Abstract
Morphogenetic proteins such as SpoVID and SafA govern assembly of the Bacillus subtilis endospore coat by guiding the various protein structural components to the surface of the developing spore. Previously, a screen for peptides able to interact with SpoVID led to the identification of a PYYH motif present in the C-terminal half of the SafA protein and to the subsequent demonstration that SpoVID and SafA directly interact. spoVID and safA spores show deficiencies in coat assembly and are lysozyme susceptible. Both proteins, orthologs of which are found in all Bacillus species, have LysM domains for peptidoglycan binding and localize to the cortex-coat interface. Here, we show that the interaction between SafA and SpoVID involves the PYYH motif (region B) but also a 13-amino-acid region (region A) just downstream of the N-terminal LysM domain of SafA. We show that deletion of region B does not block the interaction of SafA with SpoVID, nor does it bring about spore susceptibility to lysozyme. Nevertheless, it appears to reduce the interaction and affects the complex. In contrast, lesions in region A impaired the interaction of SafA with SpoVID in vitro and, while not affecting the accumulation of SafA in vivo, interfered with the localization of SafA around the developing spore, causing aberrant assembly of the coat and lysozyme sensitivity. A peptide corresponding to region A interacts with SpoVID, suggesting that residues within this region directly contact SpoVID. Since region A is highly conserved among SafA orthologs, this motif may be an important determinant of coat assembly in the group of Bacillus spore formers.
A large number of proteins produced in the mother cell of sporulating cells of Bacillus subtilis are targeted to the surface of the developing spore to form a structure known as the spore coat (11, 16, 18). Assembly of the coat confers protection against lytic enzymes and small noxious molecules and allows efficient interaction of the spore with compounds able to trigger germination (11, 16, 18). Because of the resistance it imparts to the action of the lytic enzymes, the coat also protects phagocytosed spores from digestion by predatory microorganisms (22). Structurally, the coat consists of an amorphous undercoat that contacts the underlying cortex peptidoglycan, a lamellar inner layer, and a thick electron-dense outer layer (11, 16, 18). The genes encoding the various coat components are expressed at different times during the deployment of a mother cell cascade of gene expression, and this temporal control is an important factor contributing to the ordered assembly of the coat (16). However, assembly of the coat relies to a large extent on the action of a class of so-called morphogenetic proteins that guide the assembly of the structural components (11, 16, 18, 29). One, called SpoIVA, localizes early to the mother cell side of the asymmetric septum and is responsible for recruiting another morphogenetic protein, SpoVID, to the developing spore (11, 12, 33). A third morphogenetic protein, CotE, localizes in a SpoIVA-dependent manner close to the developing spore, around a region of unknown composition called the matrix (11, 12). The localizations of SpoIVA, SpoVID, and CotE take place prior to the complete engulfment of the prespore by the mother cell when σE and the ancillary transcription factors SpoIIID and GerR govern gene expression in the mother cell (13, 41). Following engulfment completion, and the activation of the late mother cell-specific regulators σK and later of GerE, the coat layers start gaining their final appearance (11, 13, 16, 18, 41). The prior localization of CotE at the edge of the matrix region is thought to be required to nucleate assembly of the outer coat, whereas the matrix region develops into the inner coat (11, 12). CotE appears to have a modular design, specifying information for its targeting to the surface of the developing spore, for oligomerization, and also for interaction with other coat proteins (3, 28). Presumably, CotE controls outer coat assembly by means of complex network of direct or indirect interactions leading to the recruitment of many proteins to this coat layer (21, 28).
SpoVID does not influence the localization of CotE at an early stage, but following engulfment completion in cells mutant for spoVID, CotE and the rest of the coat detaches from the surface of the prespore and forms swirls of partially structured material dispersed throughout the mother cell cytoplasm (4, 12). Left with an exposed cortex, spoVID spores are sensitive to lysozyme (12). To study the function of SpoVID in the assembly process, Ozin et al. (32) used a phage display screening to identify peptides able to interact with SpoVID. They found a peptide motif, PYYH, occurring in the C-terminal half of another coat protein, SafA (387 residues long), the absence of which causes deficient formation of the coat and spore susceptibility to lysozyme (32, 40). At least three forms of SafA accumulate in sporulating cells: a full-length form (SafAFL) (45 kDa), a C-terminal form derived from internal translation of the safA mRNA at methionine 164 (SafAC30, 30 kDa) and an N-terminal form (SafAN21, 21 kDa) (31). Assays in vitro and in vivo showed that SafA and SpoVID directly interact, that SafA interacts with itself, and that each of the C- and N-terminal forms are capable of interacting with SafAFL, with themselves, and, for at least SafAN21, with SpoVID (33). Both SpoVID (at its C terminus) and SafA (at its N terminus) contain a LysM domain, which has a general peptidoglycan-binding function (2, 5, 20, 27, 35, 39), and both proteins localize to the cortex/coat interface (12, 32). Moreover, the localization of SafA depends on SpoVID (33). These results have led to a model according to which the interaction between SafA and SpoVID recruits the former to the coat region in close proximity to the cortex layer, and the two proteins function in this region by promoting attachment of the nascent coat to the underlying cortex layer (33). However, in spite of the fact that it contains the PYYH motif, the SafAC30 form is not recruited to the prespore in the absence of SafAFL, suggesting that a second region of interaction could exist in the N-terminal half of SafA (33). This second region of interaction between SafA and SpoVID was attributed to the LysM domain of both proteins, which is presumed to be involved in multimerization (2, 5, 20, 27, 35).
Here, we have analyzed the interaction between SafA and SpoVID. We present biochemical, cytological, and genetic evidence that strongly suggests that the interaction of SafA with SpoVID via a region just downstream of the LysM domain is responsible for the targeting of SafA to the surface of the developing spore and for the proper assembly and function of the coat structure.
MATERIALS AND METHODS
Strains and general techniques.
All of the B. subtilis strains used in this study are congenic derivatives of the wild-type strain MB24 (trpC2 metC3) (Table 1). The Escherichia coli strain DH5α (Bethesda Research Laboratories) was used for molecular cloning, and CC118(DE3)/pLysS was used for the overproduction of native or glutathione S-transferase (GST) fusion proteins (Table 1). Luria-Bertani medium was used for maintenance and growth of E. coli and B. subtilis strains, with appropriate antibiotic selection when needed. Sporulation was induced by growth and nutrient exhaustion in Difco sporulation medium (DSM) (30). Genetic manipulations of B. subtilis and spore resistance or germination properties were assessed as previously described (9).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype/phenotypea | Origin or reference |
|---|---|---|
| E. coli strains | ||
| CC118 | (DE3)/pLysS | Colin Manoil |
| AH2687 | CC118(DE3)/pLysS/pTC55 Cmr Ampr | This work |
| AH2688 | CC118(DE3)/pLysS/pTC49 Cmr Ampr | This work |
| AH2689 | CC118(DE3)/pLysS/pTC50 Cmr Ampr | This work |
| AH2690 | CC118(DE3)/pLysS/pTC51 Cmr Ampr | This work |
| AH2691 | CC118(DE3)/pLysS/pTC52 Cmr Ampr | This work |
| AH2718 | CC118(DE3)/pLysS/pTC61 Cmr Ampr | This work |
| AH2719 | CC118(DE3)/pLysS/pTC62 Cmr Ampr | This work |
| AH2692 | CC118(DE3)/pLysS/pOZ169 Cmr Ampr | This work |
| AH2934 | CC118(DE3)/pLysS/pTC138 Cmr Ampr | This work |
| AH2935 | CC118(DE3)/pLysS/pTC139 Cmr Ampr | This work |
| AH2937 | CC118(DE3)/pLysS/pTC141 Cmr Ampr | This work |
| AH2978 | CC118(DE3)/pLysS/pTC159 Cmr Ampr | This work |
| AH2979 | CC118(DE3)/pLysS/pTC160 Cmr Ampr | This work |
| AH2980 | CC118(DE3)/pLysS/pTC161 Cmr Ampr | This work |
| AH2985 | CC118(DE3)/pLysS/pTC165 Cmr Ampr | This work |
| AH4086 | CC118(DE3)/pLysS/pTC196 Cmr Ampr | This work |
| AH4087 | CC118(DE3)/pLysS/pTC197 Cmr Ampr | This work |
| B. subtilis strains | ||
| AOB68 | trpC2 metC3 ΔsafA::sp Spr | 32 |
| AOB90 | trpC2 metC3 safA-flag Cmr | 33 |
| AH2781 | trpC2 metC3 ΔsafA::safAΔPYYH Spr Cmr | This work |
| AH2787 | trpC2 metC3 ΔsafA::safAN1-162 Spr Cmr | This work |
| AH2788 | trpC2 metC3 ΔsafA::safAC164-387 Spr Cmr | This work |
| AH4006 | trpC2 metC3 ΔsafA::safAΔG51-S63 Spr Cmr | This work |
| AH4090 | trpC2 metC3 ΔsafA::safAΔG51-E57 Spr Cmr | This work |
| AH4091 | trpC2 metC3 ΔsafA::safAΔP58-S63 Spr Cmr | This work |
| AH4007 | trpC2 metC3 ΔsafA::safA4×AlaSpr Cmr | This work |
| AH4102 | trpC2 metC3 ΔsafA::safA+-gfp Spr Nmr | This work |
| AH4103 | trpC2 metC3 ΔsafA::safAΔG51-S63-gfp Spr Nmr | This work |
| AH4107 | trpC2 metC3 ΔspoVID::cm ΔsafA::safA+-gfp Spr Cmr Nmr | This work |
| AH4108 | trpC2 metC3 ΔspoVID::cm ΔsafA::safAΔG51-S63-gfp Spr Cmr Nmr | This work |
Spr, spectinomycin resistant; Cmr, chloramphenicol resistant; Nmr, neomycin resistant.
GST-SafA fusions.
Primer safA204D and primer safA-STOP51R, safA-STOP64R, safA-STOP78R, safA-STOP91R, or safA-STOP99R (the sequences of all primers are available upon request) were used to PCR amplify the coding regions of safA from residues 1 to 50, 1 to 63, 1 to 77, 1 to 90, and 1 to 98, respectively. The PCR fragments were digested with BamHI and XhoI and cloned between the same sites of pGex4T-3 (Pharmacia) to form pTC138 (GST-SafAE50), pTC159 (GST-SafAS63), pTC160 (GST-SafAK77), pTC161 (GST-SafAK90), and pTC139 (GST-SafAP98), respectively. The region encompassing codons 1 to 162 of safA was amplified with primers safA204D and safA693R, digested with BamHI, and cloned between the BamHI site and a filled-in XhoI site of pGex4T-3 (Pharmacia) to yield pTC141 (GST-SafAS162). pTC165 (GST-SafAΔG51-S63) was obtained after digestion with HincII and autoligation of a PCR obtained from pTC139 (see above) with primers safA50-64D and safA50-64R. pTC196 (GST-SafAΔG51-E57) and pTC197 (GST-SafAΔP58-S63) were constructed as follows. First, the safA coding regions from residues 1 to 50 and 1 to 57 were PCR amplified with primer sets safA204D/safA51-57R and safA204D/safA58-63R, respectively. Second, the regions corresponding to residues 58 to 98 and 64 to 98 of safA were PCR amplified with primer sets safA51-57D/safA-STOP99R and safA58-63D/safA-STOP99R, respectively. The fragments (codons 1 to 50 and 58 to 98 or 1 to 57 and 64 to 98) were mixed and subjected to PCR using primers safA204D and safA-STOP99R to obtain safA fragments comprising the region coding for the first 98 residues but excluding residues 51 to 57 or 58 to 63, respectively. The fragments were digested with BamHI and XhoI and introduced between identical sites of pGex4T-3 (Pharmacia) to create pTC196 and pTC197. All PCR products were sequenced. Plasmids pTC138, pTC159, pTC160, pTC161, pTC139, pTC141, pTC165, pTC196, and pTC197 were used to transform E. coli CC118(DE3)/pLysS to obtain strains AH2934, AH2978, AH2979, AH2980, AH2935, AH2937, AH2985, AH4086, and AH4087, respectively (Table 1).
GST-SpoVID fusions.
Strain AH2692 (Table 1) was obtained by transformation of E. coli CC118(DE3)/pLysS cells with pOZ169 (GST-SpoVIDFL) in which the entire spoVID coding sequence was translationally fused to the gst gene (33). We used pOZ169 and primer pairs spoVIDSTOP1498D/spoVID-STOP1498R, spoVID-STOP1198D/spoVID-STOP1198R, spoVIDSTOP907D/spoVID-STOP907R, and spoVID-STOP607D/spoVID-STOP607R to replace codons 500, 400, 303, and 203 in spoVID with nonsense codons by using the QuickChange site-directed mutagenesis system (Stratagene) to obtain plasmids pTC52 (GST-SpoVID499), pTC51 (GST-SpoVID399), pTC50 (GST-SpoVID302), and pTC49 (GST-SpoVID202), respectively. Plasmids pTC52, pTC51, pTC50, and pTC49 were used to transform E. coli CC118(DE3)/pLysS, yielding strains AH2691, AH2690, AH2689, and AH2688, respectively (Table 1). Fusion of GST to residues 201 to 575 and 201 to 399 of SpoVID was done as follows. We PCR amplified DNA fragments of spoVID (codons 201 to 575 and 201 to 399) with primers spoVID+800D and spoVID1935R and the pOZ169 and pTC51 templates, respectively. The products were digested with BamHI and XhoI and cloned between the same sites of pOZ169, creating pTC61 (GST-SpoVID201-575) and pTC62 (GST-SpoVID201-399), which were then introduced into CC118(DE3)/pLysS, yielding AH2718 and AH2719, respectively (Table 1). In order to obtain strain AH2687, producing native GST (Table 1), we excised the spoVID coding region from pOZ169 (33) by digestion with BamHI and XhoI and filled-in the ends before autoligation. The resulting plasmid, pTC55, was used to transform CC118(DE3)/pLysS, creating AH2687 (Table 1).
Mutations in regions A and B of SafA.
A PCR fragment comprising the entire safA coding region and 595 bp upstream of its start codon, generated with primers safA-595D and safA+1176R, was introduced in pCR2.1TOPO (Invitrogen) to create pTC75. Primers safA791D and safA834R were used to delete codons 203 to 206 (the PYYH motif) of safA in pTC75 by the QuickChange method, yielding pTC78. A chloramphenicol resistance (Cmr) cassette was released from pMS38 (43) with XbaI and XhoI and inserted between the same sites of pTC78, generating pTC90. A PCR fragment encompassing codons 1 to 162 of safA, including 595 bp upstream of the safA start codon, was amplified with primers safA-595D and safA693R and inserted into pCR2.1TOPO (Invitrogene) to obtain pTC93 (SafAN1-162). pTC93 was then digested with XbaI and XhoI, and a Cmr cassette released from pMS38 (43) with the same enzymes was inserted to form pTC95. To fuse the safA promoter directly to codons 164 to 387, codons 1 to 163 were eliminated by amplification of pTC88 (obtained by cloning of the Cmr cassette from pMS38 between the XbaI and XhoI sites of pTC75) with primers safA203R and safA677D, digestion of the resulting product with HincII, and religation to form pTC97 (SafAC164-387). To delete residues 51 to 63 of SafA, a product generated from pTC88 with primers safA50-64D and safA50-64R was cut with SacII and religated, to yield pTC170 (SafAΔG51-S63). pTC198 (SafAΔG51-E57) and pTC199 (SafAΔP58-S63) were constructed by cloning 143- and 146-bp fragments released from pTC196 and pTC197 (see above) with BbvII and EspI, respectively, between the same sites of pTC88 (see above). Alanine substitutions in SafA (R55A, K56A, K59A, and K62A) were obtained by the QuickChange method (Stratagene) by using pTC75 and primers safAAlamut50-64D and safAAlamut50-64R. The result was pTC172 (SafA4×Ala), from which pTC173 was made by introducing a Cmr cassette (see above) between its XbaI and XhoI sites. All PCR products were sequenced. pTC90, pTC95, pTC97, pTC170, pTC190, pTC191, and pTC173 were transformed into B. subtilis AOB68 to form AH2781, AH2787, AH2788, AH4006, AH4090, AH4091, and AH4007, respectively (Table 1).
Green fluorescent protein (GFP) fusions.
pTC204 (SafA+-GFP) and pTC205 (SafAΔG51-S63-GFP) were constructed as follows. First, the safA 3′ coding region (684 bp) was PCR amplified with primers safA677D and safA-gfpR. Second, a 714-bp fragment comprising the coding region of the gfp gene was PCR amplified with pEA18 (a gift from Alan Grossman) and primers gfp-30D and gfpmut2/749R. The resulting fragments were mixed and subjected to PCR with primers safA677D and gfpmut2/749R. The resulting 1,398-bp ′safA-gfp fragment was cleaved with SmaI and EcoRI, and the 1,110-bp fragment was gel purified. DNA fragments corresponding to the 5′ regions of safA+ or safAΔG51-S63 were obtained by digestion of pTC75 and pTC170 (see above), respectively, with SmaI and NotI. The safA+ and safAΔG51-S63 fragments were cloned together with the ′safA-gfp fragment between the NotI and EcoRI sites of pBEST501 (19) to yield pTC204 and pTC205, respectively. All PCR products were verified by sequencing. Strains AH4102 and AH4103 resulted from the integration of pTC204 or pTC205, respectively, at the safA locus of AOB68 via a single reciprocal (Campbell-type) crossover (Table 1). The spoVID null mutant AH1910 was transformed with chromosomal DNA of strains AH4102 and AH4103 to yield AH4107 and AH4108, respectively (Table 1).
Pull-down assays.
In vitro pull-down assays were performed as described previously (33), except that 60 μl of a 50% slurry of glutathione-Sepharose beads (Amersham Biosciences) were used and the interaction mixtures were resuspended in a final volume of 30 μl of loading buffer, before analysis by sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Purification of spores and analysis of the spore coat.
Spores were harvested by centrifugation of DSM cultures 24 h after the onset of sporulation, washed, and purified on step gradients of 20% to 50% Gastrografin (Schering) (8, 42). Coat proteins were extracted from purified spores at an optical density at 580 nm (OD580) of about 2 and resolved by SDS-15% PAGE (14, 15). The gels were stained with Coomassie brilliant blue R-250 or transferred to nitrocellulose for immunoblot analysis as described below.
Western blot analysis.
Pull-down samples and coat protein extracts were resolved by SDS-PAGE (12.5 and 15% gels, respectively) and immunoblot analysis conducted as previously described (8). Anti-SafA (32), anti-CotE (our unpublished work), and anti-CotG (43) antibodies were used at dilutions of 1:15,000, 1:1,000, and 1:7,500, respectively, whereas a horseradish peroxidase-conjugated secondary antibody (Sigma) was used at a dilution of 1:6,000. The immunoblots were developed with chemiluminescence reagents (Amersham Biosciences).
Microscopy.
Samples (0.6 ml) of DSM cultures were collected about 2.5 and 4 h after the initiation of sporulation, resuspended in 0.2 ml of phosphate-buffered saline supplemented with 2 μl of a 1-mg ml−1 solution of 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) for visualization of DNA. Microscopy was carried out as described previously (8, 36). DAPI staining allowed the identification of the prespore region and the ability to distinguish between cells that had just completed asymmetric division (with highly condensed prespore chromosomes) and those that had completed the engulfment process (diffuse DAPI signal in the prespore) (8, 36).
SPR experiments.
Surface plasmon resonance (SPR) experiments were conducted on a BIACORE 2000 instrument with a CM5 sensor chip (Biacore International AB). For all measurements and immobilization procedures, the temperature was set to 25°C. The running buffer was HEPES-buffered saline (150 mM NaCl, 10 mM HEPES [pH 7.2], 3 mM EDTA, and 0.005% surfactant P20). Two flow cells (Fc) of the CM5 chip were prepared for the immobilization of GST (Fc3) and GST-SpoVID202 (Fc4) by the standard amine coupling method provided by the manufacturer. Ligand immobilization was done at a flow rate of 10 μl/min with a buffer containing 10 mM sodium acetate, pH 3.9. The immobilization levels were established from 0.1 to 0.23 pmol/mm2 (about 3,000 to 13,500 resonance units [RUs]). Two peptides were used as analytes: SafA-regionA (GVPVRKEPKAGKS) and SafA-Ala (GVPVAAEPAAGAS) (GenScript Corporation). Binding assays were performed in triplicate in HEPES-buffered saline with a 0.8 mM concentration of each analyte and a flow rate of 10 μl/min. The flow cells were regenerated with 10 mM glycine (pH 2.0) after each cycle. Injection times were 3 min for binding and 30 s for regeneration.
Protein identification by peptide mass fingerprinting.
Proteins were excised from Coomassie brilliant blue R-250-stained gels and sent to the Microchemical Facility at Emory University for matrix-assisted laser desorption ionization-time-of-flight analysis.
RESULTS
The PYYH motif is involved in formation of the SafA-SpoVID complex.
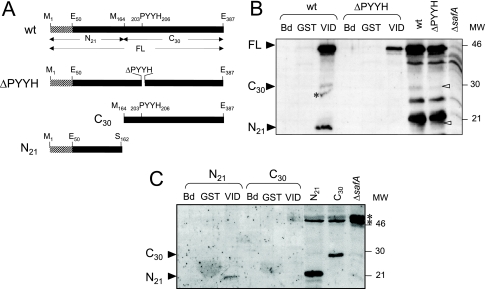
The PYYH motif, found in a phage display screen for peptides able to interact with SpoVID, is present in the C-terminal half of the SafA protein (residues 203 to 206) (Fig. 1A) (32). To investigate whether the PYYH motif is involved in the interaction of SafA with SpoVID, we constructed a mutant expressing a form of SafA lacking residues 203 through 206, or SafAΔPYYH (Fig. 1A). Then, we tested whether a GST-SpoVID purified protein was able to pull down SafAΔPYYH. GST-SpoVID and GST alone were overproduced in E. coli, bound to glutathione-Sepharose beads, and incubated with extracts from wild-type B. subtilis or the safAΔPYYH mutant prepared at hour 4 of sporulation. We also incubated the extracts with beads alone as a control. After extensive washing, retained proteins were resolved by SDS-PAGE, blotted, and probed with an anti-SafA antibody (see Materials and Methods). The three main forms of SafA that accumulate in sporulating cells, SafAFL (45 kDa), C-terminal (SafAC30, 30 kDa), and N-terminal (SafAN21, 21 kDa), were pulled down by the glutathione-Sepharose beads to which GST-SpoVID had been bound but not by beads containing GST or by the beads alone (Fig. 1B). Similar experiments showed that the full-length form of SafA deleted for the PYYH motif (SafAΔPYYH) was also retained by GST-SpoVID but not by GST or the beads (Fig. 1B). In contrast, the SafAC30 and SafAN21 forms in the SafAΔPYYH strain were not retained by the GST-SpoVID beads (Fig. 1B). Yeast two-hybrid experiments have shown that SafAN21 can directly interact with SpoVID, but the interaction seemed weaker than the interaction of SafAN21 with SafAFL (32; also see below). SafAN21 may not bind directly to SpoVID in the pull-down assay because it is out-competed by SafAFL or by SafAΔPYYH. This suggests that SafAN21 is recruited by the SpoVID-SafA complex but not by the SpoVID-SafAΔPYYH complex (Fig. 1B). This also suggests that a site able to interact with SpoVID is likely to exist in the N-terminal region of SafA (also see below). In the yeast two-hybrid analysis of Ozin et al. (32), no interaction was detected between SafAC30 and SpoVID, and GST-SafAC30 was able to pull down only itself from extracts of sporulating cells (33). Thus, it appears that the SafAC30 form interacts with the SpoVID-SafA complex but not with the SpoVID-SafAΔPYYH complex (Fig. 1B). Together, the results suggest that the PYYH motif in SafA (hereafter named region B) is not essential for the interaction of full-length SafA with SpoVID but appears to affect the SpoVID-SafA complex in a way that interferes with its capacity to recruit the SafAN21 and SafAC30 forms of SafA.
FIG. 1.
Interaction of the N terminus of SafA with GST-SpoVID. (A) The 387-residue-long wild-type (wt) SafA protein and SafA variants deleted for residues 203 to 206 (ΔPYYH), for the N-terminal first 163 residues (C164-387, corresponding to the SafAC30 form), or missing the last C-terminal 225 residues (N1-162, corresponding to the SafAN21 form). The striped pattern represents the LysM domain (first 50 residues) of SafA. Other relevant residues are also indicated. SafA accumulates in sporulating cells in three main forms: SafAFL (45 kDa), a C-terminal form (SafAC30, 30 kDa), and an N-terminal form (SafAN21, 21 kDa). (B and C) Interaction of immobilized GST-SpoVID with wild-type SafA and SafAΔPYYH (B) and with the SafAC30 and SafAN21 forms (C) present in extracts at hour 4 of sporulation. The extracts were incubated with GST-SpoVID (VID for simplicity), with GST bound to glutathione-Sepharose beads, or with the beads alone (Bd). Pulled-down proteins were resolved by SDS-PAGE and immunoblotted with an anti-SafA antibody. The positions of SafAFL, SafAC30, and SafAN21 (black arrowheads), of cross-reactive species (asterisks), and of molecular mass markers (in kilodaltons) are indicated. The last three lanes in panel C show the immunoblot analysis of the extracts from strains producing SafAN21 or SafAC30, used in the pull-down assays, and of a ΔsafA mutant, with the anti-SafA antibody.
Residues 51 to 63 of SafA are essential for its interaction with SpoVID in vitro.
The results in preceding section, together with previous results showing that SafAN21 interacts with SpoVID in a yeast two-hybrid assay (32), strongly suggest that a second site in SafA (presumably in its N-terminal region) must exist that promotes its interaction with SpoVID. To test this, we performed pull-down assays using B. subtilis mutants producing only the SafAN21 (residues 1 to 162) or SafAC30 (residues 164 to 387) forms (Fig. 1A). The results in Fig. 1C show that only SafAN21 (first 162 residues) was retained by GST-SpoVID, indicating that this region contains residues sufficient for the interaction with SpoVID. The observation that, when produced alone, “wild-type” SafAC30 (i.e., carrying region B) was not retained by GST-SpoVID (Fig. 1C) is consistent with results of previous studies in which SafAC30 and SpoVID did not interact (32, 33).
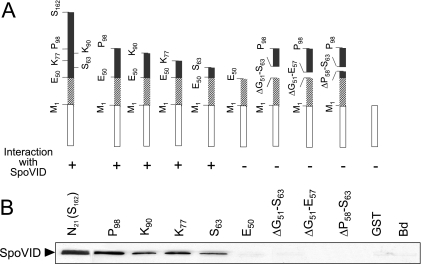
To determine whether a specific region within the first 162 residues of SafA was involved in the interaction with SpoVID, we overproduced several truncated forms of SafA as GST fusion proteins in E. coli. The forms of SafA produced contained residues 1 to 162 (herein designated S162, the letter and number referring to the identity and position of the last residue in the SafA fragment), 1 to 98 (P98), 1 to 90 (K90), 1 to 77 (K77), 1 to 63 (S63), or 1 to 50 (E50) (Fig. 2A). All of the truncated forms of SafA were able to pull down untagged SpoVID overproduced in E. coli except the form containing just the first 50 residues of SafA (E50), which correspond to its LysM domain (Fig. 2B). The results indicate that the region of 13 amino acids from residues 51 to 63 in SafA (GVPVRKEPKAGKS, hereafter referred to as region A), just downstream of the LysM domain, is sufficient for the interaction with SpoVID in vitro. In confirmation of this assumption, deletion of residues 51 to 63 (ΔG51-S63) from the P98 form prevented its interaction with SpoVID (Fig. 2A and B). In an attempt to define region A further, residues 51 to 57 (ΔG51-E57, i.e., the N-terminal half of region A) or 58 to 63 (ΔP58-S63, its C-terminal half) were removed from the P98 form (Fig. 2A). Neither of the new versions of P98 was able to pull down SpoVID (Fig. 2B), indicating that residues on both halves of region A are required for the interaction with SpoVID in vitro.
FIG. 2.
Deletion mapping of region A in SafA. (A) The various GST-SafA fusions used, with the white box representing GST (not to scale) and the striped pattern (residues 1 to 50) the LysM motif. The plus or minus symbol below each diagram indicates an interacting or noninteracting fusion protein, respectively. (B) Results of pull-down assays using truncated forms of GST-SafA and SpoVID. A cell extract prepared from an E. coli strain overproducing native SpoVID was incubated with the various purified GST-SafA truncated forms containing the following residues, as indicated: 1 to 162 (S162), 1 to 98 (P98), 1 to 90 (K90), 1 to 77 (K77), 1 to 63 (S63), 1 to 50 (E50), 1 to 98 but excluding residues 51 to 63 (ΔG51-S63), 1 to 98 but excluding residues 51 to 57 (ΔG51-E57), and 1 to 98 but excluding residues 51 to 63 (ΔP58-S63). The same extracts were incubated with GST bound to glutathione-Sepharose beads or with beads alone (Bd), as controls. Pulled-down proteins were resolved by SDS-PAGE and immunoblotted with an anti-SpoVID antibody (see Materials and Methods). The position of SpoVID is indicated by the black arrowhead.
Region A of SafA directly contacts SpoVID.
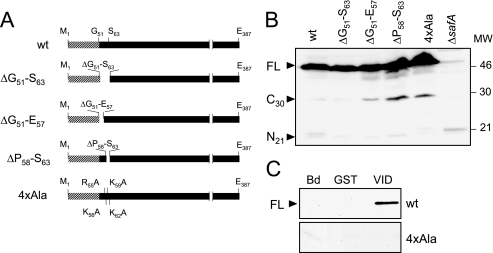
All three deletion forms of SafA (SafAΔG51-S63, SafAΔG51-E57, and SafAΔP58-S63) accumulated to wild-type levels in extracts from B. subtilis cells harvested at hour 4 of sporulation (Fig. 3B) but failed to bind to immobilized GST-SpoVID (not shown). Therefore, the complete or partial deletion of region A does not alter the stability of SafA significantly. (Note that for reasons we do not presently understand, the accumulation of the SafAN21 and SafAC30 forms varied among the three safA deletion mutants and in comparison to the wild type, as shown in Fig. 3B; however, we also note that only SafAFL is required for the formation of lysozyme-resistant spores [33].) We also tested the effect of alanine substitutions within region A on the interaction with SpoVID. The results shown in Fig. 3B indicate that the replacement of residues R55, K56, K59, and K62 in region A by alanines (R55A, K56A, K59A, and K62A), to produce the SafA4×Ala form (Fig. 3A), did not interfere with the accumulation of SafA in extracts of sporulating cells. However, SafA4×Ala was not pulled down by GST-SpoVID in vitro (Fig. 3C), suggesting that residues R55, K56, K59, and K62 in SafA are directly involved in the interaction with SpoVID.
FIG. 3.
Accumulation of SafA variants in sporulating B. subtilis and analysis of their interaction with GST-SpoVID. (A) The 387-amino-acid wild-type (wt) SafA protein and SafA variants deleted for residues 51 to 63 (ΔG51-S63), 51 to 57 (ΔG51-E57), and 58 to 63 (ΔP58-S63) or with alanine substitutions of residues R55, K56, F62, and K59 (4×Ala). The LysM domain (residues 1 to 50) in SafA is indicated by the striped pattern. (B) Accumulation of SafA variants in cell extracts prepared from cultures of B. subtilis at hour 4 of sporulation. The proteins were resolved by SDS-PAGE and immunoblotted with an anti-SafA antibody. The positions of the full-length (FL), SafAC30, and SafAN21 forms of SafA (black arrowheads), of a cross-reactive species (asterisk), and of molecular mass markers (in kilodaltons) are indicated. (C) Interaction of SafA (wild type) or the SafA4×Ala variant present in extracts from sporulating B. subtilis with GST-SpoVID (VID). Cell extracts prepared from wild-type B. subtilis and from strains producing SafA+ or SafA4×Ala were incubated with GST-SpoVID or GST immobilized on glutathione-Sepharose beads or with the beads alone (Bd), and the pulled-down proteins were resolved by SDS-PAGE and immunoblotted with an anti-SafA antibody (see Materials and Methods). The position of SafAFL is indicated by the black arrowhead.
To independently test whether region A of SafA is directly involved in the interaction with SpoVID, we conducted SPR experiments with immobilized GST or GST-SpoVID202 (a truncated form of SpoVID that binds SafA in pull-down assays as well as the wild-type protein; see below) and peptides corresponding to either region A (SafA-regionA, GVPVRKEPKAGKS) or to region A bearing the four alanine substitutions (SafA-Ala, GVPVAAEPAAGAS). GST and GST-SpoVID202 were immobilized in flow cells 3 and 4 of a CM5 sensor chip, respectively (see Materials and Methods), and the SafA-regionA or SafA-Ala peptides were tested for binding with a BIAcore system (see Materials and Methods). We obtained 27.7 ± 4.0 RUs as the response for binding of SafA-regionA to GST-SpoVID202 (in flow cell 4) but only −0.7 ± 0.4 RUs for binding of the SafA-Ala peptide (note that the negative values are caused by differences in the refractive indices between the buffer with or without the ligand). No binding was detected for either the SafA-regionA or SafA-Ala peptide (0.9 ± 0.4 RUs and −1.7 ± 0.4 RUs, respectively) to immobilized GST alone (in flow cell 3). These results strengthen our inference that residues in region A of SafA are involved in the interaction with SpoVID.
The LysM motifs in both SpoVID and SafA are not involved in their interaction.
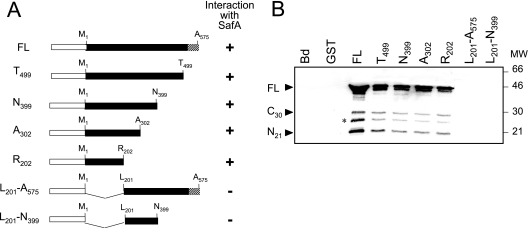
The observation that SafAN21 interacted with SpoVID in a yeast two-hybrid assay led Ozin and coworkers to propose that the two proteins could interact via their LysM domains (32). However, the results presented above indicate that SafA does not interact with SpoVID via its LysM domain (fusion E50 in Fig. 2). To examine whether the LysM domain present at the C terminus of SpoVID was involved in the interaction with SafA, we conducted pull-down assays using GST fusions to SpoVIDFL or to SpoVID deleted in its last 76 residues (T499), which encompass the LysM motif (Fig. 4A). The results shown in Fig. 4B indicate that both versions of SpoVID were able to pull down all main forms of SafA present in B. subtilis extracts, that is, SafAFL, SafAN21, and SafAC30. These experiments suggest that the LysM domain of SpoVID is not involved in the interaction with SafA. To determine whether SafA binds to a specific section of the 575-residue-long SpoVID protein, additional pull-down assays were carried out with fusions of GST to truncated forms of SpoVID corresponding to residues 1 to 202 (R202), 1 to 302 (A302), and 1 to 399 (N399) (Fig. 4A). We found that all of the truncated forms of SpoVID were able to pull down the main forms of SafA present in B. subtilis extracts (Fig. 4B). Hence, the first 202 residues of SpoVID are sufficient for the interaction with SafA. To determine whether this region is necessary for the interaction, we performed similar pull-down experiments using fusions of GST to truncated forms of SpoVID lacking either its first 200 residues (L201-A575) or comprising only residues 201 to 399 (L201-N399) (Fig. 4A). Neither of these fusion proteins retained SafA (Fig. 4B). We infer from these results that the LysM domain of SpoVID is not involved in the interaction with SafA. We further infer that the N-terminal 202 residues of SpoVID are necessary for the interaction with SafA (also see below).
FIG. 4.
SafA interacts with a defined region of SpoVID. (A) Schematic representation of fusions of the 575-residue-long SpoVIDFL protein (FL), as well as forms of SpoVID containing residues 1 to 499 (T499), 1 to 399 (N399), 1 to 302 (A302), 1 to 202 (R201), 201 to 575 (L201-A575), and 201 to 399 (L201- N399), to GST. The white box represents GST (not to scale), and the LysM motif (residues 525 to 575) corresponds to the striped pattern. The plus or minus symbol on the right side of each diagram indicates a protein interacting or not interacting with SafA, respectively. (B) Results of pull-down assays performed with the various purified truncated forms of SpoVID fused to GST and extracts from wild-type B. subtilis AOB90 (Table 1) prepared at hour 4 of sporulation. The immunoblot was probed with an anti-SafA antibody. The same extracts were also incubated with immobilized GST or with glutathione beads alone (Bd). The black arrowheads indicate the positions of the full-length, SafAC30, and SafAN21 forms of SafA, and the asterisk indicates a degradation product of SafA. Molecular mass markers (in kilodaltons) are also indicated.
Region A but not region B of SafA is required for proper coat assembly.
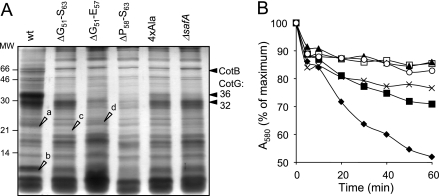
Spores of a safA null mutant have abnormal coats, from which several coat components are missing, and are susceptible to lysozyme (32, 40). Because the SafAΔG51-S63, SafAΔG51-E57, SafAΔP58-S63, and SafA4×Ala mutant forms accumulated to normal levels in B. subtilis yet failed to interact with SpoVID, we wanted to assess the impact of mutations in region A in the assembly of the spore coat. Spores of the various region A mutants, along with wild-type and safA null mutant spores, were purified, and the collection of coat polypeptides was extracted and analyzed by SDS-PAGE (Fig. 5A). safA null mutant spores lack the 36-kDa CotG protein, as judged by Coomassie staining of gels of coat protein extracts (40) (Fig. 5A). A 36-kDa band, and to a lesser extent a band of about 32 kDa, seen in wild-type coat extracts appeared reduced from the extracts of the various safA mutants (Fig. 5A). These represent 36- and 32-kDa forms of CotG (43), as verified by immunoblot analysis (not shown). In agreement with the reduction in the representation of CotG, the 66-kDa form of CotB (as verified by immunoblot analysis; data not shown), whose formation depends on CotG (43), was also reduced in all safA mutants (Fig. 5A). Two other bands that appeared reduced in all of the safA mutants are labeled a and b in Fig. 5A. Mass spectrometry analysis indicated that these bands contained CotJC and YhcN (band a [38]) and CotC sequences (band b [10]). In contrast, band c appeared more abundant in the extracts from SafAΔG51-S63 spores than from the wild type (Fig. 5A), whereas band d was more abundant in the extracts from SafAΔG51-E57 spores (Fig. 5A). We found band c to contain GerQ and band d to contain SodA, previously found in association with the spore coat and involved in the oligomerization of CotG (17, 25). The differences between the various SafA deletion mutants may reflect subtle differences in the capacity of residues in the N- and C-terminal halves of region A to promote the interaction of SafA with SpoVID (also see below). In contrast, deletion of region B did not affect in a discernible way the profile of Coomassie-stained coat polypeptides (data not shown). Together, the results show alterations of the coat structure caused by lesions in region A of SafA that are largely coincident with those caused by a safA null allele. We infer that the coat structural deficiencies seen in region A mutants are caused by the impaired interaction of SafA with SpoVID.
FIG. 5.
Spores of region A mutants have an altered coat and impaired germination. (A) Coomassie-stained gel of SDS-PAGE-resolved coat protein extracts prepared from purified spores of the wild type (wt) (lane 1) and the SafAΔG51-S63 (lane 2), SafAΔG51-E57 (lane 3), SafAΔP58-S63 (lane 4), SafA4×Ala (lane 5), and ΔsafA (lane 6) mutants. Open arrowheads point to bands (a to d) that were excised and subjected to mass spectrometry analysis (see Materials and Methods). The positions of bands corresponding to the 66-kDa form of CotB and to the 32- and 36-kDa forms of CotG are indicated by black arrowheads. (B) Germination responses to AGFK of purified spores of the wild type (closed diamonds) and the SafA4×Ala (closed squares), SafAΔG51-S63 (open squares), SafAΔG51-E57 (closed triangles), SafAΔP58-S63 (crosses), and ΔsafA mutants (open circles). Germination was monitored by following the decrease in absorbance at 580 nm over time (in minutes) and is shown as the percentage of the OD580 at the beginning of the assay.
Region A in SafA is required for the formation of a functional coat.
Spores of a safA null mutant are susceptible to lysozyme (32, 40). To examine whether the mutations in region A and the associated perturbations at the level of the coat polypeptide composition were sufficient to affect this functional property of the coat, spores of the various region A mutants were assayed for lysozyme resistance. The results in Table 2 show that spores produced by mutants lacking residues 51 to 63 (region A), 51 to 57, or 58 to 63 of SafA have a degree of susceptibility to lysozyme treatment in the range of that observed for spores of a safA null mutant (about 107 lysozyme-resistant spores/ml of culture, compared to 108 for wild-type spores). None of the mutations in region A affected spore heat resistance (Table 2), a property that relies on proper cortex formation, and hence the effects herein described are specifically related to the assembly of the spore coat. We have also found that spores of the SafAΔPYYH mutant are fully resistant to lysozyme (Table 2), an observation that is in agreement with absence of any discernible alteration in the profile of Coomassie-stained coat polypeptides (see above). This reinforces the suggestion that the PYYH motif is not a critical determinant for the SafA-SpoVID interaction (see above). Surprisingly however, spores of the SafA4×Ala mutant were also found to be resistant to lysozyme (Table 2). Presumably, the alanine substitutions within region A still permit some degree of interaction of SafA with SpoVID in vivo, sufficient to support spore lysozyme resistance. In support of this interpretation, we found that when examined for their germination in response to a mixture of asparagine, glucose, fructose, and KCl (AGFK), a property that is dependent on safA (40), spores of the SafA4×Ala mutant showed an intermediate phenotype between the wild type and the safA null mutant (Fig. 5B). In contrast, the results in Fig. 5B show that spores of the SafAΔG51-E57 and SafAΔG51-S63 mutants were as impaired in AGFK-induced germination as were spores of the safA null mutant. Spores of the SafAΔP58-E63 mutant however, showed a response to AGFK that closely paralleled that of the SafA4×Ala mutant (Fig. 5B), suggesting that residues in the N-terminal half of region A have a greater impact on the SafA-SpoVID interaction, at least as assessed by this spore property. Overall, the finding that mutations in region A led to a germination phenotype that largely mimics that of a safA null mutant strengthens the view that the function of SafA critically depends on its interaction with SpoVID.
TABLE 2.
Spore resistance properties
| Strain | Relevant genotype | Cell count (CFU/ml)a
|
||
|---|---|---|---|---|
| Viable | Heat resistant | Lysozyme resistant | ||
| MB24 | Wild type | 8.3 × 108 | 7.6 × 108 | 6.5 × 108 |
| AOB68 | ΔsafA | 5.0 × 108 | 6.1 × 108 | 3.1 × 107 |
| AH2781 | safAΔPYYH | 7.9 × 108 | 5.1 × 108 | 4.3 × 108 |
| AH4006 | safAΔG51-S63 | 4.9 × 108 | 3.8 × 108 | 7.6 × 107 |
| AH4007 | safA4×Ala | 4.4 × 108 | 4.7 × 108 | 2.2 × 108 |
| AH4090 | safAΔG51-E57 | 2.3 × 108 | 2.3 × 108 | 2.7 × 107 |
| AH4091 | safAΔP58-S63 | 4.1 × 108 | 3.4 × 108 | 7.0 × 107 |
Total (viable) and heat- and lysozyme-resistant cell counts were determined as described in Materials and Methods.
Targeting of SafA to the surface of the developing spore depends on its interaction with SpoVID.
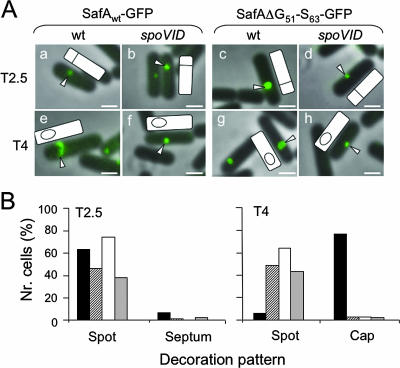
Lastly, we examined the impact of eliminating region A on the subcellular localization of SafA in developing cells. SafA is targeted to the surface of the developing spore early in the process of coat assembly, just after formation of the asymmetric septum that partitions the sporulating cell into the mother cell and prespore compartments (33). Targeting of SafA occurs in two stages, as defined by immunofluorescence microscopy: first, SafA localizes as a spot at the mother cell/prespore border in a SpoVID-independent manner, and later SafA migrates around the developing spore in a SpoVID-dependent manner (33). Both wild-type SafA and SafAΔG51-S63 were fused to GFP, and the localizations of both fusion proteins were studied by fluorescence microscopy (Fig. 6A). First, we established that spores of a strain expressing SafA-GFP were resistant to lysozyme, indicating that the fusion protein is largely functional (data not shown). Then, we studied the pattern of localization of both fusion proteins in the wild type and in a spoVID congenic mutant 2.5 and 4 h after the onset of sporulation. In wild-type cells, SafA-GFP was first targeted to the mother cell/prespore border as a spot (Fig. 6A, panel a) and later started migrating around the prespore to form a cap covering the spore end distal to the cell pole (Fig. 6A, panel e) in a spoVID-dependent mode (Fig. 6A, panel f). In the wild type, the “spot” pattern was associated with cells that had just undergone asymmetric division but had not initiated engulfment, whereas the “cap” pattern was found for cells that had initiated engulfment (the morphological stages were scored by DAPI staining, as described in Materials and Methods). The quantification of these results is shown in Fig. 6B. Note that the “spot” pattern predominated in samples at hour 2.5 of sporulation and was not affected by mutation of spoVID, whereas accumulation of the “cap” pattern, which predominated at hour 4, required expression of spoVID (Fig. 6B). These results agree with those of Ozin and coworkers (33), except that in their experiments SafA was found as caps covering both spore ends. However, the signal at the pole-proximal spore end was always weaker than the signal at the spore end away from the cell pole (33). We emphasize that our SafA-GFP fusion is functional, and we presume that the fluorescence at the pole-proximal spore end is not detected under our conditions. In any event, a strikingly different pattern was found for the SafAΔG51-S63-GFP fusion protein. SafAΔG51-S63-GFP was still targeted at an early time (hour 2.5) to the asymmetric septum, where it accumulated as a spot (Fig. 6A, panels c and d). However, for this protein the incidence of the “spot” pattern persisted at hour 4, at the expense of the “cap” pattern, whose representation was severely curtailed regardless of the presence of a functional spoVID locus (Fig. 6A, panels g and h, and Fig. 6B). We conclude that deletion of region A in SafA strongly interferes with the SpoVID-dependent stage of SafA localization, and we infer that the interaction of SafA with SpoVID via region A is required for the correct localization of SafA to the developing coat.
FIG. 6.
Region A is important for the SpoVID-dependent localization of SafA. (A) Cells expressing a functional fusion of SafA to GFP (SafA-GFP) or SafAΔG51-S63-GFP in the wild type (wt) (panels a, c, e, and g) or in a spoVID background (panels b, d, f, and h) were observed by fluorescence microscopy at 2.5 h (panels a to d) and 4 h (panels e to h) after the onset of sporulation in DSM. Overlay images of GFP fluorescence and phase-contrast microscopy are shown in all panels. White arrowheads point to the region showing fluorescence in a selected cell, and the position of the prespore (as determined by DAPI chromosomal staining; not shown) is shown in the schematic representation of the same cell included in each panel. The scale bar represents 1 μm. (B) Quantification of GFP decoration patterns. A minimum of 75 cells were scored for fluorescence patterns designed by spot, septum, or cap at hours 2.5 and 4 in cultures of both the wild type and spoVID mutant expressing either SafA-GFP or SafAΔG51-S63-GFP: AH4102 (SafA-GFP; black bars), AH4103 (SafAΔG51-S63-GFP; striped bars), AH4107 (SafA-GFP spoVID; white bars), and AH4108 (SafAΔG51-S63-GFP spoVID; gray bars).
DISCUSSION
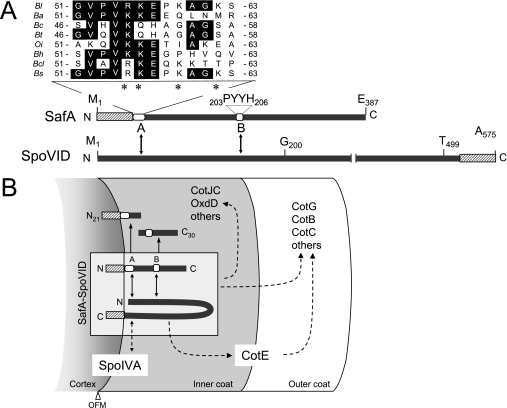
Previous work has suggested that the PYYH motif (region B) in the C-terminal half of SafA, found in a phage display screen for peptides able to interact with SpoVID, could mediate the interaction of SafA with SpoVID in vivo (32, 33) (Fig. 7A). However, a second region of interaction was postulated to exist in the N-terminal half of SafA (33). We now show that such a region indeed exists in the N-terminal half of SafA (Fig. 7A). SafAN21 (but not SafAC30) could be pulled out by immobilized GST-SpoVID, and deletion mapping indicated that a region of 13 amino acids encompassing residues 51 to 63 of SafA, or region A, is sufficient for the interaction with SpoVID in vitro (Fig. 2). Moreover, region A is required for the interaction with SpoVID and for the proper subcellular localization and function of SafA in vivo (Fig. 5 and 6). Importantly, variants of SafA with the complete or partial deletion of region A accumulated in B. subtilis but failed to promote proper assembly of the coat, lysozyme resistance, and normal spore germination, mimicking the phenotypes of a safA null mutant (32, 40). That region A of SafA is directly involved in the interaction with SpoVID is supported by two lines of evidence. First, a form of SafA with alanine substitutions in region A accumulated in vivo but could not be pulled down by GST-SpoVID. Second, a peptide consisting of the wild-type region A sequence, but not a related peptide bearing the same four alanine substitutions (above), was able to bind to immobilized GST-SpoVID.
FIG. 7.
Interaction of SafA with SpoVID and role of the complex in spore coat assembly. (A) Structure of SafA and SpoVID, with the LysM motifs (striped pattern) shown in each protein, as well as regions A (residues 51 to 63) and B (PYYH motif) in SafA (white boxes). Also shown is an alignment of the sequence of region A with the following SafA orthologs: Bs, B. subtilis 168 (accession number NP_390662); Bl, B. licheniformis ATCC14580 (YP_080055); Bt, B. thuringiensis serovar konkukian strain 97-27 (YP-038478); Ba, B. anthracis “Ames ancestor” (YP_021306); Bc, B. cereus ATCC 10987 (NP_980805); Bcl, B. clausii KSM-K16 (YP_175046); Bh, B. halodurans C-125 (NP_242087); Oi, Oceanobacillus iheyensis HTE831 (NP_692961). Identical residues are highlighted, and asterisks indicate the alanine substitutions in SafA4×Ala. (B) Model for the localization of SafA and SpoVID. Both SafA and SpoVID localize at the cortex/coat interface, delimited by the outer forespore membrane (OFM). Targeting of SpoVID to this region requires SpoIVA and may involve a direct interaction between the two proteins. SafA is initially targeted to the cortex/coat interface independently of SpoVID, a step that may involve its LysM domain. In a second stage, SafA interacts with SpoVID via regions A and B to form the SpoVID-SafA complex (shown within a box), which allows SafA to encircle the spore. SafA or the SpoVID-SafA complex controls the assembly of a subset of inner and outer coat proteins (i.e., CotE controlled), and SpoVID is essential for maintaining the coat anchored to the cortex, in part because of its requirement for keeping CotE at the inner coat/outer coat interface. Solid arrows indicate a direct interaction, whereas broken arrows indicate direct or indirect interactions.
Region B is also involved in SpoVID-SafA complex formation (33) (Fig. 7A), as its deletion did not allow immobilized SpoVID to pull down SafAN21 or SafAC30 (Fig. 1B). This suggests that SafAN21 and SafAC30 are not bound directly by SpoVID but rather are recruited by the complex formed by SafA and SpoVID. SafAN21 binds to SpoVID when made alone but may be outcompeted by SafAFL or by SafAΔPYYH, in agreement with the yeast two-hybrid analysis of Ozin and coauthors, which suggested a SafAN21-SpoVID interaction weaker than that of SafAN21 with SafAFL (33). SafAC30, in turn, was not pulled down by GST-SpoVID when made alone (Fig. 1C), also in agreement with an earlier study which found that GST-SafAC30 could not bind to SpoVID in the absence of SafAFL and that no interaction was detected between SafAC30 and SpoVID in a yeast two-hybrid assay (33). In any case, while deletion of region A causes spore susceptibility to lysozyme, deletion of region B does not (Table 2; also see below). Taken together, the results suggest that region A is essential for the interaction of SafA with SpoVID and that region B (while not essential) favors the interaction and/or allows the SafA-SpoVID complex to attain the correct topology for binding of SafAN21 and SafAC30 (Fig. 7B).
The consequences of lesions in region A were reminiscent of the effects of a safA null allele. First, spores of the various mutants had aberrant coats, from which CotG (both the 32- and 36-kDa forms), CotB, CotC, and possibly CotJC (10, 15, 37, 38) were reduced or missing (Fig. 5A). Second, as did those of a safA null mutant (40), spores of all region A mutants showed impaired germination in response to AGFK (Fig. 5B). Lastly, with the exception of the SafA4×Ala mutant, the partial or complete deletion of region A results in spores that were, like those of the safA null mutant (32, 40), susceptible to lysozyme (Table 2). We take these results as indicating that the interaction of SafA (via region A) with SpoVID is responsible for its morphogenetic function in coat assembly. The differences in the composition of the coat extracts, as well as in the germination responses among the various mutants and the lysozyme resistance of the SafA4×Ala mutant, suggest that the various mutations affect the interaction in vivo to various degrees, influencing the assembly of other downstream components.
SafA localizes to the developing spore in two stages (33). First it localizes as a dot at the mother cell/prespore border in a SpoVID-independent manner. Second, under the guidance of SpoVID, it encircles the prespore. Our results indicate that while a functional fusion of SafA to GFP undergoes assembly following the two-stage pathway described for the native protein, a SafA-GFP fusion lacking region A did not initiate the SpoVID-dependent stage (Fig. 6). These results suggest that SafA interacts with SpoVID via region A after the initial targeting of both proteins to the nascent coat and that this interaction allows SafA to follow SpoVID in its movement around the prespore as engulfment proceeds. We suggest that SpoVID directly interacts with a factor localized close to or at the prespore outer membrane that itself is pulled around the prespore during engulfment. This factor could be the morphogenetic protein SpoIVA, which localizes close to the prespore outer membrane and upon which the localization of SpoVID is dependent (12, 33) (Fig. 7B).
We do not yet know how is SafA initially targeted to the surface of the developing spore. It is possible that the initial (SpoVID-independent) targeting of SafA, which then permits the interaction with SpoVID, involves its LysM domain. For example, the tandem LysM domains of another coat protein component, YaaH, are sufficient to direct β-lactamase to the surface of the developing spore (23, 24). However, we could not yet test for the involvement of the LysM domain in the initial targeting of SafA because deletion of this region results in an unstable protein that does not accumulate in sporulating cells (our unpublished results).
Formation of the SafA-SpoVID complex involves defined regions in SafA (regions A and B) and SpoVID (first 202 residues), suggesting that SafA and SpoVID have a modular design. Other morphogenetic proteins, for example, SpoIVA and CotE, also appear to have a modular design, with regions specifically involved in their targeting, multimerization, and interactions with additional coat components (3, 7, 21, 28, 34). Both SafA and SpoVID localize to the cortex/coat interface, where they promote attachment of the coat to the cortex (4, 12, 32, 40; also this work), and both proteins and their complex are likely to interact with additional coat components (6, 10, 23-26, 33, 38) (Fig. 7B).
SpoVID and SafA orthologs are present in all Bacillus species whose genomes have been sequenced to date, as well as in other related species (not shown). Importantly, the recent study of a SafA ortholog, called ExsA, revealed its role in promoting anchoring of both the coat and exosporium layers to the spore in B. cereus (1). Together with the observation that region A is highly conserved among SafA orthologs (Fig. 7A), this suggests that the overall role and perhaps some of the mechanistic details of the functioning of SafA in coat morphogenesis are conserved in the Bacillus group of spore formers.
Acknowledgments
We thank Melissa Wilby (Emory University) for comments on the manuscript and John Pohl (Emory Microchemical Facility) for the mass spectrometry analysis.
This work was supported by European Union grant QLK5-CT-2001-01729 and by grant CONC-REEQ/692/2001 from “Fundação para a Ciência e a Tecnologia” (FCT) to A.O.H. and by NIH grant GM54395 to C.P.M. T.C. (SFRH/BD/1167/00) and A.L.I. (SFRH/BPD/8967/2002) were the recipients of fellowships from FCT.
Footnotes
Published ahead of print on 1 September 2006.
REFERENCES
- 1.Bailey-Smith, K., S. J. Todd, T. W. Southworth, J. Proctor, and A. Moir. 2005. The ExsA protein of Bacillus cereus is required for assembly of coat and exosporium onto the spore surface. J. Bacteriol. 187:3800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, T., S. Little, A. G. Stöver, and A. Driks. 1999. Functional regions of the Bacillus subtilis spore coat morphogenetic protein CotE. J. Bacteriol. 181:7043-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkeland, N. K. 1994. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi LC3: a dual lysis system of modular design. Can. J. Microbiol. 40:658-665. [DOI] [PubMed] [Google Scholar]
- 6.Bourne, N., P. C. FitzJames, and A. I. Aronson. 1991. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J. Bacteriol. 173:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalano, F. A., J. Meador-Parton, D. L. Popham, and A. Driks. 2001. Amino acids in the Bacillus subtilis morphogenetic protein SpoIVA with roles in spore coat and cortex formation. J. Bacteriol. 183:1645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, T., L. Steil, L. O. Martins, U. Völker, and A. O. Henriques. 2004. Assembly of an oxalate decarboxylase produced under σK control into the Bacillus subtilis spore coat. J. Bacteriol. 186:1462-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. V. Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 10.Donovan, W., L. B. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driks, A., S. Roels, B. Beall, C. P. Moran, Jr., and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 13.Eichenberger, P., M. Fujita, S. T. Jensen, E. M. Conlon, D. Z. Rudner, S. T. Wang, C. Ferguson, K. Haga, T. Sato, J. S. Liu, and R. Losick. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriques, A. O., B. W. Beall, and C. P. Moran, Jr. 1997. CotM of Bacillus subtilis, a member of the alpha-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J. Bacteriol. 179:1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a σE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriques, A. O., T. Costa, L. O. Martins, and R. Zilhão. 2004. Functional architecture and assembly of the spore coat, p. 34-52. In E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spores: probiotics and emerging applications. Horizon Scientific Press, London, United Kingdom.
- 17.Henriques, A. O., L. R. Melsen, and C. P. Moran, Jr. 1998. Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J. Bacteriol. 180:2285-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 19.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joris, B., S. Englebert, C. P. Chu, R. Kariyama, L. Daneo-Moore, G. D. Shockman, and J. M. Ghuysen. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70:257-264. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59:487-502. [DOI] [PubMed] [Google Scholar]
- 22.Klobutcher, L. A., K. Ragkousi, and P. Setlow. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 103:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodama, T., H. Takamatsu, K. Asai, K. Kobayashi, N. Ogasawara, and K. Watabe. 1999. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J. Bacteriol. 181:4584-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama, T., H. Takamatsu, K. Asai, N. Ogasawara, Y. Sadaie, and K. Watabe. 2000. Synthesis and characterization of the spore proteins of Bacillus subtilis, YdhD, YkuD, and YkvP, which carry a motif conserved among cell wall binding proteins. J. Biochem. (Tokyo) 128:655-663. [DOI] [PubMed] [Google Scholar]
- 25.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 26.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limpens, E., C. Franken, P. Smit, J. Willemse, T. Bisseling, and R. Geurts. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302:630-633. [DOI] [PubMed] [Google Scholar]
- 28.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 29.McPherson, D. C., H. Kim, M. Hahn, R. Wang, P. Grabowski, P. Eichenberger, and A. Driks. 2005. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 187:8278-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biology methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 31.Ozin, A. J., T. Costa, A. O. Henriques, and C. P. Moran, Jr. 2001. Alternative translation initiation produces a short form of a spore coat protein in Bacillus subtilis. J. Bacteriol. 183:2032-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 183:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radutoiu, S., L. H. Madsen, E. B. Madsen, H. H. Felle, Y. Umehara, M. Gronlund, S. Sato, Y. Nakamura, S. Tabata, N. Sandal, and J. Stougaard. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585-592. [DOI] [PubMed] [Google Scholar]
- 36.Real, G., S. Autret, E. J. Harry, J. Errington, and A. O. Henriques. 2005. Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis. Mol. Microbiol. 55:349-367. [DOI] [PubMed] [Google Scholar]
- 37.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 39.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 40.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, L. B., and R. Losick. 1990. Cascade regulation of spore coat gene expression in Bacillus subtilis. J. Mol. Biol. 212:645-660. [DOI] [PubMed] [Google Scholar]
- 42.Zilhão, R., R. Isticato, L. O. Martins, L. Steil, U. Völker, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2005. Assembly and function of a spore coat-associated transglutaminase of Bacillus subtilis. J. Bacteriol. 187:7753-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 2004. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 186:1110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]