Abstract
Promoter-lux fusions that showed rifampin-modulated transcription were identified from a Salmonella enterica serovar Typhimurium 14028 reporter library. The transformation of a subset of fusions into mutants that lacked one of six global regulatory proteins or were rifampin resistant showed that transcription modulation was independent of the global regulators, promoter specific, and dependent on the interaction of rifampin with RNA polymerase.
Antibiotics have been shown to alter global bacterial transcription patterns at concentrations below those that completely inhibit the growth of the bacterial cell (sub-MIC) (10, 29). Among the genes affected by antibiotics are those related to modes of action and bacterial stress responses. It has been suggested that characteristic gene expression patterns may be used to identify unknown small molecule inhibitors (2, 28). There have also been studies demonstrating antibiotic-induced transcription modulation of genes for accessory functions such as motility and virulence (reviewed in references 2 and 29).
Using a library of 6,528 promoter-reporter clones, we previously demonstrated dramatic up- and down-regulation of the transcription induced by the antibiotic rifampin (a transcription inhibitor [3, 14]) and the macrolide class of antibiotics on a global scale in Salmonella enterica serovar Typhimurium 14028 (10, 28). In this communication, we further examine rifampin-responsive promoter-luxCDABE fusion clones from our initial screen (∼5% of library clones [10]) to explore the effects of known global transcription regulators on rifampin-induced transcription modulation (RITM).
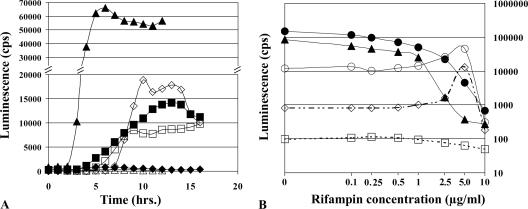
Fusions whose activity was modulated by rifampin in our initial screens (10) were screened continuously by inoculating cultures grown at 37°C in a white, opaque microtiter plate (Costar; Fisher Scientific, Ottawa, Ontario, Canada) sealed with a breathable sealing membrane (Nalge Nunc, Naperville, IL). Luminescence was followed during 12 to 16 h of growth in the presence or absence of rifampin by using a Victor II multilabel counter (PerkinElmer, Boston, MA). Clones were discarded if luminescence readings were below 1,000 cps in medium with and without rifampin. This resulted in the identification of a subset of 22 moderately to strongly affected promoters that displayed between 200-fold down-regulation and 24-fold up-regulation (Table 1). Figure 1A shows the patterns of luminescence produced from two promoters that were sensitive to rifampin and from an unaffected promoter. The strain carrying the vector alone produced very low levels of luminescence (∼100 cps [data not shown]). Note that both stimulation and inhibition of the transcription by the inhibitor were found and that the timings of the maximum response differed between promoters. As a result, promoters were screened for a minimum of 12 h to identify the maximum response. The effect of rifampin was observed to be concentration dependent and generally maximal at 5 μg/ml rifampin (Fig. 1B).
TABLE 1.
Characteristics of rifampin-responsive promoters in S. enterica serovar Typhimurium 14028
| Promoter | Luminescence (cps) witha:
|
Fold induction for LB with rif atb:
|
Putative function | ||||||
|---|---|---|---|---|---|---|---|---|---|
| LB | SD | LB with rif 2.5 μg/ml | SD | LB with rif 5.0 μg/ml | SD | 2.5 μg/ml | 5.0 μg/ml | ||
| STM2899/invF | 61,605 | 15,125 | 1,139 | 244 | 295 | 72 | ↓54.1 | ↓209.2 | Virulence, invasion |
| STM1091/sopB | 44,671 | 10,443 | 853 | 309 | 274 | 105 | ↓52.3 | ↓163.0 | Virulence, invasion |
| STM2066/sopA | 16,133 | 5,246 | 224 | 141 | 140 | 61 | ↓72.1 | ↓115.2 | Virulence, invasion |
| STM4255 to -4258 | 3,172 | 596 | 203 | 54 | 240 | 27 | ↓15.7 | ↓13.2 | Virulence |
| STM1956/fliA (σF) | 147,748 | 61,007 | 22,000 | 2,265 | 4,598 | 562 | ↓6.7 | ↓32.1 | Flagellum synthesis |
| STM1914/flhBA | 6,553 | 2,634 | 1,407 | 247 | 251 | 40 | ↓4.7 | ↓26.1 | Flagellum synthesis |
| STM1183/flgK | 93,550 | 8,524 | 25,533 | 3,462 | 5,222 | 2,221 | ↓3.7 | ↓17.9 | Flagellum synthesis |
| STM2199/cirA | 2,169 | 757 | 673 | 645 | 516 | 374 | ↓3.2 | ↓4.2 | Iron metabolism |
| STM1328 | 2,480 | 150 | 113 | 46 | 100 | 38 | ↓21.9 | ↓24.9 | Unknown |
| STM1248 | 2,806 | 386 | 150 | 72 | 148 | 54 | ↓18.7 | ↓19.0 | Unknown |
| STM1444/slyA | 1,029 | 316 | 2,558 | 787 | 5,223 | 473 | 2.5 | 5.1 | Virulence, systemic |
| STM1154 to -1155/yceE, htrB | 10,040 | 3,041 | 26,165 | 7,776 | 38,305 | 5,030 | 2.6 | 3.8 | Virulence, systemic |
| pSLT041-39/spvRAB | 3,688 | 515 | 7,017 | 5,170 | 13,230 | 3,942 | 1.9 | 3.6 | Virulence, systemic |
| STM4118/yijP | 2,633 | 619 | 3,941 | 626 | 8,688 | 1,821 | 1.5 | 3.3 | Virulence |
| STM4454/treB | 4,179 | 1,430 | 17,662 | 3,545 | 54,241 | 5,188 | 4.2 | 13.0 | Carbon metabolism |
| STM2445/ucpA | 12,867 | 3,627 | 26,283 | 6,120 | 46,325 | 11,770 | 2.0 | 3.6 | Carbon metabolism |
| STM1597/ydcW | 1,992 | 472 | 2,361 | 106 | 6,441 | 1,651 | 1.2 | 3.2 | Carbon metabolism |
| STM2473/talA | 3,149 | 660 | 4,543 | 1,054 | 9,194 | 1,340 | 1.4 | 2.9 | Carbon metabolism |
| STM0425/thiI | 1,927 | 324 | 5,245 | 2,685 | 15,918 | 2,758 | 2.7 | 8.3 | RNA modification |
| STM3595, STM3084 | 824 | 29 | 2,157 | 165 | 19,686 | 775 | 3.0 | 24.0 | Unknown |
| STM0389/yaiA | 2,590 | 506 | 4,721 | 2,093 | 10,180 | 4,030 | 1.8 | 3.9 | Unknown |
| STM2287 | 2,740 | 1,164 | 3,557 | 617 | 7,697 | 981 | 1.3 | 2.8 | Unknown |
cps, counts per second; rif, rifampin; SD, standard deviation.
Down arrows indicate RIDR.
FIG. 1.
(A) Luminescence profiles of promoter-lux reporters. Luminescence patterns for serovar Typhimurium 14028 reporters STM3595::luxCDABE (⋄), invF::luxCDABE (▵), and STM2091::luxCDABE (□) in LB supplemented with kanamycin at 25 μg/ml (for plasmid maintenance) and rifampin at 5.0 μg/ml (open symbols) or LB supplemented with kanamycin at 25 μg/ml in the absence of rifampin (filled symbols) are shown. Promoters drive expression from the luxCDABE operon, producing luminescence without any exogenous substrate (19). (B) Concentration dependence of rifampin-induced transcription modulation. Serovar Typhimurium 14028 with fliA (○), invF (▴), STM3595 (⋄), and ucpA (○) luxCDABE reporters and pCS26 without an insert (□) grown in LB supplemented with kanamycin at 25 μg/ml and the indicated concentrations of rifampin were grown for at least 14 h. Average peak luminescence values from a minimum of three time courses are plotted.
Upon sequencing the promoters sensitive to RITM (Table 1), we noticed that a number of serovar Typhimurium virulence genes were included. The affected promoters were also grouped by the involvement of the associated genes in two distinct regulons. Promoters from virulence genes associated with intracellular growth and survival in macrophages, slyA (5, 13, 16), spvAB (11, 13, 17), somA (7), htrB (15), and SPI-2 genes (4, 12, 13), showed rifampin-induced up-regulation (RIUR). Promoters from genes involved in intestinal invasion, those associated with the type III secretion system encoded on SPI-1 and its secreted effectors, showed rifampin-induced down-regulation (RIDR). These genes included invF (6, 9, 18), sopA, and sopB (8, 24). Promoters from genes in SPI-4 (STM4255 to 4258), which have been shown to be coordinately regulated with SPI-1 by HilA (1), also displayed RIDR. Furthermore, rifampin also down-regulated the promoters from three operons involved in flagellum synthesis.
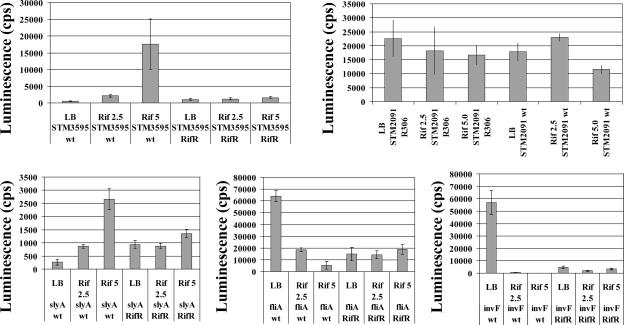
It seemed possible that RITM might be due to antibiotic effects on one of the known global transcription regulators or to the activation of one or more stress responses. To examine these possibilities, eight different representative rifampin-responsive promoter (RRP) fusions were transformed into serovar Typhimurium 14028 strains, each carrying a mutation in the gene for one of six major global regulators: CRP (25), FNR (27), FIS (22), H-NS (21), IHF (23), or σS (25). Mutant alleles were introduced into the 14028 background by using P22HTint-mediated generalized transduction (26) and confirmed by PCR or inverse PCR (20), followed by nucleotide sequencing and phenotypic analysis. The promoter activity of four rifampin-up-regulated and four rifampin-down-regulated lux fusions was examined in the presence or absence of rifampin (Table 2). The change in expression (n-fold) in response to rifampin was calculated by dividing the amount of luminescence observed with rifampin by the amount of luminescence observed without rifampin. In the majority of promoter-mutant combinations, although the magnitude of RITM may have changed, RITM levels were similar for both the wild-type and mutant host backgrounds. In four cases, the introduction of the mutations changed RITM from strong RIDR to no effect (Table 2). However, in these cases, the introduction of regulatory mutations may have reduced the basal expression of the lux reporter to the lower limit of detection, preventing a clear conclusion regarding its involvement in RIDR. Overall, in 44/48 of the combinations tested, rifampin modulation of transcription was not altered by the loss of one of the regulatory proteins. In four cases, the mutation of a global regulator either blocked RIUR or switched the response from RIUR to RIDR, indicating that the regulator was involved in RIUR for that promoter (Table 2). We note that Fis was involved in three of the four cases in which RITM was altered by the loss of a regulator. This involvement may indicate that Fis has a role in RIUR, but as it did not affect all RRPs, no model involving Fis regulation of RITM was readily apparent. We also tested the effects of rifampin in a rifampin-resistant host (resistance was conferred by a mutation in the β subunit of RNA polymerase). The mutation conferring rifampin resistance eliminated RITM in all of the RRP fusions tested (Fig. 2). Thus, RITM is a transcription-specific effect and is not due to separate effects on cell physiology. Further analysis of this novel regulation will require the identification of the specific nucleotide sequences at the RRP responsible for rifampin sensitivity.
TABLE 2.
Summary of rifampin-induced transcription modulationa
| Strain description | Modulation in indicated reporter
|
|||||||
|---|---|---|---|---|---|---|---|---|
| fliA | flhB | cirA | STM1328 | STM3595 | spvAB | ucpA | talA | |
| 14028 | ↓↓ | ↓↓ | ↓↓ | ↓ | ||||
| 14028 crp::Tn10 | ↓ | *b | ↓↓ | ↓ | **c | |||
| 14028 fis::tet | ↓ | * | ↓ | ↓ | ↓** | ↓↓** | ** | |
| 14028 hns::Tn10 | ↓↓↓ | ↓↓ | ↓ | ↓ | ||||
| 14028 ihfB::cat | ↓↓↓ | ↓↓ | ↓ | ↓ | * | |||
| 14028 fnr::Tn10 | ↓↓ | ↓↓ | ↓↓ | ↓↓↓ | ↑ | |||
| 14028 rpoS::amp | ↓↓ | ↓ | ↓↓ | ↓↓↓ | * | |||
The modulation is depicted by up arrows (RIUR) and down arrows (RIDR) as follows: a 2.5- to 5-fold effect is depicted as one arrow, a 6- to 15-fold effect by two arrows, and a 16-fold or greater effect as three arrows. No arrow indicates there was a less-than-2.5-fold difference.
In four cases, indicated by single asterisks, the introduction of the mutations changed RITM from strong RIDR to no effect.
In four cases, indicated by double asterisks, the mutation of a global regulator either blocked RIUR or switched the response from RIUR to RIDR, indicating that the regulator was involved in RIUR for that promoter.
FIG. 2.
Luminescence profiles of promoter-lux fusions in serovar Typhimurium 14028 and a rifampin-resistant isogenic mutant, R306, in response to rifampin (Rif). Two rifampin-up-regulated promoters (STM3595 and slyA), two rifampin-down-regulated promoters (fliA and invF), and one promoter unaffected by rifampin (STM2091) were assayed. Luminescence was monitored during a minimum of 14 h of growth in microtiter plates containing LB with kanamycin (25 μg/ml) and the indicated amount (μg/ml) of rifampin; the plates were sealed with a Mylar plate sealer (Thermo Labsystems, Franklin, MA) and incubated at 37°C in a Victor II multilabel counter. Average peak luminescence values from eight replicate time courses are plotted. Error bars indicate standard deviations. wt, wild type; RifR, rifampin-resistant strain.
Acknowledgments
We thank the Canadian Bacterial Diseases Network and the Natural Sciences and Engineering Research Council of Canada for providing financial support. During his sabbatical visit to the University of British Columbia, F.D.L.C. was supported by a fellowship from the Spanish Ministry of Education, Culture, and Sports (PR2003-0258). Work in the F.D.L.C. laboratory was financed by grant BFU2005-03477 from the Spanish Ministry of Education.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Brazas, M. D., and R. E. Hancock. 2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10:1245-1252. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 4.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 5.Daniels, J. J., I. B. Autenrieth, A. Ludwig, and W. Goebel. 1996. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect. Immun. 64:5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 8.Ehrbar, K., S. Hapfelmeier, B. Stecher, and W. D. Hardt. 2004. InvB is required for type III-dependent secretion of SopA in Salmonella enterica serovar Typhimurium. J. Bacteriol. 186:1215-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh, E. B., G. Yim, W. Tsui, J. McClure, M. G. Surette, and J. Davies. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc. Natl. Acad. Sci. USA 99:17025-17030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulig, P. A., T. J. Doyle, J. A. Hughes, and H. Matsui. 1998. Analysis of host cells associated with the Spv-mediated increased intracellular growth rate of Salmonella typhimurium in mice. Infect. Immun. 66:2471-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 13.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 14.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 15.Jones, B. D., W. A. Nichols, B. W. Gibson, M. G. Sunshine, and M. A. Apicella. 1997. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect. Immun. 65:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libby, S. J., M. Lesnick, P. Hasegawa, E. Weidenhammer, and D. G. Guiney. 2000. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 18.Lostroh, C. P., V. Bajaj, and C. A. Lee. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300-315. [DOI] [PubMed] [Google Scholar]
- 19.Meighen, E. A. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7:1016-1022. [DOI] [PubMed] [Google Scholar]
- 20.Nichols, B. P., O. Shafiq, and V. Meiners. 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180:6408-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 22.Osuna, R., D. Lienau, K. T. Hughes, and R. C. Johnson. 1995. Sequence, regulation, and functions of fis in Salmonella typhimurium. J. Bacteriol. 177:2021-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palacios, S., and J. C. Escalante-Semerena. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raffatellu, M., R. P. Wilson, D. Chessa, H. Andrews-Polymenis, Q. T. Tran, S. Lawhon, S. Khare, L. G. Adams, and A. J. Baumler. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect. Immun. 73:146-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmonella Genetic Stock Centre. Salmonella Genetic Stock Centre, University of Calgary, Calgary, Alberta, Canada. [Online.] http://salmonella.bio.ucalgary.ca/.
- 26.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 27.Strauch, K. L., J. B. Lenk, B. L. Gamble, and C. G. Miller. 1985. Oxygen regulation in Salmonella typhimurium. J. Bacteriol. 161:673-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsui, W. H., G. Yim, H. H. Wang, J. E. McClure, M. G. Surette, and J. Davies. 2004. Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem. Biol. 11:1307-1316. [DOI] [PubMed] [Google Scholar]
- 29.Yim, G., H. H. Wang, and J. Davies. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296:163-170. [DOI] [PubMed] [Google Scholar]