Abstract
A 17-kb scaffoldin gene cluster in Ruminococcus flavefaciens strain FD-1 was compared with the homologous segment published for strain 17. Although the general design of the cluster is identical in the two strains, significant differences in the modular architecture of the scaffoldin proteins were discovered, implying strain-specific divergence in cellulosome organization.
Ruminococcus flavefaciens is a gram-positive, anaerobic, cellulosome-producing, cellulolytic bacterium which commonly inhabits the digestive tracts of ruminants, other herbivorous animals, and humans (11, 21). R. flavefaciens FD-1 and 17 are two commonly investigated strains of this species. Although the two strains were derived from different geographical locations and time frames (6, 12), they have been classified as the same species and, accordingly, show marked similarities in their properties.
The organization of cellulases into a multienzyme cellulosome complex is one of the major paradigms of efficient bacterial degradation of cellulose and related plant cell wall polysaccharides (5, 8, 10). The incorporation of the cellulosomal enzymes into scaffoldin components is the most defining feature of the cellulosome complex and is dictated by the high-affinity protein-protein interaction between two complementary modules, the cohesin and the dockerin. Knowledge of the types and specificities of cohesins and dockerins produced by a given bacterium and how they are arranged in the parent protein (i.e., scaffoldins and enzymes, respectively) provides insight into the overall architectural scheme of a given cellulosome system.
We have previously investigated the status of the cellulosome produced by strain 17 of R. flavefaciens. This was achieved by the sequencing of the genes encoding several enzymes (2, 13, 20) and scaffoldins (9, 18, 19) isolated from clone libraries. Individual cohesins and dockerins were subcloned and overexpressed, and the interactions among the expressed modules were evaluated biochemically. Using this approach, we proposed an architectural overview of the cellulosome in strain 17 (17).
Strains of R. flavefaciens from the rumen form a well-defined phylogenetic cluster based on 16S rRNA sequencing, but they show significant genetic variations (15). Significant functional diversity between strains, with respect to the breakdown of cellulose and of different types of plant material (7, 14) and the ability to utilize degradation products from xylans (3), has been reported. In the present report, we examine the architecture and component sequences of the cellulosome system in R. flavefaciens FD-1 and compare its characteristics with those of strain 17. The results indicate a general similarity in the cellulosome organization between the two strains, with a number of novel and very surprising properties.
The sequence of the sca gene cluster from R. flavefaciens strain 17 (accession number AJ278969) was compiled from several EMBL/GenBank entries with the following accession numbers: AJ585075 (scaC), AJ278969 (scaA-scaB), AJ810898 (cttA), and AJ810899 (scaE). The corresponding gene cluster DNA sequence of strain FD-1 (accession number AM262974) was assembled from the draft genome (1) and PCR data, using the primers described in Table 1. As shown in Fig. 1A, the overall organization of the scaffoldin cluster of strain FD-1 matches well with that of strain 17 (17). The cluster consists of genes encoding four scaffoldins of different sizes and one additional gene, cttA, encoding a protein of currently unknown function.
TABLE 1.
Primers used in this studya
| Name | Nucleotide sequence | Location | Comment |
|---|---|---|---|
| ScaA-Coh1-F | AGATCTACATCAGCTCAGCCTGTTG | ScaA Coh1 | Cohesin expression |
| ScaA-Coh1-R | CTCGAGAGCAGATGTTGTTGGATCATCA | ||
| ScaB-Coh3-F | GGATCCACAGCTCCTGTAGCTAACGC | ScaB Coh3 | Cohesin expression |
| ScaB-Coh3-R | CTCGAGAGTTGTTGCCTTAGGTGTTGTTG | ||
| ScaB-Coh6-F | GGATCCGTAGTTGCTGAAGGCACAGC | ScaB Coh6 | Cohesin expression |
| ScaB-Coh6-R | CTCGAGTACAGGTGTATCACCAACAACG | ||
| ScaC-Coh-F | GGATCCACAGTGCAGATATCCGCCAG | ScaC cohesin | Cohesin expression |
| ScaC-Coh-R | CTCGAGTACTTCTGCTGAAGGAACAG | ||
| ScaE-Coh-F | AGATCTCTCACAGACAGAGGAATGACTTAC | ScaE cohesin | Cohesin expression |
| ScaE-Coh-R | CTCGAGCTCAGGCTCACCAGCCTTG | ||
| ScaA-Doc-F | GGTACCTACAACATCTGCTACAACAGC | ScaA dockerin | Dockerin expression |
| ScaA-Doc-R | GGATCCTTAGCCCTTAGCAGGGAGTGTG | ||
| ScaB-XDoc-F | GGTACCTAATTCCGGTGATAATGTATCTG | ScaB XDoc | Dockerin expression |
| ScaB-XDoc-R | GGATCCTTATGGAACGGTCAATTCAGG | ||
| ScaC-Doc-F | GGTACCTGGTACAGACAACAGCAGTATC | ScaC dockerin | Dockerin expression |
| ScaC-Doc-R | GGATCCTCAAAGTTCTGTGATGAGAGTAAGC | ||
| Cel44A-Doc-F | GGTACCTGCAAACGTAACATACGGCGATG | Cel44A dockerin | Dockerin expression |
| Cel44A-Doc-R | GGATCCTTATGCTTCGGGAAGCTTGTCG | ||
| Ce3B-Doc-F | GGTACCTAGGATCACAGGCTCGGCTTC | Ce3B dockerin | Dockerin expression |
| Ce3B-Doc-R | GGATCCTTAGGGGATATCCCTTGATGAAGG | ||
| EndB-Rf17-F | GAGCTCCAGAAGTGGTGGG | EndB-like dockerin (“Cel44A”-Doc) | Homologous gene cloning |
| EndB-Rf17-R | GACAAGCTTCCCGAA | ||
| CesA-Rf17-F | TGCCGATATCCACAGCGT | CesA-like dockerin (“Ce3B”-Doc) | Homologous gene cloning |
| CesA-Rf17-R | CTGACGCTTCCACCAGTC |
Restriction site sequences are underlined.
FIG. 1.
The scaffoldin gene cluster in R. flavefaciens FD-1 and 17. (A) The scheme shows the organization on the genome of the scaC, scaA, scaB, and scaE genes. Scaffoldins of different sizes are indicated by the filled arrows. Numbers of amino acid residues for the given genes in the respective strain are shown. (B) Modular organization of the sca genes. The scaA and scaB genes contain multiple copies of cohesin modules (numbered). (C) Phylogenetic relationship among cohesin sequences from strains FD-1 and 17. The scale bar indicates the percentage of amino acid substitutions.
The FD-1 scaA gene encodes a protein that contains two cohesins rather than three as in strain 17 (Fig. 1B). As a result, the FD-1 ScaA protein is markedly smaller in overall size than the orthologous protein of strain 17. The ScaA proteins in the two strains bear a similar type of N-terminal X module (53.9% sequence similarity) and similar dockerins (77.9% sequence similarity) at their C termini.
In the case of ScaB, however, the overall modular architecture is strikingly divergent for the two strains. ScaB of strain 17 carries seven contiguous cohesins, separated by linker sequences of various lengths. In contrast, the orthologous scaffoldin B of strain FD-1 bears nine cohesins. Both scaffoldins contain a characteristic X-module/dockerin pair (XDoc) at the C terminus. The integrity of the strain 17 ScaB XDoc has recently been shown to be critical for recognition by the ScaE cohesin (17). The differences between the two ScaB scaffoldins, however, are not only quantitative (i.e., in the respective number of cohesins) but also qualitative. Unlike the seven ScaB cohesins of strain 17, all of which emanate from a single branch on the phylogenetic tree (9, 17), those of strain FD-1 are distinctively divergent: the first four N-terminal cohesins are related (46 to 78% sequence similarity) to those of ScaA (of both strains), whereas the five C-terminal cohesins are orthologous (53 to 79% sequence similarity) to the seven ScaB cohesins from strain 17 (Fig. 1B). Although both the ScaA and ScaB cohesins of strain 17 have been classified as type III cohesins, their respective branches diverge relatively early in the phylogenetic tree (9, 17). In strain FD-1, the ScaA cohesins and the ScaB cohesins 1 through 4 all map together with the ScaA cohesins of strain 17, whereas ScaB cohesins 5 through 9 map together with all of the strain 17 ScaB cohesins (Fig. 1C). The FD-1 ScaB is but the second reported occurrence of a scaffoldin that bears divergent cohesins, the previous report being that of the ScaD scaffoldin of Acetivibrio cellulolyticus (22). Owing to the additional cohesins in the FD-1 ScaB scaffoldin, its overall size is proportionately larger than that of strain 17 (Fig. 1A).
The remaining two orthologous scaffoldins of the two strains of R. flavefaciens, ScaC and ScaE, are very similar in modular architecture and size (Fig. 1A and B). Both types of scaffoldin contain a single cohesin; the ScaC scaffoldins contain similar C-terminal dockerins (65.8% sequence similarity) and the ScaE scaffoldins contain very similar (85.7% identity) 35-residue C-terminal LPXTG motifs for sortase-mediated anchoring of the respective parent protein (17). The interstrain sequence similarities for ScaC and ScaE cohesins are 71.2% and 65.1%, respectively, and they map on respective branches of the phylogenetic tree (Fig. 1C).
In previous studies (17), we formulated an architectural scheme of the cellulosome system in strain 17. In view of the divergent cohesin types in the FD-1 ScaB scaffoldin, it was of interest to determine the specificity characteristics of the cohesin-dockerin interactions in strain FD-1 and to determine the comparative architecture of the two R. flavefaciens strains. For this purpose, we employed a recently reported matching fusion-protein approach, combined with an enzyme-linked immunosorbent assay-like assay system (4). The gene fragments coding for five putative cohesins (two of them divergent cohesins from scaB) were selected from the four designated scaffoldins of strain FD-1 and amplified by PCR using appropriate primers (Table 1). Since all seven ScaB cohesins in strain 17 were shown previously (18) to bind selectively to ScaA, we employed in the present work selected cohesins from strain FD-1 to represent their conserved “paralogues.” Thus, FD-1 ScaA cohesin 1 represented cohesins 1 and 2 from this protein, ScaB cohesin 3 was representative of ScaB cohesins 1 to 4, and ScaB cohesin 6 represented ScaB cohesins 5 to 9. The resultant products were cloned into the pETCBMCoh cassette plasmid as described by Barak et al. (4), yielding C-terminally His-tagged cohesin modules, fused at their N termini with a Clostridium thermocellum CBM3a for improved display in the immobilized state. The DNA regions encoding the dockerin modules derived from ScaA, ScaB, and ScaC of strain FD-1 were amplified using an appropriate pair of primers (Table 1). The resultant PCR fragments were cloned into the pETXynDoc cassette (4), yielding the desired His-tagged xylanase T6 fusion proteins bearing a dockerin at the C terminus. Orthologous dockerin-bearing enzyme genes in strain FD-1, encoding the equivalent of EndB (the family 44 enzyme Cel44A) and that of CesA (the family 3 carbohydrate esterase Ce3B) of strain 17, were amplified by PCR using appropriate primers designed from the sequence of either endB (GenBank accession number AJ298117) or cesA (GenBank accession number AJ238716) of strain 17 (Table 1). Both dockerins were cloned by the strategy described above for the three other dockerins. Subsequent sequence analyses indicated extensive similarity between the two strains in their corresponding dockerin modules. The recombinant cohesin and dockerin modules were expressed as the desired fusion proteins in an Escherichia coli host cell system described previously (4). The cells were sonicated, the soluble proteins were applied batchwise to nickel-iminodiacetic acid agarose resin (Rimon Biotech, Rehovot, Israel), and the adsorbed proteins were eluted with 300 mM imidazole. The purified fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) with Coomassie brilliant blue staining. In all cases, a single major protein band was consistently obtained, as reported earlier (4).
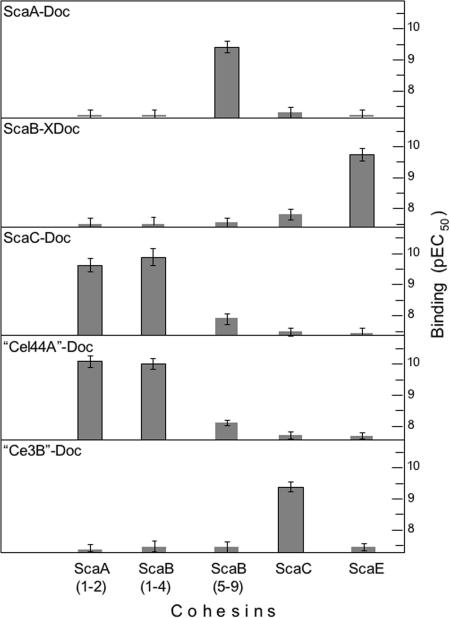
To assess the binding properties among the putative cohesin and dockerin modules, a microtiter plate analysis was performed under standardized conditions, using an efficient, matching fusion-protein system (4). The expressed CBM3 cohesins were thus adsorbed onto microtiter plates and challenged by incremented concentrations of selected xylanase T6-fused dockerins. Following extensive washing procedures, the amount of bound dockerin was assessed colorimetrically by primary anti-xylanase T6 antibody and secondary antibody-enzyme conjugate (4). The negative logarithms of the concentrations that produced the half-maximal chromogenic responses (pEC50 values) were determined from the respective binding curves (16).
As shown in Fig. 2, the ScaA-dockerin fusion protein interacts specifically with the conserved ScaB cohesins, i.e., cohesins 5 to 9 of ScaB of strain FD-1, which are orthologous to the ScaB cohesins of strain 17. The same ScaA dockerin fails to interact with the remaining ScaB cohesins (cohesins 1 to 4) of strain FD-1, which closely resemble in sequence those of ScaA in strain 17. As expected, the ScaA dockerin is unreactive toward its own cohesins, as well as those from ScaC and ScaE. The ScaB X-module/dockerin dyad also exhibited a high level of specificity for a different cohesin, the ScaE cohesin, and essentially failed to recognize the other cohesins. Conversely, the dockerins of ScaC and the enzyme equivalent of Cel44A both bound to the ScaA-like cohesins (i.e., the two reputed cohesins of ScaA and cohesins 1 to 4 of ScaB). No real preference was shown by either dockerin for the designated cohesins on the two scaffoldins, and the levels of recognition of the two dockerins were very similar. In contrast to the dockerin equivalent of Cel44A, that of the enzyme Ce3B displayed clear specificity for the cohesin of ScaC and essentially failed to recognize the cohesins of ScaA and cohesins 1 to 4 of ScaB.
FIG. 2.
Cohesin-dockerin specificity of R. flavefaciens FD-1. Wells of microtiter plates were coated with the designated CBM3 cohesin (0.1 ml; 15 nM), and incremental concentrations of the specified xylanase T6-fused dockerin were applied (4). The FD-1 enzyme-derived dockerins appear in quotation marks (“Cel44A”-Doc and “Ce3B”-Doc) to signify that the enzymes themselves remain unconfirmed. The pEC50 of the dockerin construct was determined as described by Motulsky and Christopoulos (16). Data are the means ± standard deviations for at least three independent determinations. Positive interactions were consistently in the subnanomolar range of dockerin concentration.
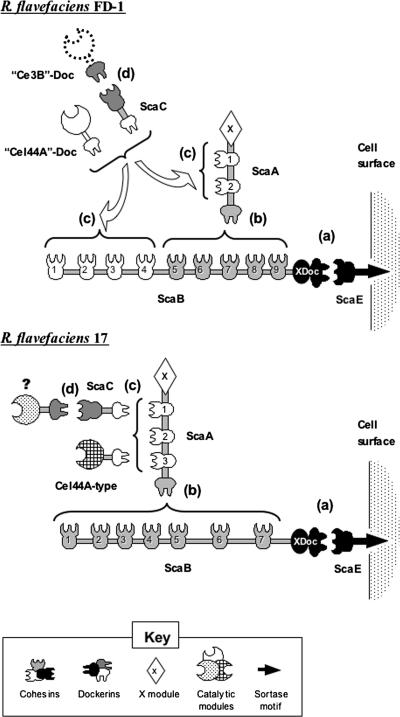
The selective nature of the cohesin-dockerin interactions derived from this study allows us to propose an overall architectural scheme for R. flavefaciens strain FD-1 and to compare it with that proposed recently for strain 17 (17). As presented in Fig. 3, the same molecular players take part in cellulosome assembly in the two strains. The orthologous ScaE scaffoldins are implanted into the respective cell surfaces via a similar sortase-mediated mechanism. The respective ScaE cohesins bind selectively to the XDoc dyad of ScaB, and ScaB cohesins serve to incorporate the primary ScaA scaffoldin. However, the major differences between the two strains are the number of confirmed cohesins in ScaA (two for strain FD-1 versus three for strain 17) and the number (nine versus seven, respectively) and nature (mixed versus homogeneous types, respectively) of the cohesins in ScaB. These differences are of apparent consequence to the respective architectures of the two cellulosome systems. In strain 17, for example, the combination of three ScaA cohesins and seven ScaB cohesins would imply the potential capacity to incorporate 21 different dockerin-containing proteins, either directly or mediated via ScaC. In strain FD-1, however, the two ScaA cohesins combined with the mixed cohesins in ScaB would presumably amount to a total of only 14 dockerin-containing proteins.
FIG. 3.
Schematic representation of the proposed cellulosome architecture in R. flavefaciens FD-1 versus strain 17. In both strains, the cellulosome is covalently linked to the cell surface via a sortase recognition motif, located at the C terminus of the ScaE sequence (17). (a) The ScaE cohesin binds to the bimodular ScaB X-dockerin. (b) In strain FD-1, cohesins 5 through 9 of ScaB bind to the ScaA dockerin, whereas in strain 17, all seven ScaB cohesins bind to the ScaA dockerin. (c) In strain 17, the ScaA cohesins bind either to Cel44A-type dockerin (representative of numerous other enzyme-borne dockerins) or to the ScaC dockerin. In strain FD-1, the “Cel44A” and ScaC dockerins similarly bind to the ScaA cohesins but are additionally recognized by the first four ScaB cohesins. (d) The ScaC cohesin of strain FD-1 binds to the “Ce3B” dockerin; in contrast, the parallel interaction has not been demonstrated for strain 17, although the ScaC cohesin is known to bind to a set of presumed dockerins different from those recognized by ScaA (19).
In both strains, the ScaA cohesins seem to recognize orthologous ScaC and homologous Cel44A-type dockerins. The adaptor role of ScaC would thus appear to be similar for the two strains, i.e., a means of increasing the component repertoire of the respective cellulosome (19). The type of dockerin recognized by the ScaC cohesin has been identified for strain FD-1 but not for strain 17. The dockerin of the family 3 carbohydrate esterase equivalent, “Ce3B”-Doc, was shown in this work to bind strongly to the ScaC cohesin. The quotation marks signify that the enzyme itself remains unconfirmed, since only its dockerin was obtained by PCR amplification of appropriate primers derived from strain 17 sequences. Interestingly, the dockerin of the presumed orthologous enzyme in strain 17 failed to interact with its cognate ScaC cohesin (19). Although the two dockerins show marked sequence similarity (59.0% identity and 74.4% similarity), the nature of the amino acid residues on homologous dockerins that interact with ScaC in either or both species has yet to be elucidated.
In summary, the general design of the scaffoldin gene cluster in strains FD-1 and 17 of R. flavefaciens is essentially identical. The two clusters include the four scaffoldin orthologues in the same positions. Nevertheless, significant variations are evident in the modular architecture of the scaffoldin proteins, which implies a divergent organization of cellulosome architecture between the two strains. It appears that the two strains have undergone a considerable extent of evolutionary divergence, but it is currently unknown whether the differences observed here have functional consequences. In this context, the two strains may conceivably represent distinct biotypes that have become adapted to subtly different niches within the rumen ecosystem. The relationship between cellulosome architecture and the variations that occur in cell wall organization between plant species, or at different stages in the process of rumen digestion, is completely unknown.
Acknowledgments
We thank Alvaro Hernandez, W. Ryan Kim, Lei Liu, and Jyothi Thimmapuram of the Keck Center for Comparative and Functional Genomics at the University of Illinois.
Parts of this research were supported by the National Research Initiative competitive grant no. 2002-35206-11634 from the USDA Cooperative State Research, Education, and Extension Service; by the Israel Science Foundation (grant no. 394/03 and 442/05); and by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel. The Rowett Research Institute is supported by the Scottish Executive Environmental and Rural Affairs Department. M.T.R. is supported by EU Grant GEMINI (QLRT-2001-02056). M.E.B. was supported by the Initiative for Future Agriculture and Food Systems grant no. 2001-52100-11527 from the USDA Cooperative State Research, Education, and Extension Service.
Footnotes
Published ahead of print on 22 September 2006.
REFERENCES
- 1.Antonopoulos, D. A., K. E. Nelson, M. Morrison, and B. A. White. 2004. Strain-specific genomic regions of Ruminococcus flavefaciens FD-1 as revealed by combinatorial random-phase genome sequencing and suppressive subtractive hybridization. Environ. Microbiol. 6:335-346. [DOI] [PubMed] [Google Scholar]
- 2.Aurilia, V., J. C. Martin, S. I. McCrae, K. P. Scott, M. T. Rincon, and H. J. Flint. 2000. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 146:1391-1397. [DOI] [PubMed] [Google Scholar]
- 3.Aurilia, V., J. C. Martin, C. A. Munro, D. K. Mercer, and H. J. Flint. 2000. Organisation and strain distribution of genes responsible for the utilization of xylans by the rumen cellulolytic bacterium Ruminococcus flavefaciens. Anaerobe 6:333-340. [Google Scholar]
- 4.Barak, Y., T. Handelsman, D. Nakar, A. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., J.-P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, M. P., N. Small, C. Bouma, and I. M. Robinson. 1958. Characteristics of ruminal anaerobic cellulolytic cocci and Cilliobacterium cellulosolvens n. sp. J. Bacteriol. 76:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehority, B. A., and H. W. Scott. 1967. Extent of cellulose and hemicellulose digestion in various forages by pure cultures of cellulolytic rumen bacteria. J. Dairy Sci. 50:1136-1141. [Google Scholar]
- 8.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 11.Flint, H. J. 1997. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 5:483-488. [DOI] [PubMed] [Google Scholar]
- 12.Flint, H. J., C. A. McPherson, and J. Bisset. 1989. Molecular cloning of genes from Ruminococcus flavefaciens encoding xylanase and β(1-3,1-4)glucanase activities. Appl. Environ. Microbiol. 55:1230-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby, J., J. C. Martin, A. S. Daniel, and H. J. Flint. 1997. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol. Lett. 149:213-219. [DOI] [PubMed] [Google Scholar]
- 14.Krause, D. O., R. J. Bunch, W. J. Smith, and C. S. McSweeney. 1999. Diversity of Ruminococcus strains: a survey of genetic polymorphisms and plant digestibility. J. Appl. Microbiol. 86:487-495. [Google Scholar]
- 15.Krause, D. O., B. P. Dalrymple, W. J. Smith, R. I. Mackie, and C. S. McSweeney. 1999. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology 145:1797-1807. [DOI] [PubMed] [Google Scholar]
- 16.Motulsky, H. J., and A. Christopoulos. 2003. Fitting models to biological data using linear and nonlinear regression: a practical guide to curve fitting. GraphPad Software, Inc., San Diego, Calif.
- 17.Rincon, M. T., T. Cepeljnik, J. C. Martin, R. Lamed, Y. Barak, E. A. Bayer, and H. J. Flint. 2005. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. J. Bacteriol. 187:7569-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rincon, M. T., S.-Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rincon, M. T., J. C. Martin, V. Aurilia, S. I. McCrae, G. Rucklidge, M. Reid, E. A. Bayer, R. Lamed, and H. J. Flint. 2004. ScaC, an adaptor protein carrying a novel cohesin that expands the dockerin-binding repertoire of the Ruminococcus flavefaciens 17 cellulosome. J. Bacteriol. 186:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rincon, M. T., S. I. McCrae, J. Kirby, K. P. Scott, and H. J. Flint. 2001. EndB, a multidomain family 44 cellulase from Ruminococcus flavefaciens 17, binds to cellulose via a novel cellulose-binding module and to another R. flavefaciens protein via a dockerin domain. Appl. Environ. Microbiol. 67:4426-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert, C., and A. Bernalier-Donadille. 2003. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane excreting subjects. FEMS Microbiol. Ecol. 46:81-89. [DOI] [PubMed] [Google Scholar]
- 22.Xu, Q., Y. Barak, R. Kenig, Y. Shoham, E. A. Bayer, and R. Lamed. 2004. A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J. Bacteriol. 186:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]