Abstract
The exosporium-defective phenotype of a transposon insertion mutant of Bacillus cereus implicated ExsY, a homologue of B. subtilis cysteine-rich spore coat proteins CotY and CotZ, in assembly of an intact exosporium. Single and double mutants of B. cereus lacking ExsY and its paralogue, CotY, were constructed. The exsY mutant spores are not surrounded by an intact exosporium, though they often carry attached exosporium fragments. In contrast, the cotY mutant spores have an intact exosporium, although its overall shape is altered. The single mutants show altered, but different, spore coat properties. The exsY mutant spore coat is permeable to lysozyme, whereas the cotY mutant spores are less resistant to several organic solvents than is the case for the wild type. The exsY cotY double-mutant spores lack exosporium and have very thin coats that are permeable to lysozyme and are sensitive to chloroform, toluene, and phenol. These spore coat as well as exosporium defects suggest that ExsY and CotY are important to correct formation of both the exosporium and the spore coat in B. cereus. Both ExsY and CotY proteins were detected in Western blots of purified wild-type exosporium, in complexes of high molecular weight, and as monomers. Both exsY and cotY genes are expressed at late stages of sporulation.
Endospores of the Bacillus cereus family, which includes Bacillus anthracis and Bacillus thuringiensis, are enveloped by a large, balloon-like layer known as the exosporium (11, 19). B. subtilis, the paradigm sporeformer, does not have this distinct layer, although the B. subtilis spore coat may have a tight-fitting outermost layer that can be visualized after extraction of coat material with urea or mercaptoethanol (23). Scanning electron microscopy has revealed that the exosporium is composed of two layers—a paracrystalline basal layer with hexagonal periodicity and a “hairy nap” outer layer (11). The exosporium is chemically complex and is composed of 53% protein, 20% amino and neutral polysaccharides, 18% lipids, and ∼4% ash (17).
Multiple proteins from the exosporium of both B. cereus and B. anthracis have been identified (22, 24, 31); the majority do not have homologues in B. subtilis. For example, the surface layers of B. cereus, B. anthracis, and B. thuringiensis spores all contain glycoproteins with characteristic collagen-like repeat regions (5, 10, 27). Of these, the B. anthracis glycoprotein BclA is an essential component of the hairy nap layer of the exosporium (27, 29).
Analysis of genome sequence information from B. cereus ATCC 14579 and B. anthracis Ames identified a cluster of genes near bclA implicated in exosporium production, including the region whose genes were designated yjbX-exsY-yjcA-yjcB-exsFA-cotY (31). The ExsY and CotY proteins are homologues (ca. 35% amino acid identity) of the B. subtilis coat proteins CotY and CotZ (37). ExsY and CotY proteins have both been detected in purified exosporium from B. anthracis endospores by N-terminal sequencing of separated peptides (22). ExsY and CotY are very similar proteins, with ca. 90% amino acid identity for most of their 152- to 156-residue sequence, after a more variable (<50% identical) N-terminal region of ca 20 amino acids. They are cysteine-rich, like their B. subtilis homologues, which have been reported to form disulfide-linked multimers (37). This study describes the effects on spore structure and properties of the inactivation of exsY and cotY genes, singly and in combination.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Oxoid nutrient broth and agar (NB and NA, respectively; Oxoid, Ltd., Basingstoke, United Kingdom) were used for routine culture of B. cereus, with antibiotics if appropriate (spectinomycin at 200 μg ml−1, kanamycin at 40 μg ml−1, erythromycin at 1 μg ml−1, and lincomycin at 25 μg ml−1). Escherichia coli strains were grown in Luria broth (LB) or L agar, containing antibiotic (ampicillin at 100 μg ml−1, spectinomycin at 100 μg ml−1, erythromycin at 150 μg ml−1, and kanamycin at 40 μg ml−1). LB was used to culture B. cereus and E. coli for conjugation experiments. NBY broth, phage assay broth (30), NBY agar, and phage assay agar (30) were used for B. cereus phage transduction. TSB medium (6) was used for preparation of chemically competent E. coli.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| TG1 | supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5 (rK− mK−) [F′ traD36 proAB lacIqlacZΔM15] | Laboratory stock |
| HB101 | supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | Laboratory stock |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| B. cereus | ||
| ATCC 10876 | trp-1 Strr | 7 |
| AM1652 | trp-1 StrrexsY::spc | This study |
| AM1653 | trp-1 StrrcotY::erm | This study |
| AM1654 | trp-1 StrrexsY::spc cotY::erm | This study |
| SJT003 | trp-1 StrrexsY::Tn917-LTV1 | This study |
| Plasmids | ||
| pUC19 | Ampr | 35 |
| pAT113 | Eryr Kanr | 34 |
| pFX113 | Spcr Kanr | 3 |
| pUC1318Erm | Eryr Ampr | 33 |
| pUC1318Spc | Spcr Ampr | 33 |
| pEY-Spc | Spcr Eryr KanrexsY::spc | This study |
| pCY-Ery | Spcr Eryr KanrcotY::erm | This study |
Spore preparation.
B. cereus strains were incubated in liquid CCY medium (7) at 30°C with shaking until the culture contained >95% free spores, and spores were washed in water, as previously described (31). The final pellet was resuspended in spore resuspension buffer (50 mM Tris-HCl, 0.5 mM EDTA, pH 7.5) and stored at −20°C.
Proteins from spore culture supernatants.
Spores were pelleted from 35 ml of CCY culture by centrifugation. Sodium deoxycholate (to 200 μg ml−1) was added to the supernatant, and after 30 min of incubation at 4°C, ice-cold trichloroacetic acid was added to 6% (wt/vol) to precipitate proteins. After incubation on ice for 60 min, samples were centrifuged at 2,500 × g at 4°C for 45 min. Pellets were resuspended in 0.5 ml spore resuspension buffer (50 mM Tris-HCl, 0.5 mM EDTA, pH 7.5), and 20 μl was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Synchronous sporulation.
A synchronized sporulation method, based on the resuspension method of Sterlini and Mandelstam (26) and modified as described in reference 1, was used. Cells were subjected to an amino acid downshift, and the appearance of phase-bright prespores was monitored by phase-contrast microscopy. Duplicate 0.5-ml samples were taken at 30-min intervals throughout sporulation and centrifuged at 20,000 × g for 5 min, and the cell pellets were stored at −20°C.
Bacterial transformation.
E. coli was transformed by electroporation (8) or by chemical methods (6). Plasmids were introduced into B. cereus by electroporation (2) or by conjugation (32).
Enrichment for exosporium mutants.
Exosporium mutants were isolated from Tn917-LTV1 transposon insertion libraries (4, 7) following enrichment for reduced surface hydrophobicity (15) and screening for an exosporium defect by phase-contrast microscopy, as described by Bailey-Smith et al. (1).
Electron microscopy.
Spore sections were prepared as described previously (16). Sections were placed on Formvar-coated copper grids and examined under a CM100 transmission electron microscope operating at an acceleration voltage of 100 kV. Micrographs were recorded using a Gatan Multiscan 794 1k × 1k charge-coupled device camera. For imaging of whole spores, suspensions were diluted to 10 mg dry weight per ml, and 3 μl of spore suspension was deposited on a carbon-coated copper/palladium grid for 2 min, washed twice in distilled water, washed rapidly in 1% uranyl formate negative stain, and negatively stained for 30 s, blotted, and air dried.
Measurement of heat resistance.
Spore dilutions in water were incubated at 90°C for 10, 20, 30, and 40 min before plating on NA at 30°C. Counts were compared to those for unheated control samples.
Lysozyme resistance of spores.
Spores were diluted in phosphate-buffered saline at 37°C to an optical density at 600 nm (OD600) of ∼2. Following addition of lysozyme (to 250 μg ml−1), the OD580 was measured every 2 minutes, after shaking, in a temperature-controlled Wallac Victor2 1420 Multilabel counter.
Chemical resistance of spores.
Spores were resuspended in water to an OD600 of ∼1. For ethanol resistance, suspensions were diluted in 9 volumes of ethanol at room temperature for 10 min before further dilution and plating. For the water-immiscible solvents chloroform and toluene, 100 μl of solvent was added to 900 μl of spore suspension, vortexed for 1 min, and left to settle at room temperature for 9 min before sampling of the aqueous layer. To test phenol resistance, 100 μl of 5% (wt/vol) neutralized phenol was added to 900 μl of the spore suspension, vortexed for 1 min, and left at room temperature for 9 min before sampling. Dilutions of all treated cultures were in 10 mM potassium phosphate buffer, pH 7.4, containing 1 mM MgSO4 and 50 mM KCl, and aliquots were plated on NA and incubated at 30°C overnight. Viable counts were compared to those for untreated samples.
Insertional inactivation of the B. cereus exsY and cotY genes.
The suicide vector pAT113 (Kanr Eryr) (34) and its derivative, pFX113 (Kanr Spcr) (3), can be introduced from E. coli by conjugation and have been used successfully for gene knockout by allelic exchange in both B. cereus (20) and B. anthracis (27). Primers EyF1 (CCCATACACCTGTACACCAATAACAGC) and EyR1 (GTCCCACATAATAAATCATTTAAGGATATG) were used with Expand HiFi polymerase (Roche) to generate a 1.1-kb PCR product that contained the B. cereus exsY gene and flanking DNA. The fragment was gel purified and blunt cloned into HincII-digested pUC19 (35). The resulting construct (pUCEXSY) was linearized with NcoI, which cuts in the exsY open reading frame (ORF) after the 24th bp, and was end filled with VENT polymerase (Roche). A 1.2-kb fragment from pUC1318Spc (33), containing a spectinomycin resistance cassette (18), was blunt cloned into the NcoI-digested pUCEXSY to generate pUCEXSY-Spc. pUCEXSY-Spc was digested with HindIII to release the fragment containing the disrupted exsY ORF, which was gel purified and cloned into HindIII-digested pAT113 (34), giving rise to pEY-Spc. This plasmid was introduced into E. coli HB101 carrying the mobilizing plasmid pRK212.1 and thence to B. cereus ATCC 10876 by filter mating (32). Spectinomycin-resistant but kanamycin-sensitive B. cereus potential exconjugant colonies were screened by colony PCR for presence of both expected junction fragments to confirm the presence of the exsY::spc allele. The region, including flanking DNA, was sequenced in one such isolate (AM1652) to check that no additional point mutations had been introduced.
The cotY gene was inactivated in a similar fashion. Primers CyF1 (GGAAGAATTAATGGAGTCTGCAACTCGTTTCACCG) and CyR1 (CCATGCTTAACTCATTATTTAGTTTTAAATAG) were used to generate a PCR fragment of 1,228 bp containing the cotY ORF and flanking DNA. The gel-purified fragment was blunt cloned into HincII-digested pUC19 (pUCCOTY). The resulting construct was digested with MfeI/SpeI to remove the portion of cotY between bases 9 and 232 of the ORF, and Vent polymerase was used to generate blunt ends. A 1.2-kb erythromycin resistance cassette, obtained by SmaI digestion of pUC1318Erm (33), was blunt cloned into the digested pUCCOTY vector to give rise to pUCCOTY-Erm. This vector was digested with HindIII/EcoRI to release the disrupted cotY gene fragment, which was cloned into HindIII/EcoRI-digested pFX113 (3) to give pCY-Erm. This was transferred to the donor E. coli strain and thence into B. cereus ATCC 10876 by conjugation in a filter mating. An erythromycin-resistant, kanamycin-sensitive exconjugant (AM1653) with the appropriate cotY::erm insertion was confirmed by colony PCR, and the flanking regions were checked for the absence of additional mutations by DNA sequencing.
To construct the ΔexsY ΔcotY double mutant AM1654, a CP51ts phage lysate (36) propagated on AM1653 (cotY::erm) was used to transduce AM1652 (exsY::spec) as described previously (7). Transductants were selected on LB agar containing erythromycin (1 μg ml−1) and lincomycin (25 μg ml−1). As the cotY and exsY genes are only 2.5 kb apart, there was significant (ca. 60%) cotransduction of the wild-type exsY allele present in the donor. Transductants that had retained the exsY::spc allele were purified, and the presence of both mutant alleles was confirmed by PCR as well as by antibiotic resistance.
Overexpression of ExsY.
The exsY ORF was amplified using primers MJ74 (CGCGGGATCCGATGAGTTGTAACGAAAATAAACAC) and MJ75 (TTTTCTCGAGGATAGTAACGTCGCGTAAGC), digested with BamHI/XhoI, and ligated into BamHI/XhoI-digested pET41b (Novagen). The resulting plasmid construct, pET-EXSY, was introduced into E. coli BL21(DE3), and ExsY, tagged at the N terminus with a glutathione S-transferase (GST) domain, was overexpressed and purified using the Novagen GST · Resin system. All buffers were supplemented with 200 mM dithiothreitol (DTT) to prevent aggregation of the overexpressed protein by disulfide bond formation.
Production of ExsY and CotY antipeptide antibodies.
Peptides were synthesized by Arthur Moir, University of Sheffield, based on the N-terminal sequences of CotY (EDHQHECDFNCV; residues 7 to 18) and ExsY (ENKHHGSSHCV; residues 5 to 15), in each case polymerized on the eight available amino groups of a C-terminal polylysine tree. Polyclonal antibodies against the GST-ExsY fusion protein and the two synthetic peptides were raised in rabbits by the Antibody Resource Centre, University of Sheffield, by subcutaneous injection using Freund's adjuvant.
Exosporium extraction and protein separation.
Exosporium was released from spores by treatment in a French pressure cell: extraction, purification, and washes were all performed as previously described (31). Protein concentrations were measured by an adaptation of the Lowry method (21), using bovine serum albumin as the protein standard. Proteins were analyzed by boiling samples in an equal volume of Tricine gel sample buffer (3 M Tris-HCl, pH 8.45, 12% glycerol, 4% SDS, 0.1% Coomassie blue G), and separating the proteins by SDS-PAGE on precast 16% Tris-Tricine gels (Novex). Gels were stained with SYPRO Ruby protein gel stain (Molecular Probes).
Glycoprotein staining.
Exosporium samples were reduced with 200 mM DTT at 4°C for 14 h prior to SDS-PAGE on Novex 10 to 20% Tris-glycine gradient gels (Novex). Proteins were then blotted onto nitrocellulose membrane (Amersham) using 10 mM CAPS (3-[cyclohexylamino-1 propanesulfonic acid]) transfer buffer. Glycoprotein staining was performed using a glycoprotein ECL enhanced chemiluminescence detection kit (Amersham).
Western blotting.
Exosporium samples were reduced with 200 mM DTT and separated by SDS-PAGE as described above for glycoprotein staining. Proteins were then blotted onto Hybond-P polyvinylidene difluoride membrane (Amersham). Extra lanes with molecular weight markers (Mark 12; Invitrogen) were removed and stained with Coomassie blue for comparison. The concentrations of primary antibody used were 1/500 for the ExsY-GST antibody and 1/100 for the antipeptide antibodies. A 1/5,000 dilution of secondary anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (anti-mouse horseradish peroxidase-conjugated immunoglobulin G for monoclonal antibody CR1) was used in the ECL-Plus Western blotting detection system (Amersham), with Hyperfilm ECL (Amersham).
Nucleotide sequence accession number.
The B. cereus ATCC 10876 sequences were submitted to GenBank under accession no. AY121973 (exsY) and AY186996 (cotY).
RESULTS
Isolation of a transposon insertion exsY mutant.
Spores from libraries of B. cereus ATCC 10876 carrying a chromosomal copy of Tn917-LTV1 (7) were subjected to a hexadecane/aqueous-phase partitioning system (1) to enrich for spores that had altered surface hydrophobicity. One of the mutants obtained, SJT003, produced spores that were not surrounded by exosporium but retained a coat layer. Generalized transduction with the phage CP51ts showed 100% linkage between the transposon insertion and the exosporium defect. The transposon Tn917-LTV1 is designed to allow recovery of flanking sequences. SJT003 chromosomal DNA was digested with EcoRI, and the excised plasmid was recovered in E. coli (4). Sequencing of the transposon-chromosome junction revealed that the transposon was inserted into the promoter region of exsY, encoding a homologue of the B. subtilis CotY and CotZ outer spore coat proteins. The ExsY protein had already been identified and named in our laboratory as a formic acid-solubilized component of B. cereus ATCC 10876 exosporium (S. J. Todd, unpublished observation).
Mutant construction.
The transposon insertion was located in the intergenic region between two divergently transcribed genes, 34 bp upstream from the start codon of exsY and 91 bp upstream of yjcA. It was therefore possible, though unlikely, that the insertion would also affect yjcA expression. A directed mutagenesis strategy was therefore adopted to generate null mutants AM1652 (ΔexsY), which carries an exsY::spc allele, and AM1653 (ΔcotY), which carries a cotY::erm allele. Inactivation of the cotY gene is not likely to affect expression of the exsFA gene 175 bases downstream, because the latter has its own promoter, as indicated by complementation studies (28). A double mutant, AM1654, was constructed using generalized transduction with phage CP51ts (36) to cross the cotY::erm mutation into the exsY::spc mutant strain.
Spore morphology is altered in exsY and cotY mutants.
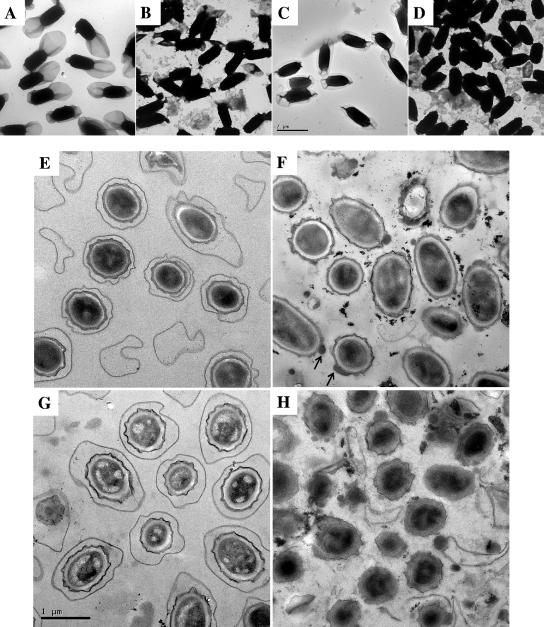
Spores of the AM1652 (ΔexsY), AM1653 (ΔcotY), and AM1654 (ΔexsY ΔcotY) mutants were compared to those of the wild type, using electron microscopy to assess any effect on gross spore morphology. Samples were viewed as both whole spores and embedded thin sections. The ΔexsY mutant whole spores (Fig. 1B) lack an intact exosporium layer: some fragments of exosporium are attached, and some are free in the spore preparation. Electron microscopy of thin sections of AM1652 (ΔexsY) confirmed the absence of an exosporium layer (Fig. 1F). The spore coat did not stain evenly, and there were occasional lumps on the coat surface. Darkly staining particulate material was also present in these preparations.
FIG. 1.
Electron micrographs showing fields of whole spores of B. cereus (A to D) and of thin sections (E to H). Images are at the same size for all of panels A to D (scale bar on panel C) and at higher magnification for panels E to H (scale bar on panel G). Panels A and E show wild-type B. cereus ATCC 10876, B and F show AM1652 (ΔexsY), C and G show AM1653 (ΔcotY), and D and H show AM1654 (ΔexsY ΔcotY). Arrows on panel F point to lumps of material on spore coats of AM1652.
In contrast, the AM1653 (ΔcotY) mutant spore has an intact exosporium that fully encloses the spore (Fig. 1C), although the exosporium appears to make a tighter fit around the poles of the spore compared with the loose “balloon-like” exosporium of the wild type. Thin sections of these spores look like wild type (Fig. 1G).
Spore preparations of AM1654 (ΔexsY ΔcotY) contained fragmented exosporium, most of which was not attached to spores (Fig. 1D). Spore sections (Fig. 1H) revealed exosporium-free spores, all with some degree of spore coat defect. Large fragments of coat or exosporium material were present in the preparation.
Spores of the ΔexsY mutant and, more dramatically, of the double mutant were prone to aggregation in suspension, as reported for B. subtilis coat-defective or chemically extracted spores (12, 37).
Hydrophobicity and resistance properties of exsY and cotY mutants.
Electron microscopy shows that the spore surface layers of the single mutants are clearly different (Fig. 1). Despite this, the individual ΔexsY and ΔcotY mutations had only small effects on the partition of spores into hexadecane (Table 2). In contrast, the spores of the double mutant were much less efficiently partitioned into hexadecane, suggesting a large reduction in surface hydrophobicity.
TABLE 2.
Spore hydrophobicity and resistance to organic solvents
| B. cereus strain | Partition into hexadecane (%) | Spore resistance/recovery (%)a
|
|||
|---|---|---|---|---|---|
| Ethanol | Chloroform | Toluene | Phenol | ||
| ATCC 10876 (wild type) | 88 ± 2 | 79 ± 3 | 42 ± 1 | 63 ± 5 | 85 ± 3 |
| AM1652 (ΔexsY) | 80 ± 2 | 94 ± 15 | 46 ± 7 | 69 ± 16 | 85 ± 13 |
| AM1653 (ΔcotY) | 82 ± 2 | 55 ± 1 | 35 ± 4 | 25 ± 4 | 55 ± 3 |
| AM1654 (ΔexsY ΔcotY) | 22 ± 2 | 25 ± 5 | 6 ± 5 | 2 ± 1 | 7 ± 3 |
Colony counts are expressed relative to untreated controls as mean ± standard deviation.
Spore coats contribute to the resistance of B. cereus spores to organic solvents (14). Spores of all three mutants were tested for resistance to chloroform, toluene, ethanol, and phenol (Table 2). The ΔexsY mutation had no effect on resistance, whereas the ΔcotY mutation affected resistance to phenol and, more dramatically, to toluene. Spores of the ΔexsY ΔcotY double mutant were much more sensitive to all four solvents.
The coats of wild-type spores are impermeable to lysozyme, which would otherwise be able to access and degrade the peptidoglycan cortex, leading to a decrease in OD and phase darkening. Spores of the ΔcotY mutant, like wild-type spores, were resistant to lysozyme, but spores of the ΔexsY mutant and ΔexsY ΔcotY double mutant were sensitive to lysozyme, with suspensions decreasing in OD between 30 and 60 min after exposure (data not shown). For comparison, a complete coat permeability defect, such as that found in an exsA mutant (1), results in a rapid and synchronous fall in OD within 3 min of exposure to lysozyme. The delayed and less synchronous decrease in OD implies that a partial barrier to passage of large molecules still remains in the ΔexsY single and double mutants. To test whether the exosporium is required for lysozyme resistance, wild-type spores were passed twice through the French press under the standard conditions used to release exosporium from the spore surface (31). Staining and microscopy of these French-pressed spores showed that their exosporium was no longer intact, although many remnants remained spore associated. Tests showed that these spores were as resistant to lysozyme as wild-type spores, so penetration through the exosporium is not sufficient to allow lysozyme access to the cortex. This suggests that the reduced resistance in the ΔexsY mutant spores is not directly due to absence of the exosporium but that coat layers must also be affected in this mutant, consistent with the observed properties of the spore coat in thin section.
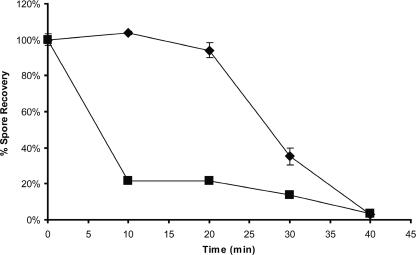
Single- and double-mutant spores showed the same resistance as wild-type spores to wet heat at 80°C, but the double mutant showed a defect in resistance to wet heat at 90°C (Fig. 2). Most of the double-mutant spores (80%) were not recoverable after 10 min, and the remainder were gradually killed over the next 30 min. This may reflect the heterogeneity in the properties of spores within the preparation.
FIG. 2.
Heat resistance of dormant spores. Spore dilutions in water were incubated at 90°C for up to 40 min. Survivors were enumerated by plating on NA. Counts are expressed as a proportion of unheated controls. ♦, wild type; ▪, AM1654 (ΔexsY ΔcotY). Error bars show 1 standard deviation.
Exosporium protein profiles of exsY and cotY mutants.
Exosporium was purified from water-washed spore preparations of the wild type and single mutants. It was collected following French press treatment of AM1652 (ΔexsY) spores, but the yield was low (ca. one-fifth that of the wild type). We did not attempt to purify exosporium from spore preparations of the double mutant AM1654 as the large amount of loose material evident in the electron micrograph sections of this spore preparation is likely to include coat as well as exosporium material.
Samples of washed exosporium preparations from B. cereus ATCC 10876, AM1652 (ΔexsY), and AM1653 (ΔcotY) were analyzed using SDS-PAGE on a 16% Tris-Tricine gel (Fig. 3). Both mutants showed some differences from wild type in exosporium protein composition. The AM1652 (ΔexsY) profile was deficient in many of the protein bands observed in the wild-type exosporium, but two prominent bands at ∼70 kDa and ∼200 kDa were retained. The AM1653 (ΔcotY) profile was more similar to the wild type, as might be expected given that a complete exosporium is present in this mutant, but there were some minor differences, including several extra bands in the 40- to 60-kDa region and the absence or reduction of several wild-type protein bands in the 6- to 14-kDa range.
FIG. 3.
Protein profiles of washed exosporium of B. cereus ATCC 10876 wild-type and mutant strains. Proteins were separated on a 16% SDS-Tris-Tricine gel and stained with SYPRO Ruby. Lane 1, wild type; lane 2, AM1652 (ΔexsY); lane 3, AM1653 (ΔcotY).
Glycoproteins in exsY and cotY mutants.
ExsY and CotY have been found in protein complexes with other exosporium proteins, including the glycoprotein BclA (22). CR1, a monoclonal antibody that detects the BclA glycoprotein, was provided by Caroline Redmond (DSTL, Porton Down, United Kingdom). CR1 was used as the primary antibody in Western blots of DTT-treated exosporium samples from B. cereus ATCC 10876 ΔexsY and ΔcotY strains (Fig. 4A). In the wild-type exosporium, the CR1 antibody detected a broad (possibly double) band of 200 kDa, which is the expected position of glycosylated BclA in B. cereus (1, 27). The 200-kDa BclA band was severely reduced in the ΔexsY sample but retained in the ΔcotY exosporium. Minor cross-reacting bands were seen at 60 kDa in the wild type and the ΔexsY mutant and at 27 kDa in the ΔcotY mutant. These are unlikely to be BclA, as the deglycosylated BclA protein from our wild-type strain of B. cereus has been reported at 40 kDa (1).
FIG. 4.
Detection of BclA and other glycoproteins in washed exosporium following SDS-PAGE on 10 to 20% gradient Tris-glycine gels. (A) Western blot of proteins from washed exosporium probed with CR1, an anti-BclA monoclonal antibody. (B) Total glycoprotein profile of the same exosporium samples. (C) Proteins concentrated from spore culture supernatants, again probed with CR1 antibody to detect BclA. Lane 1, B. cereus ATCC 10876 (wild type); lane 2, AM1652 (ΔexsY); lane 3, AM1653 (ΔcotY). In panel C; lane 4 shows AM1654 (ΔexsY ΔcotY).
There are also differences in the overall glycoprotein profiles of exosporium of the wild type and mutants (Fig. 4B), as detected by the Amersham ECL glycoprotein kit, in which metaperiodate-oxidized carbohydrates are biotinylated and then detected after binding of horseradish peroxidase-linked streptavidin. Wild-type exosporium contained strongly labeled bands at ∼200 kDa and 40 kDa as well as a smear between 60 and 70 kDa. Several fainter bands were also apparent. In both ExsY exosporium and CotY exosporium, the 200-kDa and 40-kDa bands were much reduced, but the diffuse doublet band in the 60- to 70-kDa region was predominant. The presence of additional glycoprotein bands seen in Fig. 4B, compared to the pattern for BclA in Fig. 4A, confirms previous observations that BclA is not the only glycoprotein in B. cereus ATCC 10876 exosporium (1). It is also clear that not all the glycoproteins are equally depleted in the exosporium layer of the mutant spores.
BclA is not assembled completely in the double ΔexsY ΔcotY mutant.
Spore culture supernatants were precipitated at the end of sporulation in CCY medium, following mother cell lysis, and the SDS-PAGE-separated proteins were probed in Western blots with the anti-BclA antibody CR1 (Fig. 4C). There was no significant residual free BclA in wild-type culture supernatant: only two faint cross-reacting bands at around 33 and 34 kDa were detected, which are not likely to represent a BclA monomer, as discussed above for minor bands in Fig. 4A. There was, however, a very strongly stained smear of very-high-molecular-mass material, extending from the top of the gel down to the 200-kDa region, detected in the culture supernatants from the ΔexsY mutant and the ΔexsY ΔcotY mutant. This suggests that BclA is produced by these two mutants, but that much of it is misassembled and released into the supernatant on mother cell lysis.
Detection of the ExsY and CotY proteins in exosporium.
Antibody raised against the ExsY-GST fusion protein is expected to detect both ExsY and CotY, given the high degree of amino acid identity between the two proteins. Synthetic peptide antibodies EY-N and CY-N were therefore raised against the N-terminal regions that are unique to ExsY and CotY, respectively, with the aim of distinguishing between these two proteins on Western blots. The EY-N antibody detected the 49-kDa GST-ExsY fusion protein in a Western blot of partially purified protein, as expected. The CY-N antibody did not detect this protein on the same blot, confirming its ability to distinguish the two homologues (13).
All three antibodies were used in Western blots against SDS-PAGE-separated proteins of washed, DTT-treated wild-type, ΔexsY mutant, and ΔcotY mutant exosporium (Fig. 5). When probed with anti-ExsY-GST (Fig. 5A), which would detect both proteins, bands were detected in wild-type exosporium as a large-molecular-mass smear from 60 to 200 kDa, as well as a 16-kDa monomeric form. The profile was essentially unchanged in the ΔcotY mutant, where both the large and 16-kDa bands must represent ExsY protein, in the absence of CotY. Therefore, ExsY is definitely present in the exosporium. These bands were massively reduced in intensity in the ΔexsY mutant, suggesting either that only a little CotY is present in exosporium or that the absence of ExsY reduces the assembly of CotY into the exosporium. An 8-kDa band (not shown) was sometimes seen in exosporium of the wild type and CotY mutants and may represent a proteolysed fragment, as there are also reports of smaller protein bands containing ExsY- and CotY-derived sequences in exosporium preparations of B. anthracis (22).
FIG. 5.
Detection of ExsY and CotY in exosporium fractions. Western blots are shown with primary polyclonal antibodies as follows: (A) EY-GST (raised against purified ExsY/GST, which detects both ExsY and CotY), (B) EY-N (raised against an ExsY-specific peptide), and (C) CY-N (raised against a CotY-specific peptide). Lane 1, B. cereus ATCC 10876 (wild type); lane 2, AM1652 (ΔexsY); lane 3, AM1653 (ΔcotY). ExsY and CotY monomers migrate at 16 kDa.
Western blots of exosporium proteins were probed with CY-N antipeptide antibody and then stripped and reprobed with EY-N antipeptide antibody. The EY-N antibody (Fig. 5B) detected a 16-kDa band, representing ExsY monomer, in wild-type exosporium and in a ΔcotY mutant, but not in a ΔexsY mutant. Because of the apparently low level of CotY in the ΔexsY mutant exosporium, we have no perfect control to test the discriminative ability of the anti-ExsY (EY-N) antibody. We can be confident, however, about the ability of the anti-CotY (CY-N) antibody to discriminate between the two homologues. For example, CY-N (Fig. 5C) did not detect a 16-kDa monomer in the ΔcotY mutant exosporium, where ExsY is still present at normal levels. As this antibody does detect a 16-kDa band in wild-type exosporium, but does not detect ExsY monomer or GST-ExsY, we can deduce that this 16-kDa band in wild-type exosporium is CotY protein. Other bands detected by the antipeptide antibodies in the 25- to 35-kDa range on the Western blots were variably present or absent in different wild-type exosporium preparations and are therefore likely to reflect nonspecific binding.
Synthesis of ExsY and CotY protein.
Cultures of B. cereus ATCC 10876 at 37°C were subjected to a resuspension sporulation regime in which sporulation is synchronously induced by an amino acid downshift, classically used for monitoring sporulation stages in B. subtilis (26). Phase-gray immature prespores become visible within mother cells, increasing from 7% at 4 h after resuspension to 80% at 5 h. Mature spores were released from mother cells from around 10 h. ExsY protein (16 kDa) was detected with EY-N antibody in extracts of sporulating wild-type cells from 5 h onwards (Fig. 6A). This strong 16-kDa signal was absent in the ΔexsY mutant (Fig. 6C), although a faint cross-reacting band at the same position, possibly CotY, did appear very late in sporulation. The minor band at ∼60 kDa was detected in wild-type and ΔexsY mutant extracts, so it is not ExsY related (data not shown). The membrane from Fig. 6A was stripped and reprobed (Fig. 6C) with CY-N antibody, which detected CotY monomer (16 kDa) in the wild-type sporulating cells after 6 h.
FIG. 6.
Synthesis of ExsY and CotY in sporulating cells. The times of synthesis of ExsY and CotY were determined during synchronous sporulation. Samples were taken at intervals, and proteins were separated by SDS-PAGE on 10 to 20% Tris-glycine gradient gels and probed by Western blotting with EY-N antibody (raised against an ExsY peptide) and CY-N antibody (raised against a CotY peptide). Blots represent the following: (A) B. cereus ATCC 10876 wild type, with antibody EY-N; and (B), same as panel A, but probed with antibody CY-N. The blot shown in panel C represents a culture of AM1652 (ΔexsY) probed with antibody EY-N as a control for the blot in panel A. Size markers (in kDa) are shown. For conciseness, the blots in panels B and C only show the relevant region for the protein monomers. T000 is the time of resuspension to initiate sporulation, and the numbers thereafter are in minutes.
The time of synthesis of ExsY and CotY proteins in the developing spores would correspond to late stage IV of sporulation, when expression is under the control of the mother cell sigma factor, sigma K (9). Reasonable matches to the (rather similar) consensus sequences for sigma K and sigma E promoters can be found upstream of each gene, with better similarity to K for exsY and E for cotY (13).
DISCUSSION
These experiments demonstrate a role for both ExsY and CotY proteins in B. cereus exosporium. They may be present in this layer in disulfide-cross-linked forms that can be reduced to monomers by incubation in high concentrations of dithiothreitol, as was the case for the B. subtilis proteins (37). The intensity and continuous nature of the remaining smear of 60- to 200-kDa material that contains CotY/ExsY epitopes in exosporium protein profiles suggest that they may also be cross-linked in a less-reversible fashion to other proteins in the exosporium.
Although little is known about the proteins involved in assembly of the exosporium layer, data from this study and several others (1, 14, 25, 28) are gradually elucidating dependencies between proteins that contribute to exosporium formation and integrity. BclA, ExsFA, ExsY, and CotY have all been detected in large >200-kDa aggregates in B. anthracis spores (22). Correct assembly of the BclA-containing hairy nap in B. anthracis is dependent upon the presence of ExsFA (also designated as BxpB) and its homologue, ExsFB (25, 28). Assembly of ExsY and CotY in B. cereus is not ExsFA dependent because both proteins are still present in the exosporium of an ΔexsFA mutant (data not shown). The BclA levels may be lower in exsY mutant exosporium, but preparations of purified exosporium fragments from ΔexsY or ΔcotY single-mutant strains both yielded crystalline fragments with an intact hairy nap (D. A. Ball and P. Bullough, personal communication). B. cereus ATCC 10876 has additional major exosporium glycoproteins, including ExsJ (31), that may also contribute to nap formation.
In comparison to B. subtilis cotY cotZ double mutants (37), double mutants lacking both B. cereus paralogues ExsY and CotY cause a more severe spore defect. The loss of a complete exosporium in the exsY mutant, but not the cotY mutant, led us to name these genes and their encoded proteins for the two layers, respectively, and as it is now established in the literature (31), we have not changed their names. Although they are closely related homologues, ExsY and CotY have somewhat different roles in spore coat and exosporium formation in Bacillus cereus. Both have roles in normal coat assembly, as indicated by the consequences for lysozyme or solvent resistance if either is absent. We therefore assume that both proteins are present in wild-type spore coats, but it has not been possible to generate coat fractions that are entirely exosporium free to prove this definitively.
The role of ExsA in the assembly of both the spore coat and exosporium has already indicated that the assembly of these layers is linked (1). We have now demonstrated two further exosporium and likely spore coat proteins, ExsY and CotY, that are also involved in assembly of both layers. This reinforces the relationship between spore coat and exosporium assembly in the B. cereus group of species.
Acknowledgments
We thank Caroline Redmond for providing the CR1 antibody and Michèle Mock's group for introducing M.J.J. to the technologies for B. cereus conjugation during an EMBO short-term fellowship. John Proctor assisted with electron microscopy, and undergraduate students Andrew Needham and Adam Reid helped with early transposon mutant characterization.
This work was funded by BBSRC studentships and by BBSRC project grant 50/D18848, under the Joint Grant Scheme with DSTL.
Footnotes
Published ahead of print on 15 September 2006.
REFERENCES
- 1.Bailey-Smith, K., S. J. Todd, T. W. Southworth, J. Proctor, and A. Moir. 2005. The ExsA protein of Bacillus cereus is required for assembly of coat and exosporium onto the spore surface. J. Bacteriol. 187:3800-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bone, E. J., and D. J. Ellar. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Lett. 58:171-178. [DOI] [PubMed] [Google Scholar]
- 3.Brossier, F., M. Weber-Levy, M. Mock, and J.-C. Sirard. 2000. Role of toxin functional domains in anthrax pathogenesis. Infect. Immun. 68:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camilli, A., D. A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlton, S., A. J. G. Moir, L. Baillie, and A. Moir. 1999. Characterization of the exosporium of Bacillus cereus. J. Appl. Microbiol. 87:241-245. [DOI] [PubMed] [Google Scholar]
- 6.Chung, C. T., and R. H. Miller. 1988. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 16:3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements, M. O., and A. Moir. 1998. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J. Bacteriol. 180:6729-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Patrone, M., and J. S. Tandecarz. 1995. A glycoprotein multimer from Bacillus thuringiensis sporangia—dissociation into subunits and sugar composition. Mol. Cell Biochem. 145:29-37. [DOI] [PubMed] [Google Scholar]
- 11.Gerhardt, P., and E. Ribi. 1964. Ultrastructure of the exosporium enveloping the spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James, W., and J. Mandelstam. 1985. Protease production during sporulation of germination mutants of Bacillus subtilis and the cloning of a functional gerE gene. J. Gen. Microbiol. 131:2421-2430. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, M. J. 2004. The function of ExsY and CotY, two structural proteins of the Bacillus cereus exosporium. Ph.D. thesis. University of Sheffield, Sheffield, United Kingdom.
- 14.Kim, H.-S., D. Sherman, F. Johnson, and A. I. Aronson. 2004. Characterization of a major Bacillus anthracis spore coat protein and its role in spore inactivation. J. Bacteriol. 186:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshikawa, T., M. Yamazaki, M. Yoshimi, S. Ogawa, A. Yamada, K. Watabe, and M. Torii. 1989. Surface hydrophobicity of spores of Bacillus spp. J. Gen. Microbiol. 135:2717-2722. [DOI] [PubMed] [Google Scholar]
- 16.Leatherbarrow, A. J. H., M. A. Yazdi, J. P. Curson, and A. Moir. 1998. The gerC locus of Bacillus subtilis, required for menaquinone biosynthesis, is concerned only indirectly with spore germination. Microbiology 144:2125-2130. [DOI] [PubMed] [Google Scholar]
- 17.Matz, L. L., T. C. Beaman, and P. Gerhardt. 1970. Chemical composition of exosporium from spores of Bacillus cereus. J. Bacteriol. 101:196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, E., L. Huwyler, and M. D. D. Bastos. 1985. Transposon Tn554—complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohye, D. F., and W. G. Murrell. 1973. Exosporium and spore coat formation in Bacillus cereus T. J. Bacteriol. 115:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okstad, O. A., A. Gronstad, T. Lindback, and A. B. Kolsto. 1997. Insertional inactivation of a Tet(K)/Tet(L) like transporter does not eliminate tetracycline resistance in Bacillus cereus. FEMS Microbiol. Lett. 154:181-186. [DOI] [PubMed] [Google Scholar]
- 21.Peterson, G. L. 1977. A simplification of the protein assay of Lowry which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 22.Redmond, C., L. W. J. Baillie, S. Hibbs, A. J. G. Moir, and A. Moir. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 150:355-363. [DOI] [PubMed] [Google Scholar]
- 23.Sousa, J. C. F., M. T. Silva, and G. Balassa. 1976. An exosporium-like outer layer in Bacillus subtilis spores. Nature 263:53-54. [DOI] [PubMed] [Google Scholar]
- 24.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steichen, C. T., J. F. Kearney, and C. L. Turnbough. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J. Bacteriol. 187:5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 28.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J. Bacteriol. 187:5122-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 185:1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorne, C. B. 1968. Transducing bacteriophage for Bacillus cereus. J. Virol. 2:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Todd, S. J., A. J. G. Moir, M. J. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trieu-Cuot, P., C. Carlier, P. Martin, and P. Courvalin. 1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48:289-294. [Google Scholar]
- 33.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to Gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains—nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Yelton, D. B., and C. B. Thorne. 1970. Transduction in Bacillus cereus by each of two bacteriophages. J. Bacteriol. 102:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]