex biological compounds, such as high-molecular-mass proteins, glycoconjugates, lipids, and organic compounds (for details, see references 8, 30, 31, 42, and 45). However, up to now, only a few S-layer glycoproteins have been analyzed directly by MS (1, 18).
For analysis of the three S-layer glycoprotein species of G. stearothermophilus NRS 2004/3a, different MS approaches have been pursued. In MALDI-MS sample preparation, the solubilized sample is mixed with various matrices and loaded onto a metal target. Since S-layers usually are poorly soluble in water, solubilization is frequently performed using GHCl (25). This solubilization procedure, however, is not directly compatible with MS analysis. Allmaier and coworkers have developed a method for investigations on the S-layer glycoprotein of Thermoanaerobacterium thermosaccharolyticum E207-71 in which GHCl has been almost completely removed from the target by gentle washing with water (1). However, this procedure did not work for the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a. Urea, another chaotropic agent, has also been reported for solubilization of S-layer proteins (25); notably, this agent has previously been successfully used as a matrix in IR-MALDI (28).
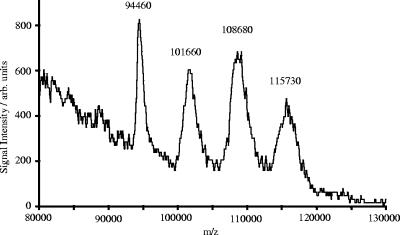
For optimizing the IR-MALDI conditions for the analysis of the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a derived from batch culture, different ratios of urea to glycerol have been tested. Glycerol is widely used as a matrix for IR-MALDI-MS, e.g., for the analysis of large proteins (2). By diluting the sample, dissolved in 8 M urea, with glycerol in a ratio of 1:5 and 1:10 (vol/vol), acquisition of spectra was possible. The other concentrations (5:1, 1:1, and 1:50 [vol/vol]) only produced at most minor analyte intensities. Figure 2 displays the mass spectrum acquired from the 1:10-diluted sample. The average masses of the four ion signals were determined to be 94.46, 101.66, 108.68, and 115.73 kDa, respectively. Taking into account that both urea and glycerol are known to form analyte-matrix adducts with the sample, which results in an increase of the apparent mass of the protein, and the reduced mass resolution in the high-mass range, the average mass of the first ion peak (94.46 kDa) is in good agreement with the calculated theoretical molecular mass of the mature S-layer protein (93.68 kDa; mass deviation, 0.83%). The average mass differences between two neighboring peaks of the singly charged ions were calculated to be 7.09 kDa, which corresponds to a glycan chain composed of 15 tri-rhamnose repeating units with an average molecular mass of 7.05 kDa (mass deviation, 0.57%). Relation of these results to the SDS-PAGE evidence, where three S-layer glycoprotein species are visible, supports the hypothesis of three glycosylation sites on the SgsE protein of G. stearothermophilus NRS 2004/3a. The first peak (94.46 kDa) would originate from the nonglycosylated protein, the second peak (101.66 kDa), differing in the mass of a single glycan chain, implies a single glycosylation site, the third peak (108.68 kDa) corresponds to SgsE with two occupied glycosylation sites, and the fourth peak (115.73 kDa) would represent triply glycosylated SgsE.
FIG. 2.
(+)IR-MALDI-orthogonal TOF mass spectrum of the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a. The spectrum shows the average mass of the nonglycosylated species of SgsE at 94.46 kDa and those of the three inherently heterogeneic glycoprotein species at 101.66 kDa, 108.68 kDa, and 115.73 kDa.
Glycosylation sites of G. stearothermophilus NRS 2004/3a.
In a previous study, only two glycosylation sites of the SgsE precursor were determined, namely, at positions threonine-620 and serine-794 (35). A third glycosylation site could not be detected, most probably because in the S-layer glycopeptides obtained after a papain digest of S-layer glycoprotein from continuous-culture biomass (35), the highest-molecular-mass band, in which three sites are proposed to be glycosylated, was of very low abundance. Thus, that material was obviously not sufficient for NMR analysis of the third glycosylation site.
In the present study, β-elimination followed by Michael addition of isopropylamine was used to replace the glycan chains at the respective glycosylation sites of the S-layer glycoprotein by an alkylamine. After proteolytic digestion, the peptides were analyzed by nano-ESI-QTOF tandem MS (MS/MS). The alkylaminylated peptides were identified by the observed mass shift in comparison to the theoretical mass of the nonglycosylated peptide. By this method, however, only the serine glycosylation site could be unambiguously identified. Threonine is known to be less reactive under alkaline conditions due to the presence of a methyl group, which might protect the β-carbon against nucleophilic attack (5, 17, 32). On the other hand, stronger alkaline reaction conditions give rise to a number of side reactions, which make the unequivocal identification of the glycosylation site(s) impossible (38).
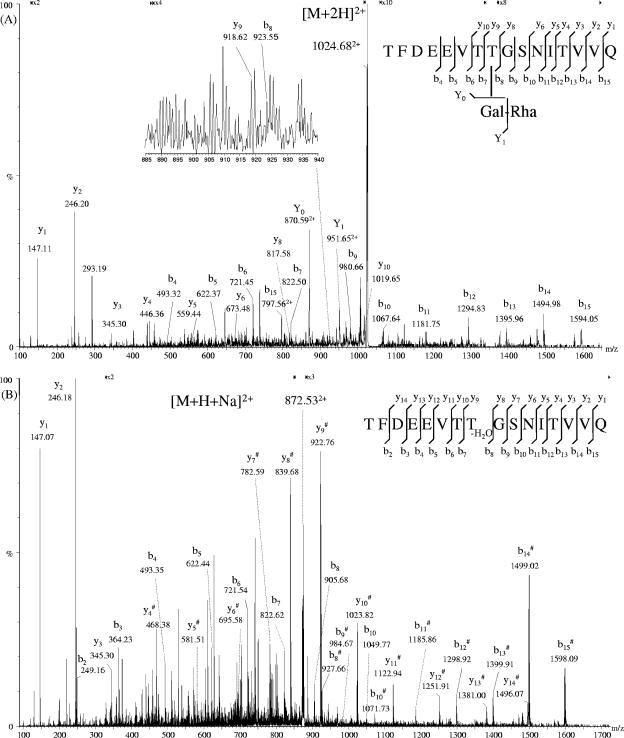
Despite these drawbacks, we succeeded in determining the position of the third glycosylation site by analyzing various glycopeptide fractions available from a previous papain digest (35) by nano-ESI-QTOF, either directly or after β-elimination. In one fraction, a new glycopeptide species was detected, but the abundance of the respective high-molecular-mass glycopeptide ions [M + 2H+Na+K]4+ with m/z = 2,102.96 and m/z = 2,212.31, corresponding to the theoretical average mass of the peptide TFDEEVTTGSNITVVQ with 14 and 15 repeating units, respectively, was too low to perform MS/MS (data not shown). When choosing cone conditions, which favor in-source fragmentation, a short glycopeptide with one galactose and one rhamnose residue attached to the peptide moiety was isolated as doubly charged ions with m/z of 1,024.68 and subjected to a virtual MS3 experiment (Fig. 3A). In the resulting spectrum, the loss of one rhamnose residue followed by the loss of one galactose residue was clearly displayed by the doubly charged ions with m/z of 951.65 and 870.59, respectively. The identity of the peptide portion was proven by a number of y and b ions and by sequence comparison with the already-known sequence of SgsE. Unfortunately, loss of the glycan is the first fragmentation event to occur on the glycopeptide. Thus, due to the presence of four threonine residues and one serine residue in the peptide, an unambiguous identification of the glycosylation site was impossible. The same glycopeptide fraction was also subjected to β-elimination with isopropylamine. Several doubly charged peptides resulting from complete and incomplete Michael addition of isopropylamine to the peptide TFDEEVTTGSNITVVQ were detected as nonsodiated and sodiated species. In Fig. 3B, the CID spectrum derived from the ion [M + H+Na-H2O]2+ with m/z of 872.53 is displayed; the additionally obtained CID spectra of the peptide ions [M + 2H+IA]2+ with m/z of 891.08 and [M + 2H-H2O]2+ with m/z of 861.55 are not shown. The position of the glycosylation site has been determined by the increment mass differences of 83.08 and 83.06 Da between ions y8# and y9# and b7 and b8, respectively. This shift corresponds to the mass of a β-methyldehydroalanine residue replacing a threonine residue. β-Methyldehydroalanine is the reaction product of the β-elimination reaction of threonine without Michael addition of isopropylamine. Except for the N-terminal amino acid, full sequence coverage of that peptide has been achieved by MS/MS. By comparing the analyses of both the glycopeptide and the β-eliminated peptide, the third glycosylation site could be unambiguously demonstrated to be threonine-590 of the SgsE precursor of G. stearothermophilus NRS 2004/3a.
FIG. 3.
(A) (+)Nano-ESI-QTOF MS/MS spectrum of the doubly charged ions at m/z = 1,024.68, representing glycopeptide TFDEEVTTGSNITVVQ with one galactose and one rhamnose attached to the peptide backbone. (B) (+)Nano-ESI-QTOF MS/MS spectrum of the doubly charged ions at m/z = 872.53 originating from glycopeptide TFDEEVTTGSNITVVQ after β-elimination. The glycosylation site is displayed by the y9 ion and the b8 ion and a loss of water from the threonine-590 in the sequence of SgsE. #, sodiated ions.
Core variability.
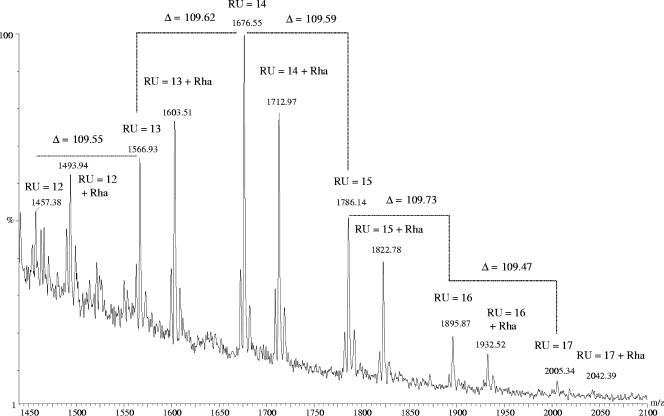
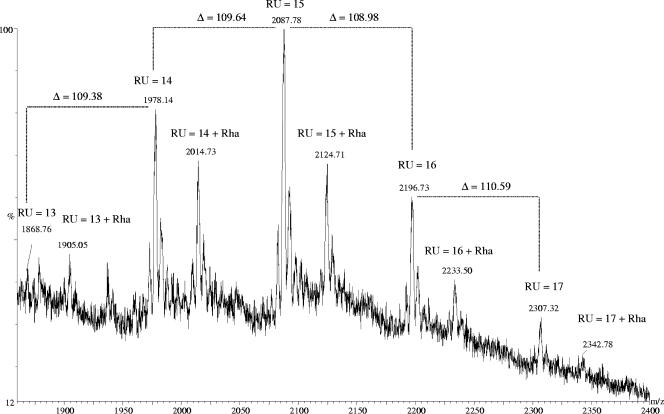
The released S-layer glycan chains from G. stearothermophilus NRS 2004/3a were analyzed by nano-ESI-QTOF-MS. The peak distribution correlates with the theoretical average masses of a repeating unit variation between 12 and 18. The most abundant peaks belong to the series [M + 2H+2K]4+ and [M + 3H+3K]6+. Both series are accompanied by two series of ions containing different numbers of Na+ and K+ ions. Most interestingly, not only the series corresponding to a glycan chain with a varied number of repeating units (designated as glycoform) with different associated adduct ions was found, but also a second series, which contained an additional rhamnose residue (Table 1 and Fig. 4). Due to the impossibility of obtaining MS/MS spectra of the second-series ions, no direct conclusion can be drawn as to whether this additional rhamnose residue is in a branched or in a linear linkage. However, considering the core structures from other S-layer glycoprotein glycans that are composed of rhamnose residues (23), it is conceivable that the additional rhamnose in the core of the G. stearothermophilus NRS 2004/3a S-layer glycoprotein glycan is a linear, α-1,3-linked sugar residue. The distribution of chain length was confirmed by the analysis of several purified glycopeptides derived from a previous papain digest of the S-layer glycoprotein (35). Nano-ESI-QTOF-MS analysis of the glycopeptides revealed charge states from +4 to +6. Different combinations of charge-specifying adducts with hydrogen, sodium, and potassium generate multiple peaks of the same charge state. Considering these different charge-specifying adducts, the masses derived from the experimentally acquired data were calculated and compared with the theoretical average masses, e.g., for the glycopeptide species with the sequence ATLTSADVIRVD (Table 2). The mass spectrum of the glycopeptide ATLTSADVIRVD carrying a single glycan chain with a distribution of 13 to 17 repeating units and furthermore with an additional rhamnose is displayed in Fig. 5.
TABLE 1.
Nano-ESI-QTOF MS analysis of oligosaccharides released from the S-layer glycoprotein of G. stearothermophilus NRS 2004/3aa
| RUb |
m/z
|
|||||
|---|---|---|---|---|---|---|
| [M + 2H+Na+K]4+
|
[M + 2H+2K]4+
|
[M + H+Na+2K]4+
|
||||
| Theorc | Exptl | Theorc | Exptl | Theorc | Exptl | |
| 12 | 1,453.43 | NDd | 1,457.46 | 1,457.38 | 1,462.96 | 1,462.77 |
| 12 + Rha | 1,489.97 | 1,489.94 | 1,494.00 | 1,493.94 | 1,499.49 | 1,499.36 |
| 13 | 1,563.04 | 1,563.07 | 1,567.07 | 1,566.93 | 1,572.56 | 1,571.78 |
| 13 + Rha | 1,599.58 | 1,599.70 | 1,603.60 | 1,603.51 | 1,609.10 | 1,609.13 |
| 14 | 1,672.65 | 1,672.65 | 1,676.68 | 1,676.55 | 1,682.17 | 1,681.90 |
| 14 + Rha | 1,709.18 | 1,709.13 | 1,713.21 | 1,712.97 | 1,718.71 | 1,718.48 |
| 15 | 1,782.26 | 1,782.22 | 1,786.28 | 1,786.14 | 1,791.78 | 1,791.82 |
| 15 + Rha | 1,818.79 | 1,818.96 | 1,822.82 | 1,822.78 | 1,828.31 | 1,827.90 |
| 16 | 1,891.86 | 1,891.47 | 1,895.89 | 1,895.87 | 1,901.39 | 1,901.15 |
| 16 + Rha | 1,928.40 | 1,928.13 | 1,932.43 | 1,932.52 | 1,937.92 | 1,937.80 |
| 17 | 2,001.47 | ND | 2,005.50 | 2,005.34 | 2,010.99 | ND |
| 17 + Rha | 2,038.01 | ND | 2,042.03 | 2,042.39 | 2,047.53 | ND |
Data show the theoretical (Theor) and experimentally determined (Exptl) average m/z values of quadruply charged ions containing different numbers of Na+ and K+ ions.
RU designates the number of trisaccharide repeating units, with the terminal rhamnose of the repeating unit at the nonreducing end being O-methylated. The core structure consists of two rhamnoses and one galactose. An additional rhamnose in the core is indicated by “+ Rha.”
Theoretical molecular masses were calculated using average masses.
ND, not determined.
FIG. 4.
(+)Nano-ESI-QTOF mass spectrum of the released glycan chains of the S-layer glycoprotein from G. stearothermophilus NRS 2004/3a. The main peaks correspond to [M + 2H+2K]4+. Chain lengths vary between of 12 and 17 repeating units (RU) for both the glycan structure with two core rhamnoses (first series; RU = X) and the one with three core rhamnoses (second series; RU = X + Rha).
TABLE 2.
NanoESI-QTOF MS analysis of the S-layer glycopeptide ATLTSADVIRVD of G. stearothermophilus NRS 2004/3aa
| RUb | Mass
|
|
|---|---|---|
| M theorc | M exptl | |
| 13 | 7,428.44 | NDd |
| 13 + Rha | 7,574.59 | ND |
| 14 | 7,866.87 | 7,866.25 |
| 14 + Rha | 8,013.01 | 8,013.41 |
| 15 | 8,305.30 | 8,305.05 |
| 15 + Rha | 8,451.44 | 8,450.85 |
| 16 | 8,743.73 | 8,743.57 |
| 16 + Rha | 8,889.87 | 8,890.21 |
| 17 | 9,182.16 | 9,181.53 |
| 17 + Rha | 9,328.30 | ND |
Data show the theoretical (Theor) and experimentally determined (Exptl) average molecular masses.
RU designates the number of trisaccharide repeating units, with the terminal rhamnose of the repeating unit at the nonreducing end being O-methylated. The core structure consists of two rhamnoses and one galactose. An additional rhamnose in the core is indicated by “+ Rha.”
Theoretical molecular masses were calculated using average masses.
ND, not determined.
FIG. 5.
(+)Nano-ESI-QTOF mass spectrum of glycopeptides belonging to the peptide ATLTSADVIRVD derived from the S-layer glycoprotein of G. stearothermophilus NRS 2004/3a. The most abundant peaks derive from overlapping peaks of the two molecular ion series [M + 3H+K]4+ and [M + 2H+2Na]4+; all glycopeptides carry a single glycan chain attached to serine-794. The number of repeating units (RU) varies between 13 and 17.
Since previous NMR analyses of the S-layer glycan chain from G. stearothermophilus NRS 2004/3a revealed that the α-1,3-linked rhamnose residue of the terminating trisaccharide repeating unit at the nonreducing end is O-methylated at carbon 2 (35), it is more likely that the newly identified additional rhamnose residue represents core variation rather than structural variation at the nonreducing end of the glycan chain. This further implies that in the S-layer glycoprotein glycan structure of G. stearothermophilus NRS 2004/3a, the already-known core region →3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→ (35) can be optionally extended by one additional rhamnose residue. Core variability has already been observed in S-layer glycoprotein glycans of Aneurinibacillus thermoaerophilus DSM 10155 (44) and A. thermoaerophilus GS4-97 (34).
Conclusions.
In general, S-layer glycoproteins are regarded as promising tools for nanobiotechnological applications (33, 37) because (i) they represent natural protein self-assembly systems, (ii) they can be tuned for distinct purposes through the addition of functional peptide or protein domains by genetic engineering methods, and (iii) naturally occurring glycosylation sites and attached S-layer glycan chains, which may be eventually rationally modified for certain applications by carbohydrate engineering, add a new and very valuable dimension to this S-layer protein-based molecular construction kit. A detailed understanding of the naturally occurring S-layer glycoprotein is a prerequisite for the envisaged nanobiotechnological applications of S-layer glycoproteins (e.g., carbohydrate vaccines or receptor mimics).
In the present study, a thorough analysis of an S-layer protein glycosylation pattern has been performed using G. stearothermophilus NRS 2004/3a as a model system. This organism has been chosen because the structural and biosynthetic knowledge about its S-layer glycoprotein is most advanced in our laboratory (20, 27, 35). So far, the lack of adequate analytical techniques has prevented a conclusive interpretation of the multiple banding pattern of this S-layer glycoprotein as observed on SDS-PA gels. Optimization and adaptation of MS methods to the water-insoluble S-layer glycoprotein (this study) allowed (i) determination of the average masses of the three inherently heterogeneic glycoprotein species of SgsE to be 101.66 kDa, 108.68 kDa, and 115.73 kDa, corresponding to SgsE with different numbers of attached glycan chains, (ii) clear assignment of nanoheterogeneity to each glycan chain, with each of them revealing the most prevalent variation between 12 and 18 trisaccharide repeating units and the possibility of extension of the already-known di-rhamnose core region by one additional rhamnose residue, and (iii) unambiguous identification of a third glycosylation site on the 93-kDa SgsE S-layer protein, namely, at position threonine-590, in addition to the known sites of threonine-620 and serine-794. These data lead to the current interpretation that in the 101.66-kDa glycoprotein species only one glycosylation site is occupied, in the 108.68-kDa glycoprotein species two glycosylation sites are occupied, and in the 115.73-kDa glycoprotein species three glycosylation sites are occupied. Future efforts will be directed towards identifying which glycosylation sites are used in the 101.66-kDa and in the 108.68-kDa glycoprotein species.
These data clearly support the high in vivo potential for diversification of bacterial S-layer glycoproteins in general.
Acknowledgments
We thank Andrea Scheberl and Sonja Zayni (Zentrum für NanoBiotechnologie) for excellent technical assistance.
This work was supported by the Austrian Science Fund, projects P15840-B10 and P18013-B10 (to P.M.), the scholarship “International Communication” of the Österreichische Forschungsgemeinschaft (to K.S.), and the scholarship “Stipendium für kurzfristige wissenschaftliche Arbeiten im Ausland” of the Universität für Bodenkultur Wien (to K.S.).
Footnotes
Published ahead of print on 8 September 2006.
REFERENCES
- 1.Allmaier, G., C. Schäffer, P. Messner, U. Rapp, and F. J. Mayer-Posner. 1995. Accurate determination of the molecular weight of the major surface layer protein isolated from Clostridium thermosaccharolyticum by time-of-flight mass spectrometry. J. Bacteriol. 177:1402-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkenkamp, S., C. Menzel, M. Karas, and F. Hillenkamp. 1997. Performance of infrared matrix-assisted laser desorption/ionization mass spectrometry with lasers emitting in the 3 μm wavelength range. Rapid Commun. Mass Spectrom. 11:1399-1406. [Google Scholar]
- 3.Bock, K., J. Schuster-Kolbe, E. Altman, G. Allmaier, B. Stahl, R. Christian, U. B. Sleytr, and P. Messner. 1994. Primary structure of the O-glycosidically linked glycan chain of the crystalline surface layer glycoprotein of Thermoanaerobacter thermohydrosulfuricus L111-69. Galactosyl tyrosine as a novel linkage unit. J. Biol. Chem. 269:7137-7144. [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Byford, M. F. 1991. Rapid and selective modification of phosphoserine residues catalysed by Ba2+ ions for their detection during peptide microsequencing. Biochem. J. 280:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christian, R., G. Schulz, J. Schuster-Kolbe, G. Allmaier, E. R. Schmid, U. B. Sleytr, and P. Messner. 1993. Complete structure of the tyrosine-linked saccharide moiety from the surface layer glycoprotein of Clostridium thermohydrosulfuricum S102-70. J. Bacteriol. 175:1250-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christian, R., G. Schulz, F. M. Unger, P. Messner, Z. Küpcü, and U. B. Sleytr. 1986. Structure of a rhamnan from the surface-layer glycoprotein of Bacillus stearothermophilus strain NRS 2004/3a. Carbohydr. Res. 150:265-272. [DOI] [PubMed] [Google Scholar]
- 8.Cristoni, S., and L. R. Bernardi. 2003. Development of new methodologies for the mass spectrometry study of bioorganic macromolecules. Mass Spectrom. Rev. 22:369-406. [DOI] [PubMed] [Google Scholar]
- 9.Dreisewerd, K., J. Müthing, A. Rohlfing, I. Meisen, Z. Vukelić, J. Peter-Katalinić, F. Hillenkamp, and S. Berkenkamp. 2005. Analysis of gangliosides directly from thin-layer chromatography plates by infrared matrix-assisted laser desorption/ionization orthogonal time-of-flight mass spectrometry with a glycerol matrix. Anal. Chem. 77:4098-4107. [DOI] [PubMed] [Google Scholar]
- 10.Hanisch, F. G., M. Jovanovic, and J. Peter-Katalinić. 2001. Glycoprotein identification and localization of O-glycosylation sites by mass spectrometric analysis of deglycosylated/alkylaminylated peptide fragments. Anal. Biochem. 290:47-59. [DOI] [PubMed] [Google Scholar]
- 11.Hart, C., B. Schulenberg, T. H. Steinberg, W. Y. Leung, and W. F. Patton. 2003. Detection of glycoproteins in polyacrylamide gels and on electroblots using Pro-Q Emerald 488 dye, a fluorescent periodate Schiff-base stain. Electrophoresis 24:588-598. [DOI] [PubMed] [Google Scholar]
- 12.Harvey, D. J. 2005. Proteomic analysis of glycosylation: structural determination of N- and O-linked glycans by mass spectrometry. Expert Rev. Proteomics 2:87-101. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., T. Konse, Y. Mechref, and M. V. Novotny. 2002. Matrix-assisted laser desorption/ionization mass spectrometry compatible beta-elimination of O-linked oligosaccharides. Rapid Commun. Mass Spectrom. 16:1199-1204. [DOI] [PubMed] [Google Scholar]
- 14.Küpcü, Z., L. März, P. Messner, and U. B. Sleytr. 1984. Evidence for the glycoprotein nature of the crystalline cell wall surface layer of Bacillus stearothermophilus strain NRS2004/3a. FEBS Lett. 173:185-190. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lechner, J., and M. Sumper. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262:9724-9729. [PubMed] [Google Scholar]
- 17.Mattila, K., J. Siltainsuu, L. Balaspiri, M. Ora, and H. Lonnberg. 2005. Derivatization of phosphopeptides with mercapto- and amino-functionalized conjugate groups by phosphate elimination and subsequent Michael addition. Org. Biomol. Chem. 3:3039-3044. [DOI] [PubMed] [Google Scholar]
- 18.Mauri, P. L., P. G. Pietta, A. Maggioni, M. Cerquetti, A. Sebastianelli, and P. Mastrantonio. 1999. Characterization of surface layer proteins from Clostridium difficile by liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 13:695-703. [DOI] [PubMed] [Google Scholar]
- 19.Messner, P. 2004. Prokaryotic glycoproteins: unexplored but important. J. Bacteriol. 186:2517-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messner, P., D. Pum, and U. B. Sleytr. 1986. Characterization of the ultrastructure and the self-assembly of the surface layer of Bacillus stearothermophilus strain NRS 2004/3a. J. Ultrastruct. Mol. Struct. Res. 97:73-88. [DOI] [PubMed] [Google Scholar]
- 21.Messner, P., R. Christian, C. Neuninger, and G. Schulz. 1995. Similarity of “core” structures in two different glycans of tyrosine-linked eubacterial S-layer glycoproteins. J. Bacteriol. 177:2188-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messner, P., F. Hollaus, and U. B. Sleytr. 1984. Paracrystalline cell wall surface layers of different Bacillus stearothermophilus strains. Int. J. Syst. Bacteriol. 34:202-210. [Google Scholar]
- 23.Messner, P., and C. Schäffer. 2003. Prokaryotic glycoproteins. Fortschr. Chem. Org. Naturst. 85:51-124. [DOI] [PubMed] [Google Scholar]
- 24.Messner, P., J. Schuster-Kolbe, C. Schäffer, U. B. Sleytr, and R. Christian. 1993. Glycoprotein nature of select bacterial S-layers, p. 95-107. In T. J. Beveridge and S. F. Koval (ed.), Advances in bacterial paracrystalline surface layers. Plenum Publishing Corp., New York, N.Y.
- 25.Messner, P., and U. B. Sleytr. 1988. Separation and purification of S-layers from gram-positive and gram-negative bacteria, p. 97-104. In I. Hancock and I. R. Poxton (ed.), Bacterial cell surface techniques. John Wiley & Sons, Chichester, United Kingdom.
- 26.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51:433-446. [DOI] [PubMed] [Google Scholar]
- 27.Novotny, R., C. Schäffer, J. Strauss, and P. Messner. 2004. S-layer glycan-specific loci on the chromosome of Geobacillus stearothermophilus NRS 2004/3a and dTDP-L-rhamnose biosynthesis potential of G. stearothermophilus strains. Microbiology 150:953-965. [DOI] [PubMed] [Google Scholar]
- 28.Overberg, A., M. Karas, U. Bahr, R. Kaufmann, and F. Hillenkamp. 1990. Matrix-assisted infrared-laser (2.94 μm) desorption ionization mass-spectrometry of large biomolecules. Rapid Commun. Mass Spectrom. 4:293-296. [Google Scholar]
- 29.Packer, N. H., M. A. Lawson, D. R. Jardine, and J. W. Redmond. 1998. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj. J. 15:737-747. [DOI] [PubMed] [Google Scholar]
- 30.Peter-Katalinić, J. 2005. O-glycosylation of proteins. Methods Enzymol. 405:139-171. [DOI] [PubMed] [Google Scholar]
- 31.Pulfer, M., and R. C. Murphy. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22:332-364. [DOI] [PubMed] [Google Scholar]
- 32.Rademaker, G. J., S. A. Pergantis, L. Blok-Tip, J. I. Langridge, A. Kleen, and J. E. Thomas-Oates. 1998. Mass spectrometric determination of the sites of O-glycan attachment with low picomolar sensitivity. Anal. Biochem. 257:149-160. [DOI] [PubMed] [Google Scholar]
- 33.Schäffer, C., and P. Messner. 2004. Surface-layer glycoproteins: an example for the diversity of bacterial glycosylation with promising impacts on nanobiotechnology. Glycobiology 14:31R-42R. [DOI] [PubMed] [Google Scholar]
- 34.Schäffer, C., N. Müller, R. Christian, M. Graninger, T. Wugeditsch, A. Scheberl, and P. Messner. 1999. Complete glycan structure of the S-layer glycoprotein of Aneurinibacillus thermoaerophilus GS4-97. Glycobiology 9:407-414. [DOI] [PubMed] [Google Scholar]
- 35.Schäffer, C., T. Wugeditsch, H. Kählig, A. Scheberl, S. Zayni, and P. Messner. 2002. The surface layer (S-layer) glycoprotein of Geobacillus stearothermophilus NRS 2004/3a. Analysis of its glycosylation. J. Biol. Chem. 277:6230-6239. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 37.Sleytr, U. B., and P. Messner. 2003. Crystalline bacterial cell surface layers (S layers), p. 286-293. In M. Schaechter (ed.), Desk encyclopedia of microbiology. Elsevier Science, Inc., San Diego, Calif.
- 38.Steiner, K. 2006. Unravelling details of the S-layer glycoprotein of Geobacillus stearothermophilus NRS 2004/3a. Ph.D. thesis. University of Natural Resources and Applied Life Sciences, Vienna, Austria.
- 39.Sumper, M., E. Berg, R. Mengele, and I. Strobel. 1990. Primary structure and glycosylation of the S-layer protein of Haloferax volcanii. J. Bacteriol. 172:7111-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 41.Upreti, R. K., M. Kumar, and V. Shankar. 2003. Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 3:363-379. [DOI] [PubMed] [Google Scholar]
- 42.Vaidyanathan, S., C. L. Winder, S. C. Wade, D. B. Kell, and R. Goodacre. 2002. Sample preparation in matrix-assisted laser desorption/ionization mass spectrometry of whole bacterial cells and the detection of high mass (>20 kDa) proteins. Rapid Commun. Mass Spectrom. 16:1276-1286. [DOI] [PubMed] [Google Scholar]
- 43.Wacker, M., D. Linton, P. G. Hitchen, M. Nita-Lazar, S. M. Haslam, S. J. North, M. Panico, H. R. Morris, A. Dell, B. W. Wren, and M. Aebi. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 44.Wugeditsch, T., N. E. Zachara, M. Puchberger, P. Kosma, A. A. Gooley, and P. Messner. 1999. Structural heterogeneity in the core oligosaccharide of the S-layer glycoprotein from Aneurinibacillus thermoaerophilus DSM 10155. Glycobiology 9:787-795. [DOI] [PubMed] [Google Scholar]
- 45.Zaia, J. 2004. Mass spectrometry of oligosaccharides. Mass Spectrom. Rev. 23:161-227. [DOI] [PubMed] [Google Scholar]