Abstract
PrfA, the master regulator of LIPI-1, is indispensable for the pathogenesis of the human pathogen Listeria monocytogenes and the animal pathogen Listeria ivanovii. PrfA is also present in the apathogenic species Listeria seeligeri, and in this study, we elucidate the differences between PrfA proteins from the pathogenic and apathogenic species of the genus Listeria. PrfA proteins of L. monocytogenes (PrfALm and PrfA*Lm), L. ivanovii (PrfALi), and L. seeligeri (PrfALs) were purified, and their equilibrium constants for binding to the PrfA box of the hly promoter (PhlyLm) were determined by surface plasmon resonance. In addition, the capacities of these PrfA proteins to bind to the PrfA-dependent promoters Phly and PactA and to form ternary complexes together with RNA polymerase were analyzed in electrophoretic mobility shift assays, and their abilities to initiate transcription in vitro starting at these promoters were compared. The results show that PrfALi resembled the constitutively active mutant PrfA*Lm more than the wild-type PrfALm, whereas PrfALs showed a drastically reduced capacity to bind to the PrfA-dependent promoters Phly and PactA. In contrast, the efficiencies of PrfALm, PrfA*Lm, and PrfALi forming ternary complexes and initiating transcription at Phly and PactA were rather similar, while those of PrfALs were also much lower. The low binding and transcriptional activation capacities of PrfALs seem to be in part due to amino acid exchanges in its C-terminal domain (compared to PrfALm and PrfALi). In contrast to the significant differences in the biochemical properties of PrfALm, PrfALi, and PrfALs, the PrfA-dependent promoters of hly (PhlyLm, PhlyLi, and PhlyLs) and actA (PactALm, PactALi, and PactALs) of the three Listeria species did not significantly differ in their binding affinities to the various PrfA proteins and in their strengths to promote transcription in vitro. The allelic replacement of prfALm with prfALs in L. monocytogenes leads to low expression of PrfA-dependent genes and to reduced in vivo virulence of L. monocytogenes, suggesting that the altered properties of PrfALs protein are a major cause for the low virulence of L. seeligeri.
In Listeria monocytogenes, a gram-positive, facultative intracellular pathogen causing systemic infections in humans and animals (25, 51), most of the known virulence genes involved in internalization and intracellular replication are under the control of the central transcription factor PrfA (3, 5, 29, 32). This listerial virulence regulator belongs to the large Crp/Fnr family of transcriptional activators (28). Like most other members of this family, it acts as a dimer and binds to a specific 14-bp-long sequence of dyad symmetry (the PrfA box) (12, 13, 33, 48). In a typical functional PrfA-dependent promoter, the PrfA box (which may possess mismatches in one or two positions in comparison to the consensus sequence) is located 22 or 23 bp apart from a sigma A (SigA)-specific −10 box. A typical −35 box is normally not found, and its presence may even inhibit the activation by PrfA. Certain sequences within the interspace region (between the −10 and the PrfA boxes) or adjacent to these sequences may positively or negatively influence the functionality of a PrfA-dependent promoter (15, 30, 31).
The expression of the prfA gene is controlled by three promoters. P1, which is under the control of SigA and thermoregulated (11, 12, 20), and P2, which is SigB regulated (37, 40, 46), are located in front of prfA, while the third one, in front of the adjacent plcA gene, initiates transcription of prfA into a bicistronic mRNA. The latter promoter is controlled by PrfA, thus causing autoregulation of prfA transcription (32). In addition to this complex transcriptional control of prfA, translation of the monocistronic prfA mRNA is thermoregulated by the formation of a secondary structure in the untranslated 5′ region of this transcript which acts as a thermoswitch (20).
Furthermore, the activity of PrfA is modulated by yet unknown factors that seem to be produced under certain environmental and nutritional conditions (7, 10, 34, 35, 41, 43). The crystal structure of PrfA suggests a potential binding domain for putative effector molecules which may stabilize the helix-turn-helix domain and, thus, could enhance the binding affinity of PrfA to its DNA target site (9). The identification of several mutations in prfA leading to PrfA proteins (PrfA*) which are constitutively active and no longer modulated in their activity by external conditions (36, 42, 49, 52-54) are in line with this assumption.
In analogy to the structurally similarity of the cAMP-binding Crp of Escherichia coli to PrfA, it has been postulated that PrfA might also be activated by similar hypothetical factor(s)(1, 2, 7, 42). Purified PrfA protein seems to be functional and initiates transcription at PrfA-dependent promoters in an in vitro transcription system in the absence of an additional factor(s) (1, 27, 31). However, the requirement of additional activators or repressors of PrfA in vivo cannot be ruled out. The activity of purified PrfA* in these in vitro transcription assays was still higher than that of wild-type PrfA (31, 53), but this increased activity may simply be caused by the structural differences in the helix-turn-helix domain of the two PrfA proteins (9, 42). The PrfA proteins used in these in vitro transcription assays carried His6 or Flag tags at the N termini, and it has been argued that these tags may cause higher binding affinity and, hence, better transcriptional activation than the untagged wild-type PrfA. Nevertheless, these findings seem to favor the idea that modulation of PrfA activity is brought about by negatively acting effector molecules whose removal may lead to full activity of PrfA more than that it is brought about by molecules activating an initially inactive PrfA. More recent data also seem to favor this assumption (10, 19; S. Mertins, unpublished data).
The occurrence of the prfA gene is not restricted to virulent L. monocytogenes isolates, as its presence together with an entire prfA gene cluster has been also demonstrated in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, an apathogenic Listeria species (4, 16, 24, 45). Evidence based on previous studies suggests that the activities of PrfA of L. ivanovii and L. seeligeri may be different from that of L. monocytogenes (4, 14, 23).
To obtain a deeper insight into the properties of these PrfA proteins, we purified the proteins of these Listeria species and determined their binding affinity to their specific target sequences (PrfA boxes) and their capacities to promote transcription initiation at various PrfA-dependent promoters of L. monocytogenes, L. ivanovii, and L. seeligeri. We also compared binding and transcriptional activation capacities of purified untagged and His6-tagged PrfA and PrfA* proteins of L. monocytogenes. The results reveal significant differences between these PrfA proteins which could shed light on the evolution of pathogenic Listeria bacteria.
MATERIALS AND METHODS
General techniques.
PCR amplifications, cloning procedures, isolation of chromosomal DNA, and DNA manipulations were carried out according to standard procedures (44). Cycle sequencing was performed using the CEQ Dye Terminator Cycle Sequencing Quick Start kit (Beckman Coulter), and sequencing reactions were run on an XL2000 Beckman Coulter sequencer. The Listeria homepage of the Institut Pasteur (http://genolist.pasteur.fr/ListiList/) and the NCBI database (http://www.ncbi.nlm.nih.gov/) were used for sequence comparison. All oligonucleotides used for PCR amplification were synthesized by Sigma-Genosys and are listed in Table 1. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining were performed according to standard protocols (26).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a | Application |
|---|---|---|
| m-prfA-Nde | ATTGGGGGATGACATATGAACGC | Purification of untagged PrfA and PrfA* proteins of L. monocytogenes |
| m-prfA-Bgl | ATGAAAATGCTTCTGTAAGATCTTTATTCG | |
| i-prfA-Bam | TAAAAACGATTGGGGGATCCAAAATGGACG | Purification of His6-tagged PrfA of L. ivanovii (PrfALi) |
| i-prfA-Kpn | TCCTGTTTTTTTTGGTACCGTAGTATTTAATTG | |
| s-prfA-Bam | GGAGGATGACGATGGATCCACTAAAGC | Purification of His6-tagged PrfA of L. seeligeri (PrfALs) |
| s-prfA-Pst | ACAATTAATAAAAAGCGCTGCAGGGTC | |
| s-prfA-rkmb2 | TGAATTTTTATACACAATGACGTTTTCTTGTTTTAG | Purification of His6-tagged hybrid sm-PrfA |
| m-prfA-rkmb | CAAGAAAACGTCATTGTGTATAAAAATTCATGCTT | |
| m-prfA-Pst | TGAAAATGCTTCTGTAAGTTCTGCAGTCG | |
| m-actA1 | GAAGATGCTTCTAGAAAAGTTGCTGA | In vitro transcription assay with L. monocytogenes actA promoter (PactALm) |
| m-actA2 | CTCCCTCCTCGGAATTCGCTAA | |
| i-actA1 | ATGGGAAGAATTCGGCGTTAGATAAGGG | In vitro transcription assay with L. ivanovii actA promoter (PactALi) |
| i-actA2 | AATCTATCTAGGATCCCTTTTTTACTCC | |
| s-actA1 | GAAGTTGTCTAGAAAATTGCCGAAGCATGGC | In vitro transcription assay with L. seeligeri actA promoter (PactALs) |
| s-actA2 | GTTAATAGAAATCAGGATCCTAAAATCTACTGAC | |
| m-hly1 | CTGTTTTTAATTGACTGCAGTTTTCCGGC | In vitro transcription assay with L. monocytogenes hly promoter (PhlyLm) |
| m-hly2 | CAGTTATTAAGCGAATAAGCTTTTCCGCC | |
| i-hly1 | TGTAATAAAAACGAGCTCTATTTTTTTCATGG | In vitro transcription assay with L. ivanovii hly promoter (PhlyLi) |
| i-hly2 | TGTCATTAAAAGTAGAATTCTTTTTTTCATGG | |
| s-hly1 | TAGATGAAAAAGCTTTCCCGTGCATAGG | In vitro transcription assay with L. seeligeri hly promoter (PhlyLs) |
| s-hly2 | AACGACATGATATCTAGACCAAATATTTTC | |
| m-actA3 | GATGCTTCTAGAAAAGTTGCTGAAGC | EMSA with PactALm |
| m-actA4 | TATTCTAGAATTATTTTAAGAATATCA | |
| i-actA3 | CATGGGAAGAAGTTGGCGTTAGAT | EMSA with PactALi |
| i-actA4 | CAATTCATGAATTGCTTCAAGAATAGCA | |
| s-actA3 | CATGGCTCGGTGTAGGTATTTC | EMSA with PactALs |
| s-actA4 | ATTCATGAATTATTTACAGAATAACAC | |
| m-hly3 | TCCTATCTAGAAGTGACTTTTATGTT | EMSA with PhlyLm |
| m-hly4 | GCTTCTCTAGATGAAACGCAATATTA | |
| i-hly3 | GCCTATCTAGAAAATAATTTCACATC | EMSA with PhlyLi |
| i-hly4 | CTTCCAAGATTTAGAAACGCAATATTA | |
| s-hly3 | GTCTTATCTAGATCAACATTAACATA | EMSA with PhlyLs |
| s-hly4 | CGATTCATTAGCTTGAAACGCAATATTA | |
| SPR-hly1 | TGAGGCATTAACATTTGTTAACGACGAT | SPR with PrfA box of PhlyLm |
| SPR-hly2 | ATCGTCGTTAACAAATGTTAATGCCTCA | |
| SPR-ctrl1 | TGAGGCAATGGCATAGGCAACCGACGAT | SPR with nonspecific DNA for control |
| SPR-ctrl2 | ATCGTCGGTTGCCTATGCCATTGCCTCA | |
| prfA_Salup> | CAATAAAGTCGACTAACATATATTATTCCTAC | Allelic replacement of prfALm with prfALs in L. monocytogenes EGD-e (upstream region) |
| <prfA_Alwup | TCTTCTGCTTGAGTGCTCACGTCTCATCC | |
| s-prfA_bnt> | GTGAGCACTAAAGCAGAGGATTTTAAAGAATA | Allelic replacement of prfALm with prfALs in L. monocytogenes (coding region) |
| <s-prfA_Bse | TTATCCCAGCTATTATGACAAGCTAAG | |
| prfA_Bsedn> | GTCCTGCTAGCTGGGATAAATTTAATTAAATC | Allelic replacement of prfALm with prfALs in L. monocytogenes (downstream region) |
| <prfA_Bamdn | GCTAAAGGATCCCGTGAAGCTGGCGCAACAAA | |
| prfA_Salup> | CAATAAAGTCGACTAACATATATTATTCCTAC | Cloning of prfALm into L. monocytogenes EGD-e ΔprfA |
| <prfA_Bgldn | GCTAAAAGATCTCGTGAAGCTGGCGCAACAAA |
Introduced restriction sites are underlined; bold letters show overlapping sequences for recombinant PCR.
Purification of RNAP.
The purification of RNA polymerase (RNAP) was achieved via heparin-Sepharose affinity chromatography as previously described (1), with the following modification: the strain used was the mutant L. monocytogenes EGD-e ΔprfA ΔsigB (50), which additionally lacked the sigB gene. The purity of RNAP (α, β, and β′ subunits plus SigA) was about 30 to 40%, as determined by silver-stained SDS-PAGE.
Purification of untagged PrfA proteins of L. monocytogenes EGD-e and L. monocytogenes P14-A.
PCR fragments carrying the prfA and prfA* genes were amplified using the primers m-prfA-Nde and m-prfA-Bgl (Table 1) and chromosomal DNA of L. monocytogenes EGD-e wild type or L. monocytogenes P14-A (42) as the template. The purified fragments were digested with NdeI and BglII and subsequently cloned into the pET-3c expression vector (Novagen), yielding the plasmids pET-3c-PrfAnt and pET-3c-PrfA*nt. The nucleotide sequences of the amplified sequences were verified by automated sequencing. The pET-3c vectors were transformed into E. coli FT1(pLysS), and expression of the native PrfA proteins was performed as described by the manufacturer (pET system; Novagen). Bacteria were lysed in sample buffer (50 mM sodium phosphate [pH 7.0], 50 mM NaCl, 10% glycerol, Complete protease inhibitor [Roche]) by using a Fast Prep FP120 shaker (Thermo Savant). Cell debris was pelleted by centrifugation for 1 h at 10,000 rpm and 4°C; subsequently, the supernatants containing soluble proteins were filtered.
Purification of nontagged PrfALm and nontagged PrfA*Lm was achieved by cation exchange chromatography on a 1-ml HiTrap SP Fast Flow Sepharose column (Amersham Biosciences) applied to an ÄKTAprime protein purification system (Amersham Biosciences). After a washing step (50 mM sodium phosphate [pH 7.0], 50 mM NaCl, and 10% [vol/vol] glycerol), the proteins were eluted with an NaCl gradient up to 500 mM. Fractions containing the nontagged PrfALm and PrfA*Lm proteins were determined by the use of Coomassie brilliant blue-stained SDS-PAGE gels. Proteins were finally stored in 50 mM sodium phosphate (pH 7.0), 500 mM NaCl, and 10% (vol/vol) glycerol at −80°C.
Purification of His6-tagged PrfA proteins of L. monocytogenes, L. ivanovii, L. seeligeri, and hybrid PrfALsm.
PrfALm and PrfA*Lm were purified as described previously (2). PCR fragments carrying the prfA genes prfALi and prfALs were amplified using the primers listed in Table 1, and chromosomal DNA of L. ivanovii (ATCC 19119) or L. seeligeri (SLCC 3954) was used as the template. The resulting fragments were cloned into the vector pQE30 (QIAGEN), generating the vectors pQE30-i-prfA and pQE30-s-prfA. To forge the hybrid PrfALsm, two PCR fragments were generated, the first using the primers s-prfA-Bam, s-prfA-rkmb2, and pQE30-s-prfA as the template and the second using m-prfA-rkmb, m-prfA-Pst, and chromosomal DNA of L. monocytogenes EGD-e as the template. The fragments were fused by recombinant PCR and reamplified with the primer pair s-prfA-Bam and m-prfA-Pst. The product was cloned into pQE30, generating the vector pQE30-sm-prfA.
The pQE vectors were transformed into E. coli M15, and protein purification was carried out following the manufacturer's instructions (QIAGEN Expressionist). Proteins were finally stored in 50 mM Tris (pH 8.0), 150 mM NaCl, 1 mM EDTA, 2.5 mM CaCl2, 1 mM dithiothreitol (DTT), and 20% (vol/vol) glycerol at −80°C.
Preparation of PhlyLm PrfA box DNA for surface plasmon resonance (SPR) measurements.
Twenty-eight-nucleotide synthetic oligonucleotides (Table 1) containing the PhlyLm PrfA box (forward, SPR-hly1; backward, SPR-hly2) or a nonspecific DNA sequence (forward, SPR-ctrl1; backward, SPR-ctrl2) were hybridized and used for analyses without further purification. Both forward 28-nucleotide oligonucleotides are biotinylated at the 5′ end. All oligonucleotides were purchased with or without modification from MWG Biotech (Ebersberg, Germany).
Surface plasmon resonance measurements.
SPR measurements were performed using a Biacore X instrument operated at 25°C. For the kinetic analyses, 100 response units of the biotinylated DNA fragment with the PhlyLm PrfA box were immobilized on a streptavidin-coated SA sensorchip (Biacore AB, Uppsala, Sweden) in flow cell two at a flow rate of 5 μl/min. Flow cell one contained the biotinylated nonspecific DNA fragment as a reference. During immobilization and interaction analyses, HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% polysorbate) purchased from Biacore AB (Uppsala, Sweden) was used as a running buffer. The mass transport limitation was tested by the alteration of flow rates. A flow rate of 40 μl/minwas suitable to minimize mass transport, except for the case with PrfA*Lm, for all experiments. Concentrations of 0.5 to 100 nM of the PrfA proteins were used for titrations of the PhlyLm PrfA box. To regenerate the chip surface, the dissociation of the PrfA protein complex was stopped by an injection of 10 μl 1 M NaCl buffer at 20 μl/min after each injection. We fitted the resulting sensorgrams according to the 1:1 Langmuir binding model or to the 1:1 Langmuir binding model with mass transfer for PrfA*Lm by using the BIAevaluation 3.1 software. The fits showed chi-square values between 1 and 10. The titrations for the kinetic measurements were carried out twice for each PrfA protein, and the mean values and the deviations of all constants were calculated.
Electrophoretic mobility shift assay (EMSA).
The double-stranded DNA probes were obtained by PCR amplification using the primers listed in Table 1. After gel purification, the amounts of the PCR fragments were quantified (Ultrospec 2100 Pro photometer; Amersham). A total of 0.8 μg of DNA was labeled with 60 μCi of [γ-32P]ATP (6,000 Ci mmol−1; Amersham) by using the T4 polynucleotidkinase (Fermentas).
The binding reactions contained different amounts of different PrfA proteins, 5 nM of 32P-labeled DNA, 4.5 μl of 5× binding buffer (100 mM HEPES [pH 7.9], 250 mM KCl, 5 mM EDTA [pH 8.0], 5 mM DTT, 15% Ficoll), 1.5 μl poly(dI-dC) (1 μg/μl), 1.5 μl bovine serum albumin (1 μg/μl), and 3 μl MgSO4 (100 mM) in a final volume of 22.5 μl. Reaction mixtures were preincubated for 3 min at 37°C following 22 min on ice. For CI complex formation, partially purified RNAP (1.5 nM) from L. monocytogenes or an RNAP-buffer mix were added to this mixture and further incubated for 5 min at 37°C. The DNA-protein complexes were separated on native 5% polyacrylamide gels in low-ionic-strength buffer (0.4× Tris-borate-EDTA) at 250 V and 100 mA for 3.5 h at room temperature. Following electrophoresis, gels were vacuum dried and visualized by phosphorimaging (Molecular Dynamics Typhoon 9200; Amersham).
For supershift assays, anti-PrfA antiserum (with a final dilution of the antibodies of 1:15) was added after incubation with RNAP, and the binding reaction was continued for an additional 25 min on ice before the samples were loaded onto the gel.
In vitro transcription assay.
For the in vitro transcription assay, the promoter region and the adjacent sequences of the actA and hly genes of L. monocytogenes EGD-e, L. ivanovii (ATCC 19119) and L. seeligeri (SLCC 3954) were amplified by PCR using the primer pairs listed in Table 1. The purified DNA was cloned into the vector pUC18, which, after linearization, served as a template in runoff transcription assays, resulting in transcript sizes of about 135 bp.
Each reaction mix was prepared with 11 μl of a master mix (86 mM Tris, 1.15 mM EDTA, 32.5 mM MgCl2, 3.25 mM K3PO4, 0.9 mM DTT, 21% [vol/vol] glycerol, 1.1 g/liter bovine serum albumin), 1.5 μl NTP mix (3 mM [each] ATP, GTP, and UTP; no CTP), 0.5 μl of RNase inhibitor (40 U/μl, Fermentas), and 2 μl of promoter DNA template (200 nM). Fifteen microliters of this premix was mixed with 8 μl of the PrfA dilution or deionized H2O before 2 μl of the RNA polymerase dilution (∼1.5 nM) was added. After 5 min of incubation at room temperature, the reaction was started by the addition of 2 μl [α-32P]CTP (5 μCi, 3,000 Ci mmol−1; Amersham) and incubated at 37°C for 5 min before 2 μl of a heparin solution (10 g/liter) was added. After 10 min at 37°C, 2 μl of nonradioactive CTP (0.75 mM) was supplemented, and the incubation continued for another 5 min before 40 μl of the stop mix (urea and bromophenol blue) was added. The mRNA was separated on acrylamide (6% [wt/vol]) urea (8 M) gels without the samples being previously heated. Subsequent to electrophoresis, gels were vacuum dried and transcripts were visualized by phosphorimaging (Molecular Dynamics Typhoon 9200; Amersham).
Bacterial strains and growth conditions.
The E. coli strain DH5α was used for cloning and construction of the mutagenesis vectors. L. monocytogenes EGD-e strains were grown in brain heart infusion (Difco), in Luria-Bertani medium, or in chemically defined minimal medium (39) supplemented with carbohydrates for Listeria monocytogenes at 37°C. Erythromycin was used at concentrations of 5 μg/ml for L. monocytogenes and 300 μg/ml for E. coli.
Fresh stock solutions of carbohydrates (cellobiose and glycerol) were filter sterilized and added to the culture medium at a final concentration of 50 mM.
Allelic replacement of prfALm with prfALs in Listeria monocytogenes.
In-frame allelic exchange of prfA genes was performed in parental strain L. monocytogenes EGD-e ΔprfA (6) as described previously (21). To construct EGD-e prfALs, three fragments of 306 bp, 704 bp, and 317 bp were amplified using the oligonucleotide pairs prfA_Salup>/<prfA_Alwup, s-prfA_bnt>/<s-prfA_Bse, and prfA_Bsedn>/<prfA_Bamdn with chromosomal DNA of L. monocytogenes EGD-e and L. seeligeri (SLCC 3954) as templates. Fragments were then ligated via the introduced BsiHKAI and BseYI sites. After reamplification using the oligonucleotides prfA_Salup> and <prfA_Bamdn with the ligation mixture as a template, the resulting 1,288-bp fragment was cloned into pLSV101 (21) via SalI and BamHI sites, giving rise to pLSV101_s-prfA. This vector was transformed into L. monocytogenes EGD-e ΔprfA by electroporation, and erythromycin-resistant bacteria growing at 42°C that harbored the chromosomally integrated plasmid were selected. After subsequent cultivation at 30°C, erythromycin-sensitive clones were screened by PCR to identify mutants for which the second recombination step resulted in an insertion of prfALs.
For a control, the original prfA gene of L. monocytogenes EGD-e (prfALm) was inserted into the genome of L. monocytogenes EGD-e ΔprfA to get a revertant with an identical genetic background. For this purpose, a 1,288-bp fragment was amplified using the oligonucleotide pairs prfA_Salup> and <prfA_Bgldn and chromosomal DNA of L. monocytogenes EGD-e as a template. The resulting fragment was cloned into pLSV101 via SalI and BglII/BamHI, giving rise to pLSV101_m-prfA. Subsequent procedures were performed as described above.
The allelic exchanges of the two mutants were confirmed by PCR analysis and sequencing.
Determination of hemolytic activity.
Listeria monocytogenes strains were grown in minimal medium with 50 mM cellobiose or glycerol at 37°C to an optical density of 0.6 at 600 nm. The hemolytic activities in the supernatants were determined as described previously (43).
Briefly, 20 μl of culture supernatant was incubated in 1 ml of a 4% horse erythrocyte suspension for 30 min at 37°C. After incubation, the tubes were centrifuged at a relative centrifugal force (RCF) of 500 for 2 min at room temperature. The hemolytic activity was estimated by measuring absorption at 543 nm using an Ultrospec 2100 Pro photometer (Amersham).
Preparation of cellular proteins of Listeria monocytogenes strains.
Overnight cultures of L. monocytogenes were diluted 1:25 into defined minimal medium and grown to an optical density of 0.6 at 600 nm. Each culture was then centrifuged for 5 min at an RCF of 5,500 at 4°C.
For the preparation of cellular proteins (containing ActA), the pellet was washed in phosphate-buffered saline, resuspended in cold lysis buffer (1× phosphate-buffered saline with additional protease inhibitor [Roche]), and transferred into a 2 ml BLUE TUBE (Q-Biogene) filled with silica-sand. The tube was shaken six times for 30 s each at the speed setting 6.5 in a bead beater (FP120 Fast Prep cell disrupter; Thermo Savant). The cell debris was removed by centrifugation at an RCF of 20,000 for 30 min at 4°C.
Total protein concentrations were determined using a protein microassay (Bio-Rad).
SDS-PAGE and immunoblotting.
SDS-PAGE was performed according to standard protocols (26). After SDS-PAGE, cytoplasmatic proteins were blotted onto nitrocellulose membranes, and equivalent loading of the gels was controlled by PonceauS staining of the blotted membranes. Proteins were immunodetected using the following antibodies: rabbit anti-ActA (1:1,000) (38) and goat anti-rabbit horseradish peroxidase (1:10,000; Dianova).
Animals.
Female C57BL/6 mice were purchased from Harlan Winkelmann GmbH, Germany, and were used when they were between 6 and 10 weeks old. All animals were housed under specific-pathogen-free conditions at the Biocenter of the University of Würzburg. All animal experiments were approved by the government of Unterfranken (Lower Franconia) and were performed according to the German animal protection guidelines.
Infection of animals.
C57BL/6 recipient mice (groups of five animals) were intravenously infected with 5 × 103 bacteria resuspended in 100 μl endotoxin-free 0.9% NaCl. Three days postinfection, spleens and livers were collected and homogenized and dilutions (10−0 to 10−4) were plated on brain heart infusion agar to determine the amount of Listeria bacteria in the tested organs.
RESULTS
Purification of His6-tagged and untagged PrfA proteins from L. monocytogenes, L. ivanovii, and L. seeligeri and determination of their binding to the PrfA box of Phly.
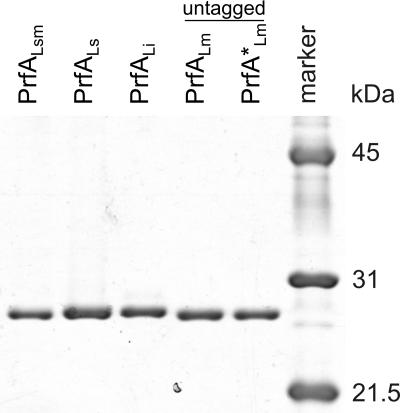
To compare the properties of the PrfA proteins of L. monocytogenes, L. ivanovii, and L. seeligeri (termed PrfALm, PrfALi, and PrfALs), we purified these three PrfA proteins and a constitutively active mutant PrfA protein (PrfA*Lm) carrying a G145S exchange (42) as recombinant proteins with an N-terminal His6 tag as previously described (2). To test whether the positively charged His6 tag may alter the binding affinity and the transcriptional activation capacity of PrfA, nontagged versions of the wild-type PrfALm protein and the mutant PrfA*Lm protein were overexpressed in E. coli by using the pET-3c vector system and also purified (Fig. 1).
FIG. 1.
Purification of His6-tagged and untagged PrfA proteins from different Listeria strains. Coomassie blue staining of an SDS-PAGE (12%) gel loaded with 1 μg of the freshly purified His6-tagged PrfA proteins from L. seeligeri (PrfALs), L. ivanovii (PrfALi), the hybrid PrfALsm, and the untagged L. monocytogenes PrfALm and PrfA*Lm proteins. A protein molecular mass marker is shown to the right.
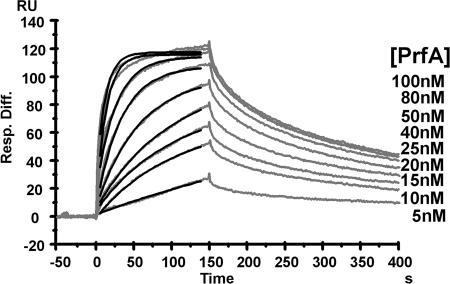
With these PrfA proteins, the kinetic and equilibrium constants of binding to target DNA were determined using a Biacore SA chip. In one flow cell, an immobilized 28-mer oligonucleotide that contained the 14-bp PrfA box of the L. monocytogenes hly promoter (PhlyLm) flanked by 7 bp of the 5′ and 3′ regions of this box was coupled to the SA chip. As a reference, in the other flow cell of the same chip, a 28-bp nonspecific DNA fragment was used. The DNA binding abilities of all purified PrfA proteins (nontagged PrfALm, nontagged PrfA*Lm, PrfALm, PrfA*Lm, PrfALi, and PrfALs) were analyzed. The kinetics of the interaction of PrfA with the PrfA box of the hly promoter could be precisely measured for nontagged PrfALm, nontagged PrfA*Lm, PrfALm, and PrfALi (Table 2). The sensorgrams and the fits of the association rate of nontagged PrfALm with the PhlyLm PrfA box are shown in Fig. 2. The equilibrium constants (KD) indicate that binding to the PrfA box of PhlyLm is basically very strong for all investigated PrfA proteins. Furthermore, the results shown in Table 2 demonstrate that nontagged PrfA*Lm DNA binding is about 50-fold stronger than that of nontagged PrfALm and that PrfALm binding to the PhlyLm PrfA box is about eightfold stronger than binding of nontagged PrfALm. As the association rate constant (ka) of nontagged PrfA*Lm DNA binding is marginally higher than the range for kinetic analysis with the Biacore X instrument, we state only an approximate value for this equilibrium constant. The KD values of PrfALm and PrfALi DNA binding are almost equal. The equilibrium constant of PrfALs could not be determined exactly, since this protein partially precipitates after dialysis in the applied running buffer. The obtained (still preliminary) data suggest, however, that the equilibrium constant of PrfALs is at least 10-fold lower than that of PrfALm. The analysis of PrfA*Lm binding to the PhlyLm PrfA box showed that the association rate is far above 107 M−1 s−1 and therefore not reliably quantifiable with the Biacore X instrument. Nevertheless, these measurements showed that the PrfA*Lm interaction with the PhlyLm PrfA box is clearly stronger than the interaction with the nontagged PrfA*Lm. Therefore, it seems that the presence of an N-terminal His6 tag increases PrfA binding to the PhlyLm PrfA box.
TABLE 2.
Surface plasmon resonance analysis: binding of different PrfA proteins to the PhlyLm PrfA boxa
| Protein | Rate constant
|
Equilibrium constant KD (M) | |
|---|---|---|---|
| ka (M−1 s−1) | kd (s−1) | ||
| Nontagged PrfALm | (6.1 ± 0.9) × 105 | (2.7 ± 0.1) × 10−3 | (4.6 ± 0.6) × 10−9 |
| Nontagged PrfA*Lm | (∼1.9 ± 0.3) × 107 | (1.6 ± 0.3) × 10−3 | (∼8.2 ± 0.6) × 10−11 |
| PrfALm | (2.2 ± 0.1) × 106 | (1.3 ± 0.2) × 10−3 | (6 ± 1) × 10−10 |
| PrfALi | (3.7 ± 0.1) × 106 | (1.2 ± 0.1) × 10−3 | (3.6 ± 0.1) × 10−10 |
Rate and equilibrium constants of nontagged PrfALm, nontagged PrfA*Lm, PrfALm, and PrfALi binding to the PhlyLm PrfA box. KD is the ratio of the dissociation constant (kd) to the association rate constant (ka).
FIG. 2.
SPR analysis of nontagged PrfALm-PhlyLm PrfA box binding. The graph shows sensorgrams (gray) from a titration of coupled PhlyLm PrfA box DNA and nontagged PrfALm. The applied PrfA concentrations are listed to the right; corresponding response units (RU) are shown on the y axis. The black lines represent the fits of the association phase of each sensorgram. Resp. Diff., response difference.
Determination of the binding affinity of the PrfA proteins with and without RNA polymerase to the PrfA-dependent promoter PhlyLm by EMSA.
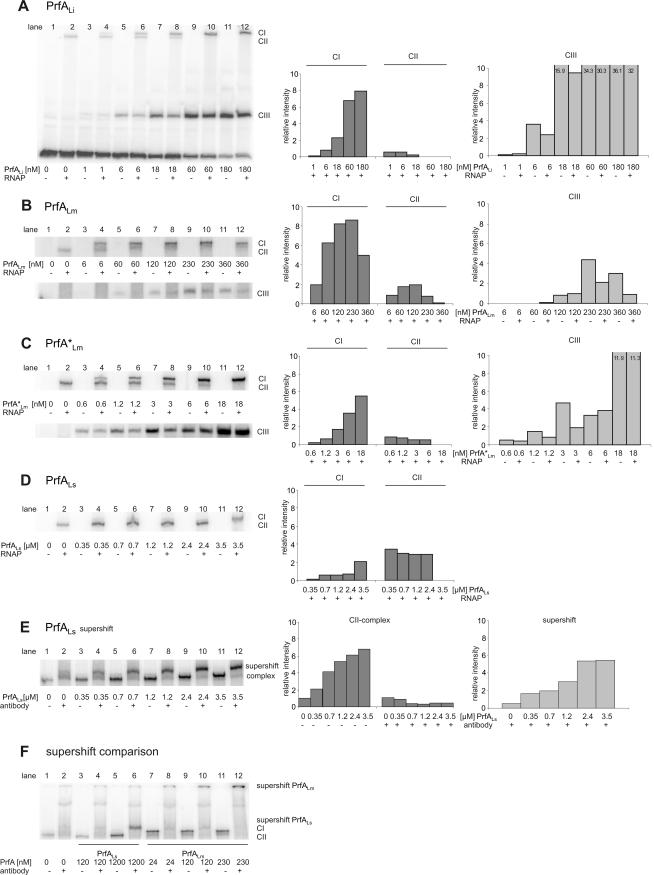
As previously described (1, 2), the CIII complex consists of PrfA bound to the PrfA box within the promoter sequence, and the ternary CI complex is composed of RNAP, PrfA, and the promoter sequence, while the CII represents a complex with RNAP bound to the promoter sequence in the absence of PrfA. The capacities of all purified PrfA proteins to form CIII and CI complexes were determined by EMSA.
For these studies, the complete hly promoter of L. monocytogenes (PhlyLm) had to be used as the target sequence, since the CI complex is formed only with the entire promoter, and previous results have shown that CIII complex formation is more efficient with the full promoter sequence than with the PrfA box alone (7). In the presence of constant amounts of PhlyLm promoter fragment (about 5 nM), CIII complex formation was readily observed with the His6-tagged PrfALi and PrfA*Lm (Fig. 3A and C, lanes 3, 5, 7, 9, and 11). With increasing concentrations of these PrfA proteins, saturation of CIII complex formation was reached at 60 nM for PrfALi and 18 nM for PrfA*Lm. The efficiency of CIII complex formation was considerably lower for His6-tagged PrfALm (230 nM) (Fig. 3B). Under these conditions, no CIII complex was detected with PrfALs, even when the PrfA protein concentration was raised up to 3.5 μM (not shown). The results obtained for the binding of PrfALm and PrfALi to Phly in the EMSA and Biacore X experiments show similar trends. However, the difference in binding affinity between PrfALm and PrfALi is less pronounced in the Biacore X assays than in the EMSAs, and this may be due to the different running buffers that had to be used for these measurements. This assumption is supported by the observation that PrfALs precipitated in the Biacore running buffer (see above) but not in the buffer used for the EMSAs.
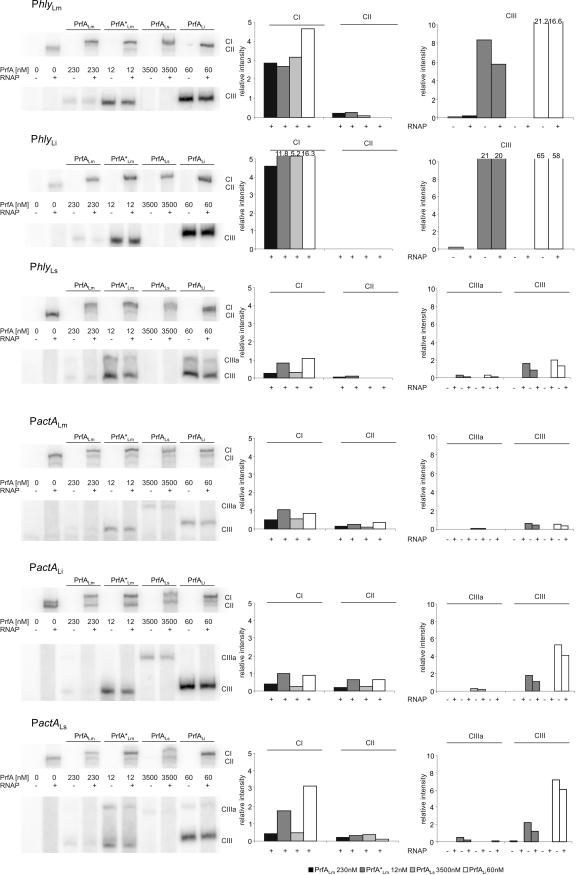
FIG. 3.
Determination of the binding affinity of the PrfA proteins with and without RNA polymerase to the PrfA-dependent promoter PhlyLm by EMSA. EMSAs were performed with purified PrfA proteins of L. ivanovii (PrfALi), L. monocytogenes (PrfALm and PrfA*Lm), and L. seeligeri (PrfALs) at various concentrations, 5 nM hly DNA promoter fragment of L. monocytogenes (PhlyLm), and 1.5 nM RNAP. CI (complex of DNA, RNAP, and PrfA), CII (complex of DNA and RNAP), and CIII (complex of DNA and PrfA) were quantified by using the ImageMaster program (Total Lab software, version 1.11; Amersham) and are shown in the graphs to the right. Lane 2 in each panel shows a PhlyLm control with L. monocytogenes RNAP. The intensity of this band is taken as 1, and all other values are normalized to it (values shown on y axis). The data shown here represent the results of one of three independently performed experiments. Supershift assays were performed with purified anti-PrfA antibodies (in a final concentration of 1:15) and increasing concentrations of PrfA protein (0.35, 0.7, 1.2, 2.4, and 3.5 μM PrfALs) (E), and a supershift comparison of PrfALs (120 and 1,200 nM) with PrfALm (24, 120, and 230 nM) was performed (F).
In parallel to the CIII complex formation, the formations of CI and CII complexes were also determined by the addition of a constant amount (1.5 nM) of partially purified L. monocytogenes RNAP to the binding assays (Fig. 3A to D, lanes 4, 6, 8, 10, and 12). At this RNAP concentration, saturation of CI complex formation (defined as the point where the entire CII complex, indicative of free RNAP bound to the promoter fragment, was completely shifted to CI) with PrfALi, PrfALm, and PrfA*Lm was reached at concentrations of 60 nM, 230 nM, and 6 nM, respectively, i.e., at roughly the same concentrations observed for the saturation of the corresponding CIII complexes (Fig. 3A to C). Especially with PrfALm, we repeatedly observed at concentrations above saturation a decrease in the amount of CI and CIII complexes, which may be due to the formation of higher PrfA aggregates. These PrfA oligomers may no longer bind to the PrfA box and/or to RNAP.
With increasing PrfALs concentrations, a band whose migration rate did not differ from that of the CII band (formed with the added RNAP alone) but whose intensity increased with increasing amounts of PrfALs (Fig. 3D) was formed, suggesting that PrfALs is involved in the formation of this band. We therefore performed supershift assays using purified anti-PrfA antibodies and observed an up-shifted band which was now clearly separated from the CII band (Fig. 3E). Based on this supershifted CI band, we conclude that saturation of CI formation with PrfALs was reached at 3.5 μM s-PrfA protein (Fig. 3D), i.e., at a concentration almost 500-fold higher than that of PrfA*Lm.
Interestingly, the supershifted band obtained with PrfALs migrates at a significantly higher rate than the supershifted band obtained with PrfALm (both formed under the same conditions) (Fig. 3F), which suggests that less PrfALs molecules may be bound in the PrfALs-mediated CI complex than in the corresponding CI complex formed with PrfALm.
To determine whether the applied RNAP concentration in the CI complex formation assays described above is indeed at a saturating level, we performed EMSAs by adding increasing concentrations of RNAP to the four PrfA proteins at the concentrations which showed saturation in the CIII formation assays described above. As shown in Fig. 4A and B, at the indicated PrfA concentrations, concentrations of RNAP lower than 1.5 nM (Fig. 3) were quantitatively shifted into CI (no CII complex observed) with PrfALm, PrfA*Lm, PrfALi, and PrfALs, while a higher RNAP concentration (3 nM) yielded only slightly more CI complex, indicating that the RNAP concentration for which results are shown in Fig. 3 is near saturation.
FIG. 4.
Effects of increasing concentrations of RNA polymerase on formation of CI, CII, and CIII complexes. Electrophoretic mobility shift assays were performed with RNAP concentrations of 0.38, 0.75, 1.5, and 3 nM, with 0.23 μM PrfALm, 3.5 μM PrfALs (A), 6 nM PrfA*Lm, and 60 nM PrfALi (B) at 5 nM hly DNA promoter fragment of L. monocytogenes (PhlyLm). The data shown here represent the results of one of three independently performed experiments.
There were still large amounts of free CIII complex, especially in cases of PrfA*Lm and PrfALi (Fig. 4B) under these conditions, that apparently could not be shifted to CI even in presence of the higher RNAP concentration. The opposite seems to be the case for PrfALm and especially PrfALs. With PrfALm, CI complex is already formed under conditions where CIII complex formation is not yet observed (Fig. 3B) and no CIII complex is detected at all with PrfALs (Fig. 4A).
These data confirm that PrfA*Lm and PrfALi bind more strongly to their specific PrfA boxes (forming CIII complexes) than PrfALm and PrfALs. But more importantly, these data suggest that preformed CIII complexes with PrfA*Lm and PrfALi (and, to some extent, even with PrfALm) are not easily shifted to CI, even in the presence of high RNAP concentrations. It is therefore likely that the formation of the transcriptionally active ternary CI complex is formed by the simultaneous interaction of all three partners. The binding efficiency of a given PrfA to its binding site alone (CIII formation) would then not necessarily indicate its potential to initiate transcription at a PrfA-dependent promoter.
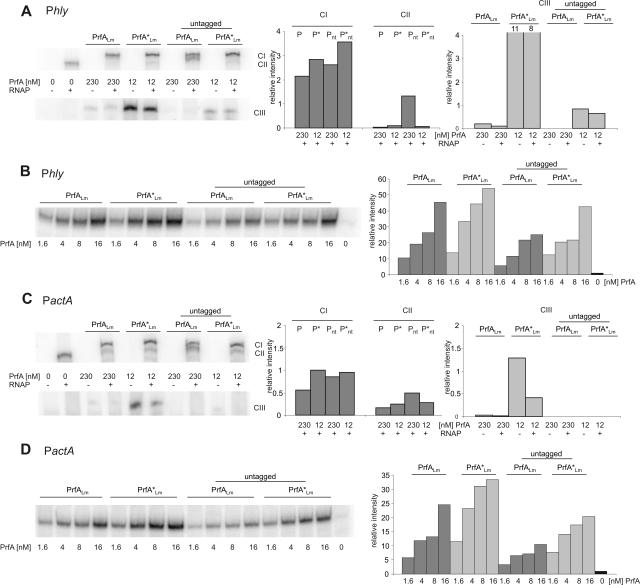
The N-terminal His6 tag of PrfA enhances CIII complex formation but not the efficiency of CI complex formation and of in vitro transcription initiation.
The difference between the His6-tagged PrfALm and PrfA*Lm and their untagged counterparts was determined in similar binding assays as described in the legend to Fig. 3 by using constant amounts of PhlyLm fragment (Fig. 5A) or PactALm fragment (Fig. 5C), each 5 nM, and RNAP (1.5 nM). The applied PrfA concentration was always chosen close to saturation for the His6-tagged versions determined as described in the legend to Fig. 3.
FIG. 5.
Comparison of His6-tagged and untagged PrfA proteins and the effect of a His6 tag on formation of CI, CII, and CIII complexes and in vitro transcription. Binding affinity of 230 nM PrfALm (P), 230 nM nontagged PrfALm (Pnt), 12 nM PrfA*Lm (P*), and 12 nM nontagged PrfA*Lm (P*nt) to 5 nM of the hly and the actA DNA promoter fragments of L. monocytogenes PhlyLm (A) and PactALm (C). The RNAP concentration is 1.5 nM. The graphs to the right show the band intensities relative to that of the CII band in lane 2. Transcriptional activity of 1.75 nM RNAP with increasing concentrations (1.6, 4, 8, and 16 nM) of the different PrfA proteins was detected with 19 nM of the two promoter fragments PhlyLm (B) and PactALm (D). The mRNA was labeled with [α-32P]CTP during transcription. The graphs to the right show the increasing transcriptional initiations compared to that from the lane without PrfA. The data shown here represent the results of one of three independently performed experiments.
As depicted in Fig. 5A and C (lower lanes), PrfA*Lm and PrfALm showed higher efficiencies of CIII formation than the untagged PrfA*Lm and untagged PrfALm proteins (almost 10-fold in the cases of PrfA*Lm and untagged PrfA*Lm at PhlyLm). At the applied PrfA concentrations, CIII complex formation with the Phly and PactA fragments could not even be observed in the case of untagged PrfALm, and CIII formation was weak with PrfALm. These data indicate that the His6 tag enhances binding of PrfA to the PrfA target sequence considerably, in agreement with the Biacore X determinations (Table 2).
Interestingly, the N-terminal His6 tag of PrfA did not show a corresponding increase in CI formation. As shown in the upper lanes of Fig. 5A and C, the difference between the His6-tagged and corresponding untagged PrfA proteins was at most a factor of two.
We next determined whether the efficiency of CI formation corresponds to the efficiency of initiating transcription at the PrfA-dependent promoters by using the previously described runoff in vitro transcription assay (1, 31). As shown in Fig. 5B and D, this indeed appears to be the case. At constant concentrations of RNAP and promoter fragments similar to those used in the assays whose results are shown in Fig. 3, we obtained, with increasing amounts of the His6-tagged and untagged versions of PrfALm and PrfA*Lm, runoff transcripts which differed in their amounts at most by, again, a factor of two. Moreover, there was not much difference between the abilities of PrfA and PrfA* to initiate in vitro transcription at Phly and PactA.
Binding of PrfALm, PrfA*Lm, PrfALi, and PrfALs to the hly and actA promoters of L. monocytogenes, L. ivanovii, and L. seeligeri and their in vitro transcription activation.
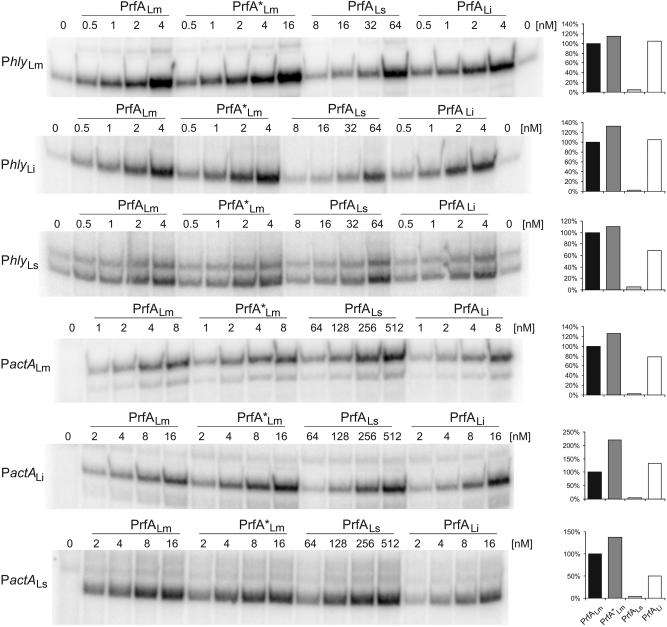
Since there are significant species-specific differences in the promoter sequences of the hly and actA genes (affecting less the PrfA box and −10 sequences and more the adjacent sequences) (Fig. 6), we tested whether the PrfA proteins of the three Listeria species may be more adapted to their specific PrfA-dependent promoters. For this goal, we measured again binding affinity and transcriptional activation of His6-tagged PrfALm, PrfA*Lm, PrfALi, and PrfALs with the hly and actA promoters of each of the three species (termed PhlyLm, PhlyLi, PhlyLs, and PactALm, PactALi, and PactALs). In all cases, the optimal concentrations for the PrfA proteins were chosen as determined in the assays described in the legend to Fig. 3, while a constant concentration of the promoter fragment (5 nM) was used. As shown in Fig. 7, similar binding efficiencies (determined by the formation of the CIII complex), determined as described above for binding to PhlyLm, were observed between the different PrfA proteins and the two other species-specific Phly and PactA promoter fragments. A slower-moving CIII complex (designated CIIIa) (Fig. 7) was observed with the high concentration of PrfALs (and sometimes with PrfALi and PrfA*Lm) and the studied Phly fragment and, particularly, the PactA promoter fragment. Since the used concentration of PrfALs was much higher than those of the other PrfA proteins, it is possible that these higher complexes formed with PrfALs are more stable than the normal CIII complex, which seems to consist of a PrfA dimer bound to the promoter. It is also noticeable that the CIII complex formed with PrfALi always migrated (for an unknown reason) slightly slower than those formed with PrfALm and PrfA*Lm (Fig. 7). Binding of PrfALi and binding of PrfA*Lm to all tested promoters were again the most efficient, and the binding of PrfALi to these promoters was comparable to that of PrfA*Lm. The CI formation efficiencies observed with PrfALm, PrfA*Lm, PrfALs, and PrfALi were quite similar in all analyzed promoter fragments. Note that CI formation with PrfALs required amounts of this protein that were at least 20-fold larger than than the amounts of the other PrfA proteins (Fig. 7). In vitro transcription initiation induced by the different PrfA proteins at these promoters was also examined. In each case, PrfA*Lm showed an activity that was only slightly higher (<2-fold) than those of PrfALm and PrfALi, whereas more than 20-fold-larger amounts of PrfALs were needed for a transcription efficiency similar to that with PrfALm (Fig. 8). These results indicate that there are no species-specific preferences for the different PrfA proteins to the homologous and heterologous actA and hly promoters used in this study.
FIG. 6.
ClustalW alignment of the hly and actA promoter regions of L. monocytogenes (mono), L. ivanovii (ivan), and L. seeligeri (seel). The PrfA box, the −10 box, the translational start, and identical base pairs in all three sequences (*) are marked.
FIG. 7.
Comparison of the hly and actA promoters of L. monocytogenes, L. ivanovii, and L. seeligeri in EMSA experiments. Specific binding affinity of 230 nM PrfALm, 12 nM PrfA*Lm, 3.5 μM PrfALs, and 60 nM PrfALi to the hly and actA promoter fragments of L. monocytogenes (PhlyLm and PactALm), L. ivanovii (PhlyLi and PactALi), and L. seeligeri (PhlyLs and PactALs). Quantification of the CI, CII, and CIII complexes, shown in the graphs to the right, was performed using ImageMaster (Amersham). The intensity of the band in lane 2 was again set to 1, and all other values are normalized to it. The data shown here represent the results of one of three independently performed experiments.
FIG. 8.
In vitro transcription assays with the hly and actA promoters of L. monocytogenes (PhlyLm and PactALm), L. ivanovii (i-Phly and i-PactA), and L. seeligeri (s-Phly and s-PactA). Results are from an in vitro transcription assay with increasing concentrations of the PrfA proteins PrfALm, PrfA*Lm, PrfALs, and PrfALi using 16 nM promoter template DNA and 1.3 nM (when using Phly) or 1.9 nM (when using PactA) RNA polymerase. The runoff transcripts were radioactively marked by adding [α-32P]CTP to the in vitro assay. The transcription-activating potentials of the different PrfA proteins compared to that of PrfALm are given in the graphs to the right. Values represent the relative ratios of the measured radioactivities and the molar concentrations of PrfA protein in the range of linear dependency. The data shown here represent the results of one of three independently performed experiments.
Replacement of the C-terminal 38 amino acids of PrfALs with those of PrfALm leads to a significantly higher transcription activation at PhlyLs and PactALm.
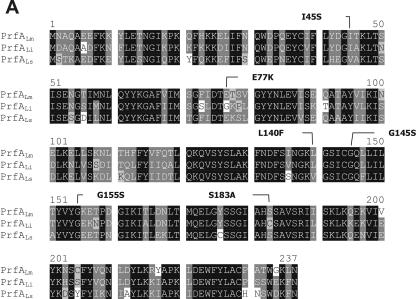
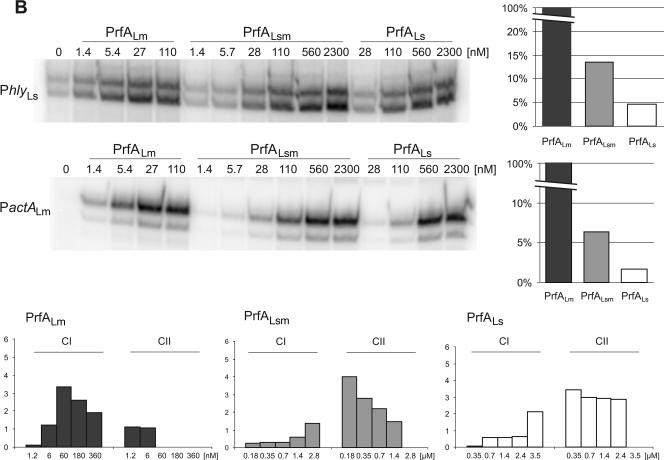
As shown in Fig. 9A, there is a clustering of amino acid exchanges in the C-terminal region of PrfALs (amino acids 200 to 237) compared to that of PrfALm or PrfALi, whereas the other differences in the amino acid sequence between PrfALs and PrfALi or PrfALm are rather randomly distributed. The C-terminal region has been shown to be critical for PrfA function (18). We therefore replaced this region of PrfALs with that of PrfALm, purified the hybrid protein PrfALsm (Fig. 1), and determined the binding affinity and the transcriptional activation capacity in comparison with those of PrfALm and PrfALs. As shown in Fig. 9B, both properties of this hybrid, PrfALsm, were improved compared to those of PrfALs but did not reach those of PrfALm, suggesting that some of the other amino acid exchanges in PrfALs may be even more decisive for the low activity of PrfALs than the amino acid differences in the C-terminal region.
FIG.9.
Replacement of the C-terminal 38 amino acids of PrfALs with those of PrfALm. (A) ClustalW alignment of the PrfA proteins of L. monocytogenes (PrfALm), L. ivanovii (PrfALi), and L. seeligeri (PrfALs). Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Amino acid substitutions leading to a constitutively active PrfA are marked. (B) In vitro transcription assay with increasing concentrations of PrfALm, the hybrid PrfALsm, and PrfALs using 16 nM promoter template DNA and 1.9 nM RNA polymerase. The mRNA was marked with [α-32P]CTP during transcription. The transcription-activating potentials of the different PrfA proteins compared to that of PrfALm are given in the graphs to the right. The graphs in the lower part of the panel below show the amounts of CI and CII measured in EMSAs. The components were 5 nM 32P-marked promoter DNA (PhlyLm) and 1.5 nM RNA polymerase, and the PrfA concentration is given in the figure. Quantification of CI and CII complexes was performed using ImageMaster (Amersham). The data shown here represent the results of one of three independently performed experiments.
Allelic replacement of prfALm with prfALs in L. monocytogenes leads to a reduction in virulence.
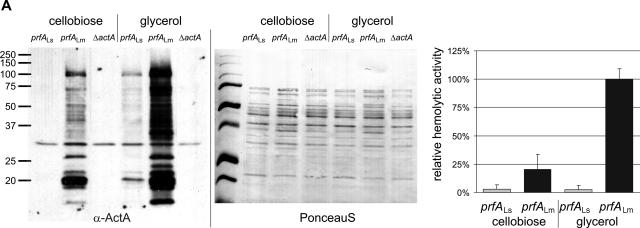
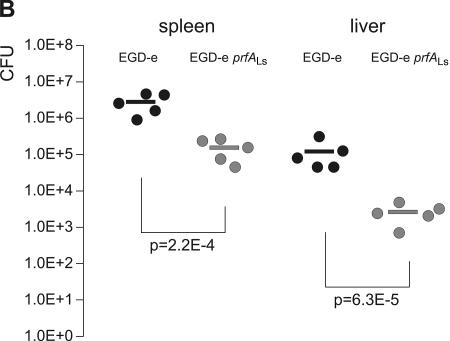
The observed low in vitro transcriptional activity of PrfALs compared to PrfALm and PrfALi suggests that the observed low virulence of L. seeligeri might be, at least in part, caused by PrfALs. To provide further experimental evidence for this assumption, we replaced the coding region of prfALm in L. monocytogenes EGD-e with the corresponding prfALs region. This new L. monocytogenes strain was less hemolytic, produced smaller amounts of the PrfA-dependent ActA protein (Fig. 10A), and showed a significantly reduced virulence in the mouse sepsis model (Fig. 10B), indicating that the PrfALs protein contributes to the low virulence of L. seeligeri.
FIG. 10.
Allelic replacement of prfALm with prfALs in Listeria monocytogenes. (A) Expression of the PrfA-regulated genes actA and hly in the strains L. monocytogenes EGD-e prfALs and L. monocytogenes EGD-e prfALm. The strains were grown to an optical density at 600 nm of 0.6 in minimal medium supplemented with 50 mM cellobiose or glycerol. Cytoplasmic proteins were prepared, and equal amounts (5 μg) were separated by SDS-PAGE and analyzed by Western blotting. After use of a loading control with PonceauS, immunodetection of ActA protein (α-ActA) was performed. Protein preparations of the strain L. monocytogenes EGD-e ΔactA (S. Pilgrim and S. Bauer, unpublished data) are also shown as a negative control. The hemolytic activities of L. monocytogenes EGD-e prfALs and L. monocytogenes EGD-e prfALm grown in minimal medium to an optical density at 600 nm of 0.6 are shown to the right. The hemolytic activity was determined in three independently performed experiments; the error bars indicate standard deviations of the means for the three experiments. (B) Viable bacterial counts in spleens and livers of C57BL/6 mice infected intravenously with 5 × 103 CFU of wild-type L. monocytogenes EGD-e or L. monocytogenes EGD-e prfALs (EGD-e s-prfA). Bacterial loads in spleens and livers of infected animals are shown. Each symbol represents a single animal. The lines indicate the means for the experimental groups (n = 5). For each organ, the P value determined with Student's t test of the log-transformed CFU amounts is given. L. monocytogenes EGD-e prfALs was highly significantly (P ≤ 0.01) attenuated compared to the wild-type strain.
DISCUSSION
PrfA, first identified as the key regulator of virulence genes of L. monocytogenes (29), was later also found in the animal pathogenic species L. ivanovii and in the nonpathogenic species L. seeligeri (16, 28) but not in the strictly environmental saprophytes Listeria innocua and Listeria welshimeri (8, 17). Recent phylogenetic studies of the various Listeria species suggest that the prfA gene, which is part of a gene cluster (now termed LIPI-1) (24), is inserted together with LIPI-1 into the listerial chromosome rather early in evolution but is again deleted together with LIPI-1 in L. innocua and L. welshimeri (4, 45).
The genes which are transcriptionally activated by PrfA in L. monocytogenes and L. ivanovii encode primarily virulence factors enabling these bacteria to enter nonphagocytic mammalian cells and to survive and replicate efficiently in the host cell's cytosol. The limited data available from previous studies on the expression of L. seeligeri genes that are presumably under PrfALs control (23, 51) show low expression of these genes when L. seeligeri is grown in culture media or in mammalian cells (22). This reduced gene expression could be caused by altered regulation of the prfALs gene expression, by an altered mechanism of activation of PrfALs compared to PrfALm and PrfALi, or by the altered amino acid composition of PrfALs.
Since in vivo PrfA activity is regulated by complex, not yet fully understood additional factors and metabolic conditions (7, 10, 34, 35, 41, 43) which may be different in the three listerial species (23), we have chosen in the present study to purify the three PrfA proteins and to compare their binding affinities to homologous (deriving from the same species) and heterologous (deriving from other listerial species) PrfA-dependent promoters. Additionally, we determined their capacities to initiate transcription at these promoters in vitro.
Purified PrfALm has been previously shown to be active in vitro, exhibiting a transcriptional initiation capacity at PrfA-controlled promoters which is quite similar to that of the purified constitutively active PrfA*Lm (31, 53). Different from the situation in vitro, wild-type PrfA activity in vivo is positively or negatively modulated under certain physiological conditions (15, 23), while PrfA* remains constitutively active independent of the culture conditions (43). This difference between in vitro and in vivo activity suggests that wild-type PrfA activity in vivo is modulated by a still unknown cellular factor(s).
Binding affinities of the various PrfA proteins to their specific binding sites (the PrfA boxes) was determined by two methods: (i) by measuring the equilibrium constants of PrfA to an oligonucleotide that contains the binding site of PhlyLm only and (ii) by measuring the binding affinity of PrfA to the entire PrfA box-containing promoter by EMSA. The latter was done, as previous experiments had shown that binding affinity of PrfA is highly inefficient when one uses the PrfA box oligonucleotide only and complex formation (CIII) could be observed only with a high PrfA concentration (7).
The data obtained by the two different assays show similar trends with respect to the binding of the PrfA proteins and confirm the earlier notion that purified PrfA without any additional factor is already in an (at least partially) active conformation. The nanomolar equilibrium constants suggest that already one to five molecules of purified PrfALm protein, as well as nontagged PrfALm, per cell could bind efficiently to the PrfA boxes, which seems to be the initial step of PrfA-mediated transcription activation. It should be noticed that the equilibrium constants of PrfALm and PrfA*Lm determined in this study are considerably lower than those determined by Eiting and coworkers (9); this may be due to the different PrfA preparations. Both experiments showed binding affinities of PrfA* that were higher than those of PrfA, which, in our measurements, were 18-fold higher in the case of His6-tagged PrfA proteins (9) and 50-fold higher with untagged PrfA proteins.
Although the Biacore X and EMSA experiments lead to the conclusion that the His6-tagged PrfA proteins bind significantly better to their target sites than their untagged counterparts, the in vitro transcription studies show that the transcriptional activation capacity of wild-type PrfA is not due to the applied N-terminal His6 tag. Both untagged PrfALm proteins show only a slightly reduced activation to initiate transcription at the PrfA-dependent promoters Phly and PactA compared to the His6-tagged PrfA proteins despite the much greater difference (up to 10-fold) between the abilities of the His6-tagged and the untagged PrfA proteins to bind to the target sites (PrfA boxes). These data suggest that PrfA binding to the PrfA box alone does not necessarily reflect the efficiency of transcription initiation at PrfA-dependent promoters.
This conclusion is also supported by the comparative binding and transcriptional activation exerted by the other PrfA proteins analyzed. The purified (His6-tagged) PrfA proteins of L. monocytogenes, L. ivanovii, and L. seeligeri show remarkable differences in binding affinity to PrfA-dependent homologous and heterologous listerial promoters when measured by EMSA. The PrfALi shows (in the absence of RNA polymerase) a binding affinity to the hly and actA promoters of L. monocytogenes, L. ivanovii, and L. seeligeri, which is similar to that of PrfA*Lm and more than 2 logs higher than that of wild-type PrfALm. It has not been experimentally clarified what causes the high binding affinity of PrfALi compared to that of PrfALm. Among the various amino acid exchanges which were previously shown to lead to PrfA*Lm activity (42, 47, 49, 52-54) (these mutations are also indicated in Fig. 9A), an amino acid exchange at position 183 is also present in PrfALi (S183C) compared to the amino acid sequence of PrfALm (S183A) (47) and may contribute to the enhanced binding of PrfALi compared to PrfALm.
Contrary to the different binding efficiencies of PrfA*Lm and PrfALi on one hand and PrfALm on the other to the various PrfA box-containing promoters in the absence of RNAP, the efficiencies of all three PrfA proteins to form the ternary transcription complex between PrfA, RNAP, and the PrfA promoter sequence (CI complex) are rather similar. The CI complex formation capacity also parallels the abilities of these PrfA proteins to initiate in vitro transcription at the PrfA-dependent promoters; these abilities are almost equal for PrfALi and PrfALm, and that of PrfA*Lm is at most twofold higher. These data are thus in accord with the conclusion given above, that the efficiency of the binding of PrfA to its specific site alone does not determine its ability to form a transcriptionally active initiation complex. The data rather suggest that the simultaneous interaction of PrfA with RNAP and the PrfA box-containing promoter sequence may be decisive for the formation of a transcriptionally active complex, and this property seems to be quite similarly expressed among all three PrfA proteins.
The correlation of the efficiency of PrfA binding to its target site and transcription activation at PrfA-dependent promoters is most evident in case of PrfALs. This PrfA protein has a drastically lower binding affinity to all studied PrfA boxes than the other PrfA proteins and, in parallel, an equally reduced ability to form the ternary CI complex with RNAP and to initiate transcription at all studied PrfA-dependent promoters. Furthermore, the supershift experiments performed with the PrfALs- and PrfALm-mediated CI complexes (using purified polyclonal anti-PrfA antibodies) suggest that less PrfALs molecules may be bound to the PrfALs-mediated CI complex than are bound to that formed with PrfALm, which may be due to a limited capacity of PrfALs to form functional dimers. These impaired properties of PrfALs are in part due to amino acid differences in the C-terminal region compared to PrfALm, as shown by replacing this region of PrfALs with that of PrfALm. The C-terminal region of PrfA was previously shown to be crucial for binding to its target site, probably by influencing dimer formation of PrfA (18). However, one or more of the other amino acid exchanges, distributed throughout the PrfALs protein (compared to PrfALm), must also be responsible for the low PrfALs activity, since the enhancement in activity of the hybrid PrfALsm does not reach the level of PrfALm.
Contrary to the low activity of PrfALs, the PrfA-dependent promoters of L. seeligeri are still fully functional. The binding activities of PrfALi and PrfALm to the two L. seeligeri PrfA-dependent promoters, PhlyLs and PactALs, are comparable to those of the corresponding L. monocytogenes and L. ivanovii promoters, and transcription initiation efficiencies at these L. seeligeri promoters by PrfALm and PrfALi are as good as those at the corresponding promoters of L. monocytogenes and L. ivanovii. These data lead us to conclude that the low expression of PrfA-dependent genes in L. seeligeri is mainly caused by the low activity of the PrfALs protein, which is probably due to the multiple mutations in the prfA gene. The replacement of the open reading frame of prfALm with that of prfALs in L. monocytogenes strongly supports this view, as this replacement leads to a reduced expression of PrfA-dependent genes in L. monocytogenes and a reduced virulence of L. monocytogenes in vivo.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB473 “Mechanisms of Transcriptional Regulation” and SFB479-B1) and the Fonds der Chemischen Industrie.
We thank Nico Marr for purification of untagged PrfALm and PrfA*Lm. Biju Joseph is thanked for critical reading of the manuscript.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Böckmann, R., C. Dickneite, W. Goebel, and J. Bohne. 2000. PrfA mediates specific binding of RNA polymerase of Listeria monocytogenes to PrfA-dependent virulence gene promoters resulting in a transcriptionally active complex. Mol. Microbiol. 36:487-497. [DOI] [PubMed] [Google Scholar]
- 2.Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 3.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty, T., T. Hain, and E. Domann. 2000. Genome organization and the evolution of the virulence gene locus in Listeria species. Int. J. Med. Microbiol. 290:167-174. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickneite, C., R. Böckmann, A. Spory, W. Goebel, and Z. Sokolovic. 1998. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol. Microbiol. 27:915-928. [DOI] [PubMed] [Google Scholar]
- 8.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiting, M., G. Hagelüken, W. D. Schubert, and D. W. Heinz. 2005. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol. Microbiol. 56:433-446. [DOI] [PubMed] [Google Scholar]
- 10.Ermolaeva, S., S. Novella, Y. Vega, M. T. Ripio, M. Scortti, and J. A. Vázquez-Boland. 2004. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol. Microbiol. 52:601-611. [DOI] [PubMed] [Google Scholar]
- 11.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 12.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitag, N. E., P. Youngman, and D. A. Portnoy. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J. Bacteriol. 174:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1989. Production of thiol-dependent haemolysins by Listeria monocytogenes and related species. J. Gen. Microbiol. 135:481-487. [DOI] [PubMed] [Google Scholar]
- 15.Goebel, W., S. Müller-Altrock, and J. Kreft. 2006. Regulation of virulence genes in pathogenic Listeria spp., p. 499-506. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 16.Gouin, E., J. Mengaud, and P. Cossart. 1994. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 62:3550-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hain, T., C. Steinweg, and T. Chakraborty. 2006. Comparative and functional genomics of Listeria spp. J. Biotechnol. 126:37-51. [DOI] [PubMed] [Google Scholar]
- 18.Herler, M., A. Bubert, M. Goetz, Y. Vega, J. A. Vázquez-Boland, and W. Goebel. 2001. Positive selection of mutations leading to loss or reduction of transcriptional activity of PrfA, the central regulator of Listeria monocytogenes virulence. J. Bacteriol. 183:5562-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herro, R., S. Poncet, P. Cossart, C. Buchrieser, E. Gouin, P. Glaser, and J. Deutscher. 2005. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J. Mol. Microbiol. Biotechnol. 9:224-234. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 21.Joseph, B., K. Przybilla, C. Stühler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunasagar, I., R. Lampidis, W. Goebel, and J. Kreft. 1997. Complementation of Listeria seeligeri with the plcA-prfA genes from L. monocytogenes activates transcription of seeligerolysin and leads to bacterial escape from the phagosome of infected mammalian cells. FEMS Microbiol. Lett. 146:303-310. [DOI] [PubMed] [Google Scholar]
- 23.Kreft, J., and J. A. Vázquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 24.Kreft, J., J. A. Vázquez-Boland, S. Altrock, G. Domínguez-Bernal, and W. Goebel. 2002. Pathogenicity islands and other virulence elements in Listeria. Curr. Top. Microbiol. Immunol. 264(2):109-125. [PubMed] [Google Scholar]
- 25.Kuhn, M., and W. Goebel. 1995. Molecular studies on the virulence of Listeria monocytogenes. Genet. Eng. (New York) 17:31-51. [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lalic-Mülthaler, M., J. Bohne, and W. Goebel. 2001. In vitro transcription of PrfA-dependent and -independent genes of Listeria monocytogenes. Mol. Microbiol. 42:111-120. [DOI] [PubMed] [Google Scholar]
- 28.Lampidis, R., R. Gross, Z. Sokolovic, W. Goebel, and J. Kreft. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol. Microbiol. 13:141-151. [DOI] [PubMed] [Google Scholar]
- 29.Leimeister-Wächter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, Q., M. Herler, S. Müller-Altrock, and W. Goebel. 2005. Supportive and inhibitory elements of a putative PrfA-dependent promoter in Listeria monocytogenes. Mol. Microbiol. 55:986-997. [DOI] [PubMed] [Google Scholar]
- 31.Luo, Q., M. Rauch, A. K. Marr, S. Müller-Altrock, and W. Goebel. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39-52. [DOI] [PubMed] [Google Scholar]
- 32.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vázquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 33.Mengaud, J., M. F. Vicente, and P. Cossart. 1989. Transcriptional mapping and nucleotide sequence of the Listeria monocytogenes hlyA region reveal structural features that may be involved in regulation. Infect. Immun. 57:3695-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 35.Milenbachs Lukowiak, A., K. J. Mueller, N. E. Freitag, and P. Youngman. 2004. Deregulation of Listeria monocytogenes virulence gene expression by two distinct and semi-independent pathways. Microbiology 150:321-333. [DOI] [PubMed] [Google Scholar]
- 36.Mueller, K. J., and N. E. Freitag. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 73:1917-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeuffer, T., W. Goebel, J. Laubinger, M. Bachmann, and M. Kuhn. 2000. LaXp180, a mammalian ActA-binding protein, identified with the yeast two-hybrid system, co-localizes with intracellular Listeria monocytogenes. Cell Microbiol. 2:101-114. [DOI] [PubMed] [Google Scholar]
- 39.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauch, M., Q. Luo, S. Müller-Altrock, and W. Goebel. 2005. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J. Bacteriol. 187:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renzoni, A., A. Klarsfeld, S. Dramsi, and P. Cossart. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 65:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripio, M. T., G. Domínguez-Bernal, M. Lara, M. Suárez, and J. A. Vázquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ripio, M. T., G. Domínguez-Bernal, M. Suárez, K. Brehm, P. Berche, and J. A. Vázquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schmid, M. W., E. Y. Ng, R. Lampidis, M. Emmerth, M. Walcher, J. Kreft, W. Goebel, M. Wagner, and K. H. Schleifer. 2005. Evolutionary history of the genus Listeria and its virulence genes. Syst. Appl. Microbiol. 28:1-18. [DOI] [PubMed] [Google Scholar]
- 46.Schwab, U., B. Bowen, C. Nadon, M. Wiedmann, and K. J. Boor. 2005. The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol. Lett. 245:329-336. [DOI] [PubMed] [Google Scholar]
- 47.Sheehan, B., A. Klarsfeld, R. Ebright, and P. Cossart. 1996. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 20:785-797. [DOI] [PubMed] [Google Scholar]
- 48.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48:1537-1551. [DOI] [PubMed] [Google Scholar]
- 50.Stritzker, J., C. Schoen, and W. Goebel. 2005. Enhanced synthesis of internalin A in aro mutants of Listeria monocytogenes indicates posttranscriptional control of the inlAB mRNA. J. Bacteriol. 187:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001.Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vega, Y., C. Dickneite, M. T. Ripio, R. Böckmann, B. González-Zorn, S. Novella, G. Domínguez-Bernal, W. Goebel, and J. A. Vázquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vega, Y., M. Rauch, M. J. Banfield, S. Ermolaeva, M. Scortti, W. Goebel, and J. A. Vázquez-Boland. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 52:1553-1565. [DOI] [PubMed] [Google Scholar]
- 54.Wong, K. K., and N. E. Freitag. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186:6265-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]