Abstract
The UspA2 protein has been shown to be directly involved in the serum-resistant phenotype of Moraxella catarrhalis. The predicted 5′-untranslated regions (UTR) of the uspA2 genes in several different M. catarrhalis strains were shown to contain various numbers (i.e., 6 to 23) of a heteropolymeric tetranucleotide (AGAT) repeat. Deletion of the AGAT repeats from the uspA2 genes in the serum-resistant M. catarrhalis strains O35E and O12E resulted in a drastic reduction in UspA2 protein expression and serum resistance. PCR and transformation were used to construct a series of M. catarrhalis O12E strains that differed only in the number of AGAT repeats in their uspA2 genes. Expression of UspA2 was maximal in the presence of 18 AGAT repeats, although serum resistance attained wild-type levels in the presence of as few as nine AGAT repeats. Increased UspA2 expression was correlated with both increased binding of vitronectin and decreased binding of polymerized C9. Real-time reverse transcription-PCR analysis showed that changes in the number of AGAT repeats affected the levels of uspA2 mRNA, with 15 to 18 AGAT repeats yielding maximal levels. Primer extension analysis indicated that these AGAT repeats were contained in the 5′-UTR of the uspA2 gene. The mRNA transcribed from a uspA2 gene containing 18 AGAT repeats was found to have a longer half-life than that transcribed from a uspA2 gene lacking AGAT repeats. These data confirm that the presence of the AGAT repeats in the 5′-UTR of the uspA2 gene is necessary for both normal expression of the UspA2 protein and serum resistance.
Among the bacteria that can colonize the human nasopharynx, three different organisms (Streptococcus pneumoniae, nontypeable Haemophilus influenzae, and Moraxella catarrhalis) are noted for their ability to cause otitis media in infants and very young children (15). M. catarrhalis, an unencapsulated, gram-negative coccobacillus previously termed either Neisseria catarrhalis or Branhamella catarrhalis (12), is the third most common cause of otitis media in the pediatric population (17). More recent clinical studies have reaffirmed the pathogenic potential of this bacterium in adults, indicating that M. catarrhalis may cause as many as four million exacerbations of chronic obstructive pulmonary disease annually in the United States (45).
Although the ability of this organism to produce disease is now well established, information about the M. catarrhalis gene products essential for pathogenesis remains relatively limited. Among the various putative virulence factors that have been identified to date (reviewed in references 28, 42, and 53), several proteinaceous antigens have been shown to protrude from the outer membrane of M. catarrhalis. These include the UspA1 and UspA2 proteins (2, 25, 47), the Hag (MID) hemagglutinin (19, 47), and type IV pili (32). Interestingly, expression of both the UspA1 protein and the Hag protein appear to be subject to phase variation, although the relevant mechanisms are different. A homopolymeric nucleotide repeat containing 10 guanine residues [i.e., a poly(G) tract] occurs in the 5′-untranslated region (UTR) of the uspA1 gene of most wild-type isolates of M. catarrhalis (14). These relatively long homopolymeric tracts can undergo transitory base-pair misalignment (i.e., slipped-strand mispairing) (23). Upon deletion of a single G residue from this tract, presumably as the result of slipped-strand mispairing, the level of transcription of the uspA1 gene is reduced significantly with a consequent reduction in the level of expression of the UspA1 protein (30). A poly(G) tract is also present inside the hag open reading frame (ORF), just downstream from the translation initiation codon for this gene. When the number of G residues in this hag poly(G) tract is divisible by three, the Hag protein is expressed. If the number of G residues is changed to a number not divisible by three, then a frameshift occurs, resulting in a premature translational termination codon downstream of the poly(G) tract (40, 47).
Similarly, it has been reported that the region immediately upstream from the translation initiation codon of the M. catarrhalis uspA2 gene contains a tetranucleotide (AGAT) repeat (3, 14, 20). The UspA2 protein has been shown to be directly involved in the expression of serum resistance by M. catarrhalis strain O35E (4). In addition, it was recently demonstrated that UspA2-mediated binding of vitronectin from normal human serum (NHS) was responsible for the ability of three M. catarrhalis strains to resist serum killing (5). In a recent survey of nasopharyngeal isolates from young children in Greece, a uspA2 gene or the closely related uspA2H gene was detected in 100% of the 108 strains tested (38). UspA2 appears to be a target for antibodies that develop in patients with chronic obstructive pulmonary disease (43, 44), and antibodies to this protein can also be detected in very young children colonized with M. catarrhalis (37) and in serum and saliva from healthy individuals (13, 51).
In the present study, we show that the presence of the AGAT nucleotide repeat in the 5′-UTR of the uspA2 gene is essential for wild-type expression of the UspA2 protein. Elimination of the AGAT repeat resulted in very low levels of synthesis of UspA2 and converted the serum-resistant M. catarrhalis O12E wild-type strain to a serum-sensitive phenotype. Increasing the number of AGAT repeats resulted in wild-type levels of synthesis of the UspA2 protein, and this could be correlated with an increase in the level of uspA2 mRNA, increased binding of vitronectin, and restoration of the serum-resistant phenotype. Finally, the uspA2 mRNA transcribed from a uspA2 gene lacking AGAT repeats was shown to have a shorter half-life than that transcribed from a uspA2 gene containing these repeats.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The M. catarrhalis strains used in the present study are listed in Table 1 and were grown in brain heart infusion (BHI) broth (Difco/Becton Dickinson, Sparks, MD) at 37°C or on BHI plates solidified with 1.5% (wt/vol) agar in an atmosphere containing 95% air-5% CO2 at 37°C. When needed, BHI was supplemented with kanamycin (15 μg/ml), spectinomycin (15 μg/ml), or dihydrostreptomycin sulfate (750 μg/ml).
TABLE 1.
Bacterial strains used in this study
| M. catarrhalis strain | Genotype or description | Source or reference |
|---|---|---|
| O35E | Wild-type disease isolate; serum resistant | 3 |
| O12E | Wild-type disease isolate; serum resistant | 1 |
| O35E-Smr | rpsL mutant of O35E; streptomycin resistant; serum resistant | 4 |
| O12E.2 | uspA2 mutant of O12E; serum sensitive; spectinomycin resistant | 5 |
| O35E.2 | uspA2 mutant of O35E; serum sensitive; kanamycin resistant | 3 |
| O12EΔAGAT | O12E construct that lacks AGAT nucleotide repeats in the 5′-UTR of its uspA2 gene; serum sensitive; streptomycin resistant | This study |
| O35EΔAGAT | O35E construct that lacks AGAT nucleotide repeats in the 5′-UTR of its uspA2 gene; serum sensitive; streptomycin resistant | This study |
| O12EΔAGAT.2 | uspA2 mutant of O12EΔAGAT; spectinomycin resistant | This study |
| O12E-Smr | rpsL mutant of O12E; streptomycin resistant; serum resistant; has 19 AGAT repeats in the 5′-UTR of its uspA2 gene | This study |
| O12E-2rpts | O12E construct with two AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-6rpts | O12E construct with six AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-8rpts | O12E construct with eight AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-9rpts | O12E construct with 9 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-10rpts | O12E construct with 10 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-11rpts | O12E construct with 11 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-12rpts | O12E construct with 12 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-15rpts | O12E construct with 15 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-18rpts | O12E construct with 18 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| O12E-23rpts | O12E construct with 23 AGAT repeats in the 5′-UTR of its uspA2 gene; streptomycin resistant | This study |
| 7169 | Wild-type disease isolate; serum resistant | 10 |
| ATCC 25238 | Wild-type isolate; serum resistant | ATCC |
| ATCC 43617 | Wild-type isolate; serum sensitive | ATCC |
| ETSU-5 | Wild-type isolate; serum resistant | S. Berk |
| FIN2344 | Wild-type isolate; serum resistant | M. Helminen |
| V1118 | Wild-type isolate; serum resistant | F. Henderson |
| V1120 | Wild-type isolate; serum resistant | F. Henderson |
| V1145 | Wild-type isolate; serum resistant | F. Henderson |
| V1171 | Wild-type isolate; serum resistant | F. Henderson |
Recombinant DNA techniques.
Standard molecular biology and recombinant DNA techniques were performed as described previously (48). PCR-based amplification of DNA fragments was carried out with either ExTaq DNA polymerase (PanVera, Madison, WI) or Pfu DNA polymerase (Stratagene, La Jolla, CA). The oligonucleotide primers used here are listed in Table 2. Isolation of chromosomal DNA from M. catarrhalis was carried out by using the Easy-DNA kit (Invitrogen, Carlsbad, CA), and plasmid DNA was isolated by using the Qiaprep Spin Miniprep kit (QIAGEN, Valencia, CA). Gel purification of DNA fragments was done by using the Geneclean Turbo kit (Q.Biogene, Vista, CA).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| AA26-Rev | GCACCCAAGCCAACAATCATGGCA |
| AA52-Rev | GGGAGAAGTTTCATGGTTTTC |
| AA-54-Rev | TGCCTAGGAAAGCTTTTATCCATCACTCAC |
| AA63-Fw | TAAGATCTCATAGATAGCCACATCAATC |
| AA68-Fw | CTCTCATCAAAGACACACCAA |
| AA69-Fw | GAGATTTTTCCATTTATGCCAGCAAAAG |
| AA69-Rev | CTTTTGCTGGCATAAATGGAAAAATCTC |
| AA70-Fw | AAAACTCTGTCTTTTATCTGTCC |
| AA70-Rev | GGACAGATAAAAGACAGAGTTTT |
| AA71-Fw | ATGCCAGCAAAAGAAAACTCTGTCTTTTATCTGTCCGCTGATG |
| AA74-Fw | AGATAGATAAAACTCTGTCTTTTATCTGTCC |
| AA75-Rev | ATCTATCTCTTTTGCTGGCATAAATGGAAAAATCTC |
| AA9-Rev | TCGCAGTAGATGCCATACCC |
| CopB-807F | CAATCGTGCCTTGACGCTAGA |
| CopB-915R | GCCAAGTTTGTAACCCTTGCCT |
| Rps-3′ | ACGCCACCAACAGCACAATAAACC |
| Rps-5′ | TGGCGAACTCAAGCAAACAGC |
| U2-1886F | GTAAGTTTAATGCGACCGCTGC |
| U2-1987R | CAGCTTTAAACGCCAGATTTGG |
MAbs and Western blot analysis.
M. catarrhalis whole-cell lysates were prepared as described previously (4), and the proteins present in these lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% (wt/vol) polyacrylamide gels. These proteins were electrophoretically transferred to nitrocellulose membranes (Schleicher & Schuell BioScience, Keene, NH) and probed with the appropriate primary antibody, followed by goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Jackson Immunoresearch, West Grove, PA). The antigen-antibody complexes were detected by using the Western Lightning Chemiluminescence Reagent Plus (New England Nuclear, Boston, MA). Monoclonal antibody (MAb) 17C7, which binds an epitope present in both the M. catarrhalis UspA1 protein and the UspA2 protein, has been described elsewhere (3). The M. catarrhalis CopB protein-reactive MAb 10F3 (21) was used to standardize antigen loads when whole-cell lysates were used.
Serum bactericidal assay.
Killing of M. catarrhalis strains by NHS was measured as described previously (4) using either 10% (vol/vol) or 30% (vol/vol) NHS. NHS was prepared from blood drawn from adult human volunteers as described previously (4).
Detection of NHS components bound to M. catarrhalis.
The analysis of the binding of NHS components to M. catarrhalis cells was carried out as described previously (5) with slight modifications. Briefly, M. catarrhalis strains grown into mid-logarithmic phase were resuspended in Veronal-buffered saline (VBS) containing 5 mM MgCl2 and 1.5 mM CaCl2 (VBS++) to a final optical density at 600 nm of 0.6. Portions (500 μl) of these bacterial suspensions were incubated with 10% (vol/vol) NHS in a final volume of 1 ml for 30 min at 37°C. Complement activation was stopped by incubating the reaction tubes on ice for 5 min. The bacterial pellets were washed three times with ice-cold VBS containing 0.1% (wt/vol) gelatin, lysed, and analyzed by Western blotting to detect polymerized C9 and vitronectin as described previously (5).
Identification of a streptomycin-resistant mutant of M. catarrhalis strain O12E.
A spontaneous streptomycin-resistant mutant of M. catarrhalis strain O12E (O12E-Smr) was obtained by using a method described previously (4). Nucleotide sequence analysis showed that this streptomycin-resistant mutant has a single nucleotide change (A→G) in the rpsL gene at residue 128 that resulted in a single amino acid change (i.e., K43R). The mutated rpsL gene, together with flanking sequences was PCR amplified using the oligonucleotide primers Rps-5′ and Rps-3′ to generate a 3-kb amplicon that was used in congression experiments (4).
Construction of M. catarrhalis strains with uspA2 genes that lack AGAT repeats.
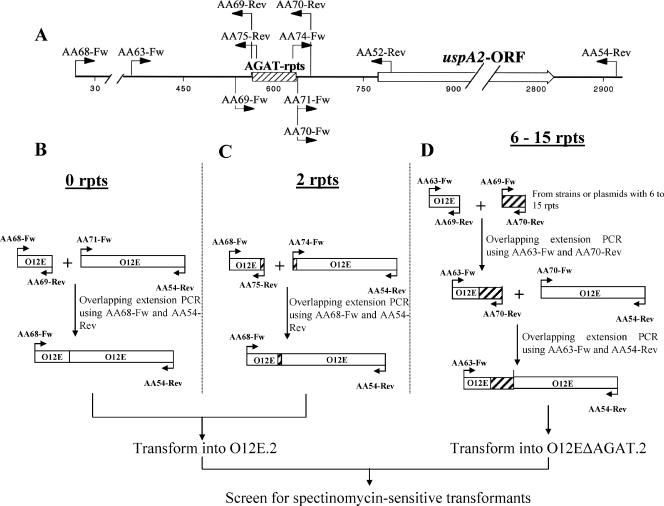
A method similar to overlapping extension PCR (26) was used to delete the AGAT repeats from the uspA2 gene. Chromosomal DNA from either M. catarrhalis strain O12E or M. catarrhalis strain O35E was used as the template for two PCRs (Fig. 1B). For the first reaction, the oligonucleotide primers AA68-Fw and AA69-Rev were used to amplify a ∼0.5-kb region directly upstream of the AGAT repeat region. For the second PCR, the region immediately downstream of the AGAT repeats was PCR amplified using the oligonucleotide primers AA70-Fw and AA54-Rev. This fragment contained the remaining putative 5′-UTR, the entire uspA2 open reading frame (ORF), and 100 nucleotides (nt) downstream of this ORF. After gel purification, the latter fragment was used as a template for another PCR with the oligonucleotide primers AA71-Fw and AA54-Rev. AA71-Fw binds to the same region as AA70-Fw except that it has an additional 13 nt in its 5′ end that are complementary to the first 13 nt of the 5′ end of AA69-Rev. The AA68-Fw-AA69-Rev and AA71-Fw-AA54-Rev fragments were used together as the template for another PCR with AA68-Fw and AA54-Rev as the primers. The final PCR product was gel purified, its nucleotide sequence was verified, and then it was used in a plate transformation-based congression experiment (4), together with the 3-kb amplicon containing the mutated rpsL gene described above. Two different strains were transformed in these experiments; the spectinomycin-resistant uspA2 mutant strain O12E.2 (Spec) and the kanamycin-resistant uspA2 mutant O35E.2. Streptomycin-resistant, spectinomycin-sensitive O12E.2 transformants and streptomycin-resistant, kanamycin-sensitive O35E.2 transformants were identified, and nucleotide sequence analysis was used to verify that the AGAT repeat region had been deleted and that the uspA2 ORF was intact. One transformant was selected from each strain, and these were designated O12EΔAGAT and O35EΔAGAT, respectively.
FIG. 1.
PCR and transformation strategies used to construct M. catarrhalis O12E strains with various numbers of AGAT repeats. (A) Chromosomal locus containing the uspA2 gene and flanking DNA. The locations of oligonucleotide primers used for PCRs are shown. (B to D) The different PCR strategies used to construct the strains containing no AGAT repeats (B), 2 AGAT repeats (C), and 6 to 15 AGAT repeats (D) are illustrated.
Construction of M. catarrhalis O12E strains with various numbers of AGAT repeats in their uspA2 genes.
The wild-type O12E strain has 19 AGAT repeats upstream of its uspA2 gene, and we were able to isolate two naturally occurring variants of this strain that have either 18 or 23 AGAT repeats. These three strains were transformed with the 3-kb amplicon containing the mutated rpsL gene to obtain streptomycin-resistant strains. These transformants were confirmed as having the original number of AGAT repeats (i.e., 18, 19, or 23) in their uspA2 genes. For the construction of an O12E strain with two AGAT repeats, the same procedures described above for the construction of the ΔAGAT deletion constructs were used except the oligonucleotide primer AA75-Rev was used instead of AA69-Rev and the oligonucleotide primer AA74-Fw was used instead of AA70-Fw (Fig. 1C). AA75-Rev and AA74-Fw have the same sequences as AA69-Rev and AA70-Fw, respectively, except AA75-Rev and AA74-Fw have an additional two AGAT repeats at their 5′ ends. For the construction of the rest of the O12E strains with different numbers of AGAT repeats (Fig. 1D), oligonucleotide primers AA69-Fw and AA70-Rev were used to amplify the AGAT repeat regions from both different M. catarrhalis isolates that had various numbers of AGAT repeats in the 5′-UTR of their uspA2 genes and from a cloned DNA fragment containing the uspA2 5′-UTR from O12E. These PCR-amplified fragments containing different numbers of AGAT repeats were used in two-step PCRs. The first involved the PCR fragment obtained by using the AA63-Fw/AA69-Rev primer pair with O12E chromosomal DNA as a template. The resultant amplicon was then used in a PCR with the PCR fragment obtained by using the AA70-Fw/AA54-Rev primer pair with O12E chromosomal DNA as a template. The final PCR products were sequence verified and then transformed by congression into the spectinomycin-resistant strain O12EΔAGAT.2. Streptomycin-resistant, spectinomycin-sensitive transformants were screened first by using a colony-PCR system with the oligonucleotide primers AA63-Fw and AA52-Rev to identify transformants whose PCR products exhibited a size shift relative to that obtained from strain O12EΔAGAT. Each selected transformant was sequence verified to ensure that it possessed the desired number of AGAT repeats and that the uspA2 ORF was intact.
Measurement of UspA2 protein levels by using flow cytometry.
The amount of UspA2 protein expressed by the O12E constructs with different numbers of AGAT repeats was measured by using flow cytometry with MAb 17C7 as the primary antibody. To avoid interference from the UspA1 protein, which is also reactive with MAb 17C7 (3), isogenic uspA1 mutants were generated by transforming each of these O12E constructs with the plasmid pUSPA1KAN as described previously (2). The 5′-UTR of the uspA2 gene of each of these uspA1 mutants was sequenced to confirm that no change had occurred in the number of the AGAT repeats during the transformation process. These constructs were then subjected to flow cytometric analysis to compare the levels of the UspA2 protein. Briefly, bacterial cells grown overnight on BHI agar plates were suspended in phosphate-buffered saline (PBS) to an optical density at 600 nm of 0.35. Portions (100 μl) of these suspensions were spun down and resuspended in 100 μl of PBS containing 1% (wt/vol) bovine serum albumin (PBS-BSA) in which purified MAb 17C7 (3 μg/μl) had been diluted 1:100. These tubes were incubated at room temperature for 20 min and then washed three times with 500 μl of PBS-BSA. The bacteria were then incubated with 1 μg of fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Abcam, Cambridge, MA) for 20 min at room temperature, followed by three washes with 500 μl of PBS-BSA. The final pellet was suspended in 1 ml of PBS and analyzed by flow cytometry using a FACScan flow cytometer (Becton Dickinson).
Measurement of uspA2 transcription by real-time RT-PCR.
RNA was isolated from bacterial strains grown to mid-logarithmic phase and then treated with DNase I (MessageClean Kit, GenHunter Corp, Nashville, TN) to remove any DNA contamination. The RNA was then washed by ethanol precipitation. For real-time reverse transcription-PCR (RT-PCR) analysis, the copB gene was used as an endogenous control to normalize the results obtained with the uspA2 gene. Primers were designed by using Primer Express software (Applied Biosystems, Foster City, CA). Primers CopB-807F and CopB-915R were used to amplify a 109-bp fragment of the copB gene, and the primers U2-1886F and U2-1987R were used to amplify a 102-bp fragment of the uspA2 gene. A master mix for each gene was prepared so that each well would contain 12.5 μl of 2X SYBR Green Master Mix (Applied Biosystems), 1 μl of a 2.5 μM stock solution of each primer (forward and reverse), 0.1 μl of MultiScribe reverse transcriptase (50 U/μl; Applied Biosystems), and RNase-free water (QIAGEN) in a final volume of 20 μl. Portions (5 μl) of the RNA samples (20 ng/μl) were added to the wells in triplicate. As a negative control, wells that had the master mix but lacked the reverse transcriptase enzyme were used to detect any DNA contamination. The data analysis was carried out by using the 7500 System SDS software v.13 (Applied Biosystems) applying the relative quantification ΔΔCT method. The amount of the uspA2 message was normalized according to the amount of the copB message, and the data are presented as the fold increase using the normalized level of the uspA2 gene of M. catarrhalis O12EΔAGAT as the calibrator.
Primer extension analysis.
A 10-μg quantity of RNA freshly isolated from either strain O12E-9rpts or O12E-18rpts was subjected to RT using SuperScript II reverse transcriptase (Invitrogen) and the primer AA52-Rev, AA26-Rev, or AA9-Rev in the presence of [α-32P]dCTP (Perkin-Elmer, Boston, MA) for 1 h at 42°C. The labeled RT product was then subjected to electrophoresis in a 6% (wt/vol) polyacrylamide-urea sequencing gel together with a sequencing ladder that had been generated by using the same primers with the AmpliCycle sequencing kit (Perkin-Elmer) according to the manufacturer's instructions. The gel was fixed by using a solution containing 5% (vol/vol) acetic acid and 5% (vol/vol) methanol, exposed to a storage phosphor intensifying screen (GE Healthcare, Piscataway, NJ) for a few hours, and then scanned using a STORM 820 scanner (GE Healthcare). The image was analyzed by using ImageQuant v.5.2 software (Molecular Dynamics, Sunnyvale, CA).
Assessment of mRNA stability.
M. catarrhalis strains O12-0rpts and O12E-18rpts were grown to mid-logarithmic phase in 40-ml culture volumes. Portions (5 ml) were then removed and mixed with an equal volume of 50 mM sodium azide and kept on ice. Rifampin was added to the rest of the bacterial cultures to a final concentration of 150 μg/ml. Additional 5-ml portions of the broth cultures were then removed after 1, 2, 3, 5, and 10 min; mixed with equal portions of 50 mM sodium azide; and kept on ice. RNA was then isolated from these samples and used for real-time RT-PCR analysis as described above except that the data analysis was carried out by using the absolute quantitation method. The amount of both uspA2 and copB mRNA transcripts was determined for each strain at the six time points. This experiment was carried out three times independently, and each RNA sample was analyzed at least in duplicate. Half-life determination was done by plotting the percentage of mRNA left (i.e., the amount of mRNA at t = 0 was considered 100%) versus the time elapsed after rifampin addition. Microsoft Excel 2003 software (Microsoft Corp., Redmond, WA) was used to determine the best-fitting line. The equation describing this line was then used to determine the time at which 50% of the original mRNA had been degraded.
Nucleotide sequence analysis.
Nucleotide sequence analysis was accomplished by using a 3730xl DNA analyzer (Applied Biosystems). Analysis of DNA sequences involved the use of SeqEd (Applied Biosystems) and MacVector (version 6.5.3; Oxford Molecular Group, Campbell, CA) software.
RNA structure prediction.
The Mfold program (version 3.2 [www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi]) of Zuker and Turner (36, 56) was used to predict RNA secondary structure and to calculate free energies.
Statistical analysis.
Statistical analysis was carried out by applying the Student t test using Microsoft Excel 2003 software. P values of <0.05 were considered significant.
RESULTS
M. catarrhalis strains vary in the number of AGAT repeats in the putative 5′-UTR of their uspA2 genes.
The nucleotide sequences of the putative 5′-UTR of the uspA2 gene from 11 different M. catarrhalis strains were compared. Nucleotide sequence analysis showed that this region was very highly conserved among the tested strains. However, there was considerable variation in the number of the AGAT nucleotide repeats (Fig. 2). This AGAT repeat region was located approximately 130 nt upstream of the translation initiation codon of the uspA2 gene. In the case of the wild-type strain O12E, the number of the AGAT repeats was 19; however, two other naturally occurring variants were isolated that contained either 18 or 23 repeats. No morphological differences were observed among colonies of these three strains. The smallest number of AGAT repeats that was observed among the tested M. catarrhalis isolates was six, and this occurred in strain ATCC 43617. It is worth mentioning that the latter strain is serum sensitive because it does not express a UspA2 protein due to the presence of a premature translation termination codon within its uspA2 ORF (data not shown).
FIG. 2.
Alignment of the nucleotide sequences of the putative 5′-UTR of the uspA2 genes from 11 different M. catarrhalis strains. Identical nucleotides are shaded in dark gray. This figure was generated by using the CLUSTAL W Alignment program in MacVector (v6.5). The dotted line encloses the AGAT repeats. The small box at the end of these sequences enclosed the ATG translation start codon of the uspA2 gene. The nucleotide sequences of the 5′-UTR from strains O35E, ATCC 25238, and V1171 were derived from a previous study (14).
Deletion of the AGAT repeats affects both UspA2 protein expression and serum resistance.
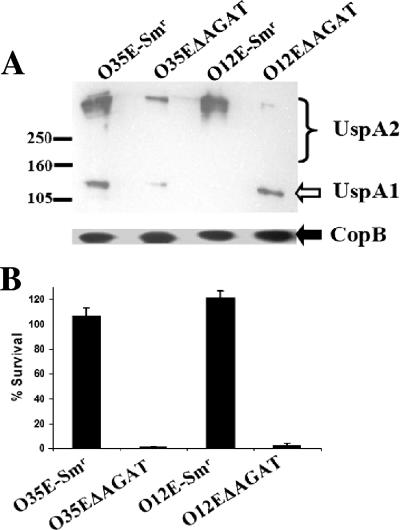
The presence of AGAT repeats upstream from the translation initiation codon in every uspA2 gene examined in this laboratory and by others (20) suggested that these repeats might be involved in the expression of the uspA2 gene. To address this possibility, a combination of PCR and transformation was used to delete the entire AGAT repeat region from the putative 5′-UTR in the uspA2 genes of M. catarrhalis strains O35E and O12E (Fig. 1B); the resultant transformants were designated O35EΔAGAT and O12EΔAGAT, respectively. No morphological differences were observed between colonies of these mutants and those of their respective wild-type parent strains. Western blot analysis revealed that these two transformants expressed much lower levels of UspA2 than did the streptomycin-resistant transformants of their respective parent strains (Fig. 3A).
FIG. 3.
Deletion of the AGAT repeats from the uspA2 gene causes a decrease in both UspA2 protein expression and serum resistance. (A) Western blot analysis of whole-cell lysates of M. catarrhalis strains O35E-Smr, O35EΔAGAT, O12E-Smr, and O12EΔAGAT. The nitrocellulose membrane was probed with MAb 17C7, which recognizes both UspA2 (bracket) and UspA1 (white arrow) (3). The amount of CopB outer membrane protein, as determined by binding of the CopB-reactive MAb 10F3 (21), was used for loading standardization. Protein molecular mass markers (in kilodaltons) are present on the left side of the panel. (B) Serum bactericidal assay with the four strains described in panel A. Bacterial cells were incubated in 10% NHS at 37°C for 30 min. Bacterial aliquots were plated at both t = 0 and t = 30 min. The percent survival was calculated with respect to the original inoculum. These results represent the mean of three independent experiments and the error bars represent the standard deviation.
We previously used mutant analysis to establish that expression of the UspA2 protein is necessary for serum resistance of both strains O35E and O12E (4, 5). Therefore, it was likely that a reduced level of expression of the UspA2 protein by these strains would be accompanied by a decrease in the level of serum resistance. To test this hypothesis, the two streptomycin-resistant parent strains (O35E-Smr and O12E-Smr) were tested together with the two mutants lacking AGAT repeats (O35EΔAGAT and O12EΔAGAT) in serum bactericidal assays with 10% (vol/vol) NHS. The two mutants lacking the AGAT repeats were found to be exquisitely serum sensitive (Fig. 3B). It was also observed that the O12E-Smr strain showed reduced expression of the UspA1 protein; this was found to be the result of phase variation [from 10(G) to 9(G) residues] in the poly(G) tract located upstream of the uspA1 ORF. However, comparison of the O12E-Smr strain with the wild-type O12E parent strain showed no significant difference between these two strains in the level of serum resistance (data not shown).
Increasing the number of AGAT repeats results in increased levels of UspA2 protein.
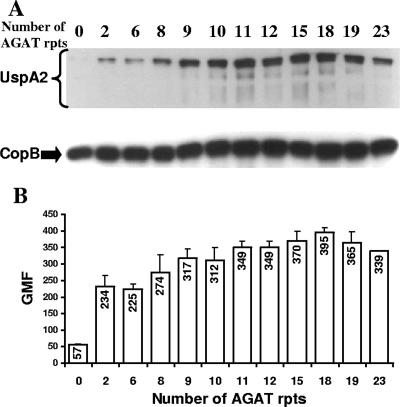
The dramatic decrease in UspA2 protein expression that accompanied the deletion of the AGAT repeats (Fig. 3A) raised a question as to how many AGAT repeats were necessary for wild-type levels of synthesis of UspA2. To address this issue, PCR and transformation were used to construct a series of M. catarrhalis O12E constructs that had various numbers of AGAT repeats in the putative 5′-UTR of their uspA2 genes (Fig. 1C and D). These different constructs contained from 2 to 15 AGAT repeats and were analyzed together with the O12EΔAGAT construct and streptomycin-resistant transformants of the wild-type O12E parent strain with 19 repeats and the two naturally occurring variants of strain O12E possessing 18 and 23 repeats. No morphological differences were apparent among colonies of these 12 strains (data not shown). Western blot analysis of these 12 strains (Fig. 4A) showed that there was an apparent increase in the level of expression of UspA2 with the gradual increase in the number of the AGAT repeats. The two constructs with 15 and 18 AGAT repeats appeared to have the highest level of expression of UspA2 among the tested strains (Fig. 4A). Next, a more quantitative analysis was performed to confirm this observation. Flow cytometry was used to compare the levels of UspA2 protein expressed by uspA1 mutants of these 12 strains with various numbers of AGAT repeats (Fig. 4B). Inactivation of the uspA1 gene was necessary because the UspA1 protein binds the MAb (17C7) used to detect UspA2 (3). There was about a fourfold increase in the geometric mean fluorescence (gmf) value obtained when the number of AGAT repeats was increased from zero to two. There was a slight and gradual increase in gmf values with the gradual increase in AGAT repeat number, with the maximum gmf value being obtained with the construct containing 18 repeats. This was followed by a very slight decrease in the gmf values obtained with the two constructs with 19 and 23 repeats (Fig. 4B).
FIG. 4.
Effect of increasing numbers of AGAT repeats on expression of the UspA2 protein. (A) Western blot analysis of O12E-derived constructs with various numbers of AGAT repeats in their uspA2 genes. Proteins present in whole-cell lysates of these strains were resolved by SDS-PAGE under nonreducing conditions and transferred to nitrocellulose membranes. The membranes were probed with MAb 17C7, which binds the M. catarrhalis UspA1 and UspA2 proteins (3). The region of the gel containing the UspA1 protein is not present in this image. As a loading control, membranes were probed with the CopB-reactive MAb 10F3 (21). (B) Flow cytometric analysis of the reactivity of MAb 17C7 with O12E constructs with various numbers of AGAT repeats in the 5′-UTR of their uspA2 genes. Whole cells of uspA1 mutants of these O12E constructs were probed with MAb 17C7, followed by washing and incubation with a fluorescein isothiocyanate-conjugated antiserum to mouse immunoglobulin G. After a washing step, the cells were analyzed by flow cytometry, and the gmf values were recorded. These results represent the mean of three independent experiments, and the error bars represent the standard deviation.
Increasing the number of AGAT repeats also results in increased serum resistance.
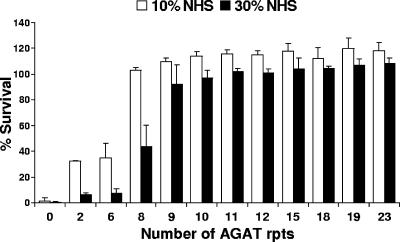
The observed increase in expression of the UspA2 protein with the increase in the number of AGAT repeats (Fig. 4) and the demonstrated serum sensitivity of the O12EΔAGAT strain (Fig. 3B) allowed us to determine whether there was a minimal or threshold level of UspA2 expression necessary for serum resistance. To accomplish this, the 12 O12E constructs with various numbers of AGAT repeats in their uspA2 genes were tested in a series of serum bactericidal assays. When NHS was used at a concentration of 10% (Fig. 5), the construct with no AGAT repeats was again exquisitely serum sensitive. When the number of AGAT repeats was increased to either two or six, there was a moderate increase in serum resistance, with ca. 30% of the initial inoculum of these two constructs surviving for 30 min under the conditions of this assay. However, when the number of AGAT repeats was increased to eight, there was a striking increase in serum resistance, with essentially 100% of the initial inoculum surviving. Further increases in the number of AGAT repeats did not cause any substantial increases in the level of serum resistance. To test whether increasing the concentration of NHS used in this assay would cause a shift in the minimum number of AGAT repeats required to obtain full serum resistance, the 12 constructs were also tested with 30% NHS (Fig. 5). Again, the deletion construct with no AGAT repeats was exquisitely serum sensitive, whereas the two constructs with either two or six AGAT repeats showed a very slight increase in serum resistance. For the construct with eight AGAT repeats, only a moderate increase in serum resistance was observed, and wild-type serum resistance was achieved in the presence of nine AGAT repeats. Further increases in the number of AGAT repeats did not appear to result in substantial increases in serum resistance in this assay.
FIG. 5.
Serum bactericidal assay performed with O12E constructs with various numbers of AGAT repeats in their uspA2 genes. Bacterial cells were incubated in 10% NHS (□) and 30% NHS (▪) at 37°C for 30 min. Bacterial aliquots were plated at both t = 0 and t = 30 min. The percent survival was calculated with respect to the original inoculum. These results represent the mean of three independent experiments, and the error bars represent the standard deviation.
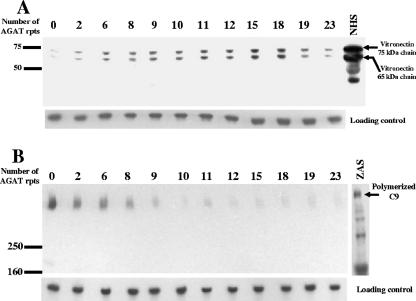
Analysis of the levels of vitronectin and polymerized C9 bound to O12E strains with different numbers of AGAT repeats.
It has been shown that M. catarrhalis strain O12E resists killing by NHS partially through its ability to bind the complement regulator vitronectin via its UspA2 protein (5). Also, it has been shown that this vitronectin binding was associated with the ability of wild-type O12E to interfere with the late stages of complement activation involving C9 polymerization (5). We next examined whether the changes in the level of UspA2 protein expression that accompanied changes in the number of AGAT repeats could be correlated with vitronectin binding and inhibition of polymerized C9 binding. The 12 O12E strains with different numbers of AGAT repeats were incubated with 10% NHS, and whole-cell lysates were analyzed by Western blotting to assess the levels of vitronectin and polymerized C9 bound to each strain. When vitronectin binding was examined, there was a gradual increase in the level of vitronectin binding to O12E constructs with the increase in the number of the AGAT repeats, reaching a maximum level with 15 and 18 repeats, and then there was a slight reduction in the amount of bound vitronectin with further increases in the number of AGAT repeats (Fig. 6A). The pattern of the vitronectin binding correlated very well with the pattern of UspA2 protein expression seen before in Fig. 4. The largest amount of polymerized C9 was associated with the serum-sensitive construct O12EΔAGAT (Fig. 6B). A gradual decrease in the amount of polymerized C9 was observed bound to the strains with two, six, and eight repeats, and very low and similar amounts of polymerized C9 were observed bound to the constructs that had nine or more AGAT repeats (Fig. 6B). This pattern of binding of polymerized C9 correlated very well with the serum resistance pattern that was observed with the same constructs (Fig. 5).
FIG. 6.
Binding of serum components from NHS to O12E constructs with various numbers of AGAT repeats in their uspA2 genes. Bacterial cells were incubated in 10% NHS at 37°C for 30 min. The cells were then washed and whole-cell lysates were prepared and analyzed by Western blotting. (A) Vitronectin bound to M. catarrhalis cells. Proteins present in the samples were resolved by SDS-PAGE under reducing conditions, transferred to nitrocellulose membranes, and probed with an MAb against human vitronectin. The last lane contains a sample of NHS diluted 1:200. (B) Polymerized C9 bound to M. catarrhalis cells. Proteins present in the samples were resolved by SDS-PAGE under nonreducing conditions, transferred to polyvinylidene difluoride membranes, and probed with an MAb to SC5b-9 that recognizes a neoepitope in polymerized C9 in the membrane attack complex. The last lane of panel A contains a control sample of zymosan-activated NHS (ZAS) that was probed with the same MAb to detect polymerized C9 (5). As a loading control, membranes were probed with M. catarrhalis CopB-reactive MAb 10F3 (21). Protein molecular mass markers (in kilodaltons) are presented on the left side of each panel.
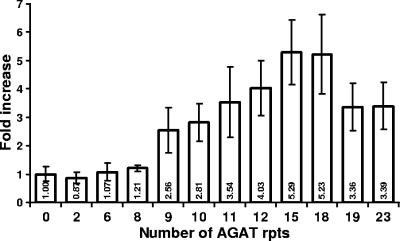
Changes in the number of AGAT repeats affect the level of uspA2 mRNA.
The previous experiments established that the number of the AGAT repeats in the putative 5′-UTR of the uspA2 gene affected the level of the expression of the UspA2 protein. Since these repeats are located upstream of the uspA2 ORF, it was likely that they affected expression of the uspA2 gene at the level of transcription. To test this hypothesis, total RNA extracted from the 12 O12E strains described above was analyzed by using real-time RT-PCR. The level of the transcripts of the copB gene (21) was used to normalize the level of the uspA2 message among the 12 tested strains. Analysis of the data obtained from the real-time RT-PCR experiments by using the ΔΔCT method (Fig. 7) showed that the increase in the number of AGAT repeats from zero to eight had a very little if any effect on the detectable level of the uspA2 transcript. However, an obvious increase (i.e., 2.5-fold) was observed upon increasing the number of the AGAT repeats to nine (Fig. 7). Further increases in the number of AGAT repeats were associated with a gradual increase in the detectable level of the uspA2 transcript, reaching a maximum with the strains possessing 15 and 18 AGAT repeats (Fig. 7). This was followed by a slight decrease in the level of the uspA2 transcript obtained with the two constructs that had 19 and 23 AGAT repeats (Fig. 7). These data indicate that the number of AGAT repeats does affect the level of detectable uspA2 gene mRNA.
FIG. 7.
Real-time RT-PCR analysis of uspA2 gene expression by O12E constructs with various numbers of AGAT repeats in their uspA2 genes. Total RNA isolated from O12E constructs with various numbers of AGAT repeats in their uspA2 genes was used for real-time RT-PCR with primers specific for the uspA2 and copB genes. The data analysis was carried out by using the 7500 System SDS software v.13, applying the relative quantification ΔΔCT method. The level of the uspA2 message was normalized according to the level of the copB message, and the data are presented as a fold increase using the normalized level of the uspA2 gene of O12EΔAGAT as the calibrator. These data represent the mean of three independent experiments (each performed with samples in triplicate), and the error bars represent the standard deviations.
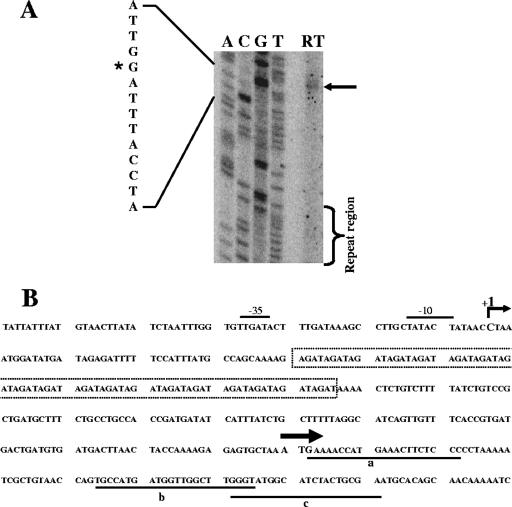
The AGAT repeat region is part of the uspA2 transcript.
Primer extension analysis was performed with two constructs (i.e., O12E-9rpts and O12E-18rpts). The oligonucleotide primers used in these experiments included AA52-Rev, AA26-Rev, and AA9-Rev, which bind to three different locations within the 5′-end of the uspA2 ORF (the locations of these primer binding sites are labeled a, b, and c, respectively, in Fig. 8B). The results of experiments using O12E-9rpts RNA with primer AA52-Rev showed that the uspA2 transcript starts at a C residue located 43 nt upstream of the AGAT repeat region (Fig. 8A). The same results were obtained using RNA from O12E-18rpts with the primers AA26-Rev, AA52-Rev, and AA9-Rev (data not shown). Analysis of the nucleotide sequences upstream of this transcriptional start point showed the presence of sequences that are similar to other prokaryotic −10 and −35 sequences (Fig. 8B). These results confirmed that the AGAT repeats are included in the uspA2 transcript and that changes in the number of AGAT repeats do not affect the location of the transcriptional start point, at least for the constructs tested here.
FIG. 8.
Determination of the uspA2 transcriptional start point by using primer extension analysis. (A) Primer extension results obtained with RNA isolated from M. catarrhalis O12E-9rpts and AA52-Rev. The arrow indicates the position of the RT product, and the repeat region is indicated with the bracket. The asterisk marks the position from which transcription starts. (B) Nucleotide sequence of the 5′ end of the M. catarrhalis O12E uspA2 gene. The C nucleotide marked with “+1” shows the predicted transcriptional start point determined from the data in panel A. Sequences similar to the −10 and −35 consensus sequences seen in bacterial gene promoters are marked with black bars. The start of the uspA2 ORF is marked with the black arrow, and the AGAT repeat region is contained within the dotted box. The locations of binding sites for the oligonucleotide primers AA52-Rev, AA26-Rev, and AA9-Rev are underlined and labeled as a, b, and c, respectively.
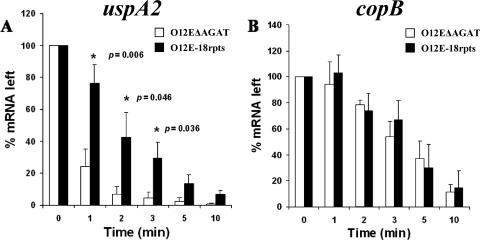
The presence of the AGAT repeats increases the stability of the uspA2 transcript.
One possible explanation for the ability of the AGAT repeats to affect the level of detectable uspA2 mRNA (Fig. 7) would involve the stability of the uspA2 transcript. To address this, mRNA stability assays were performed on RNA isolated from M. catarrhalis strains O12EΔAGAT and O12E-18rpts at different time points after the addition of rifampin to stop de novo transcription. Quantitative real-time RT-PCR showed that there was a significant difference between these two strains with respect to the relative amount of uspA2 mRNA remaining 1, 2, and 3 min after the addition of rifampin, with P values of 0.006, 0.0046, and 0.036, respectively (Fig. 9A). At the same time, there were no significant differences in the percentage of copB mRNA left after the addition of rifampin at all of the time points analyzed (Fig. 9B). There was a significant difference in the half-life of the uspA2 transcripts between the two tested strains. The half-life of the uspA2 transcript from O12EΔAGAT was 1.07 ± 0.36 min, whereas that for the uspA2 transcript from O12E-18rpts was 2.23 ± 0.27 min (P = 0.01). In contrast, the half-life of the copB mRNA from O12EΔAGAT was not significantly different from that obtained from O12E-18rpts; 3.31 ± 0.92 min for O12EΔAGAT and 3.60 ± 1.6 min for O12E-18rpts. These results indicated that the presence of the AGAT repeats does affect, in a positive manner, the stability of the uspA2 transcript.
FIG. 9.
Stability of the uspA2 and copB transcripts from strains O12EΔAGAT and O12E-18rpts. RNA isolated from M. catarrhalis strains O12E-0rpts and O12E-18rpts at different time points after the addition of rifampin was analyzed by using quantitative real-time RT-PCR to determine the amount of both uspA2 (A) and copB (B) transcripts. The percentage of mRNA left at each time point was determined and plotted. These results are the mean of three experiments, and the error bars indicate the standard deviation. The asterisk indicates that the difference between the two tested strains in the percentage of remaining uspA2 mRNA was statistically significant, with the indicated P value.
AGAT repeats may affect RNA secondary structure.
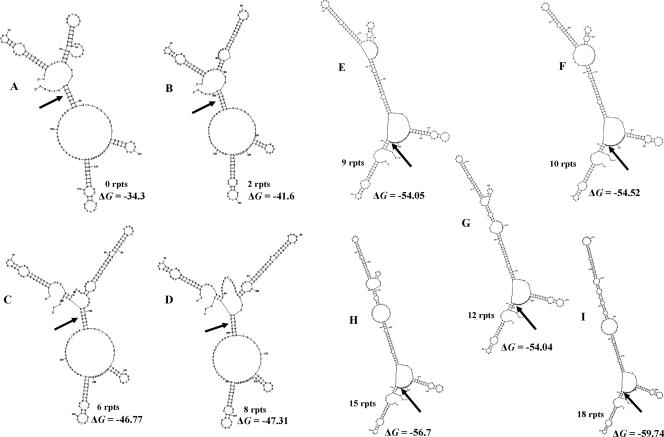
In a preliminary effort to determine how the AGAT repeats could affect mRNA stability, the Mfold program was used to predict the secondary structure of the 5′-UTR of mRNA from uspA2 genes containing increasing numbers of these repeats. In the presence of zero to eight repeats (Fig. 10A to D), the predicted ribosome-binding site was predicted to exist in a helical element. In contrast, in the presence of 9 to 18 repeats (Fig. 10E to I), the predicted ribosome-binding site could be located in an open region of a three-helical junction. Whereas alternative predicted secondary structures were obtained for each 5′-UTR (data not shown), the presence of the predicted ribosome binding site in a helical element occurred predominantly with the smaller numbers of repeats (i.e., 0 to 12).
FIG. 10.
Predicted secondary structure of the RNA transcribed from the 5′-UTR of uspA2 genes with various numbers of AGAT repeats. The Mfold program (36, 56) was used to predict these structures. The arrows indicate the position of the predicted ribosome-binding site in each structure. The ΔG of each secondary structure is listed in kcal/mol.
DISCUSSION
Alteration of the length of a nucleotide repeat motif to effect gene regulation has been described for numerous prokaryotes (6, 23). Whether the nucleotide repeat is homopolymeric [e.g., a poly(G) tract] or heteropolymeric (e.g., AGAT) in composition, there are both cis- and trans-acting factors that can cause an increase or decrease in the numbers of the repeat unit (6). The mechanism whereby these changes consequently affect expression of the encoded product is dependent primarily on the location of the nucleotide repeat. If the nucleotide repeat is located within an ORF, then phase variation occurs because of the presence of a premature translation termination codon. Examples of this phenomenon include the lgtABE locus of N. meningitidis in which the lgtA ORF has a poly(G) tract containing 14 G residues and which readily gives rise to phase variants with fewer G residues that cannot express LOS containing lacto-N-neotetraose (27). Examples of genes that are regulated on the translational level by variation in the number of heteropolymeric nucleotide repeats include the well-studied lic loci in H. influenzae which contain genes involved in the expression of LOS epitopes (24, 54, 55). Tandem repeats of the tetramer CAAT occur at the 5′ end of some of the genes in the lic loci, and these can readily be lost, resulting in frameshifts (54). Similarly, the region of the N. gonorrhoeae gene that encodes the signal peptide for outer membrane P.II has a pentanucleotide repeat that allows phase-variable expression of this gonococcal surface protein (41).
When the nucleotide repeat is located upstream of the ORF, then changes in repeat numbers affect expression at the level of transcription. Control at this level is usually not an ON-OFF switch but instead functions like a rheostat, typically affecting promoter activity (23). One example of this type of phase variation involves a homopolymeric repeat associated with the N. gonorrhoeae fetA gene that encodes the gonococcal ferric enterobactin receptor. Phase variation between high- and low-level expression of FetA was shown to be controlled by the number of C residues in a poly(C) tract located between the predicted −10 and −35 consensus sequences (11). Variation in the level of expression of the meningococcal NadA protein was caused by changes in the number of TAAA repeats located in front of the nadA promoter; this affected the interaction of this promoter with the transcriptional regulator factor IHF (33). In an exception to the usual “up-or-down” effect on transcription, changes in the number of GAA trinucleotide repeats upstream from the transcriptional start site for the Mycoplasma gallisepticum pMGA genes can ablate expression of these genes (31).
Nucleotide repeats and their potential role in contingency loci have probably been most thoroughly examined in Neisseria meningitidis and Haemophilus influenzae (7, 33-35, 49). The availability of completely sequenced genomes for both pathogens (18, 46, 52) has greatly facilitated these types of analyses. The MC58 strain of N. meningitidis has been reported to possess at least 65 genes which have or are associated with nucleotide repeats (35). Other bacterial genes that affect phase variation frequencies of homopolymeric nucleotide repeats in N. meningitidis (34) and those of heteropolymeric nucleotide repeats in H. influenzae (8) have been identified.
There are four previous examples of nucleotide repeats that affect gene expression in M. catarrhalis. Three of these nucleotide repeats are located within the ORF of the affected gene and have the potential to cause frameshift mutations when they undergo slipped-strand mispairing. The first of these is a CAAC tetranucleotide repeat found near the 5′ end of two very similar ORFs encoding predicted methylases in M. catarrhalis ATCC 23246 (50). The second is a poly(G) tract positioned at the 5′ end of the ORF encoding the Hag (MID) hemagglutinin protein (40, 47). The third is a poly(A) tract located near the 5′ end of the ORF encoding the UspA2H protein (W. Wang, M. M. Pearson, A. S. Attia, R. J. Blick, and E. J. Hansen, submitted for publication) expressed by some M. catarrhalis strains in place of a UspA2 protein (29). To date, there is only one report of a nucleotide repeat that affects transcription of a M. catarrhalis gene. This involves the uspA1 gene in which a poly(G) tract located in the 5′-UTR has been shown to undergo presumed slipped-strand mispairing that results in reduced expression of the UspA1 adhesin protein in M. catarrhalis strain O35E (30).
The UspA2 protein has been predicted to be an autotransporter (22) and forms a dense layer of relatively short, filamentous projections on the surface of M. catarrhalis (25, 47). Most M. catarrhalis strains express a UspA2 protein (29, 38) or possess a uspA2 gene (9), although a minority of M. catarrhalis strains express the very similar UspA2H protein (29). Both UspA2 and UspA2H can confer serum resistance on M. catarrhalis, and it was recently shown that several M. catarrhalis strains use UspA2 to bind vitronectin in NHS and thereby inhibit complement-mediated killing (5).
The data contained in the present study indicate that the presence of the multiple AGAT tetranucleotide repeats located between the transcriptional start of the uspA2 gene and the uspA2 ORF is necessary for normal or wild-type expression of the UspA2 protein. Elimination of the AGAT repeats resulted in very low levels of both uspA2 mRNA (Fig. 7) and UspA2 protein (Fig. 3). Reintroduction of the AGAT repeats resulted in increased levels of detectable uspA2 mRNA (Fig. 7) and UspA2 protein (Fig. 4), achieving maximal levels when 15 or 18 repeats were present. This ability to regulate the level of expression of UspA2 protein also allowed, for the first time, determination of the relative level of expression of this protein necessary to achieve resistance to killing by NHS (Fig. 5). These levels of UspA2 protein that afforded serum resistance in the bactericidal activity assay used in the present study were shown to correlate with both increased binding of vitronectin and reduced binding of polymerized C9 (Fig. 6).
It is interesting that, among the 11 wild-type strains included in the strain survey (Fig. 2), all of these strains except ATCC 43617 had at least 12 AGAT repeats in their uspA2 genes. Strain ATCC 43617 is serum sensitive because it does not express a UspA2 protein; this is a direct result of the presence of a premature translational termination codon in its uspA2 gene (data not shown). The other 10 strains all had sufficient numbers of AGAT repeats (i.e., ranging from 12 to 23) to express resistance to killing by NHS in a bactericidal activity assay. It can be inferred from these findings that, in vivo, serum-resistant M. catarrhalis strains may need to maintain a certain number of the AGAT repeats in their uspA2 genes to maintain the threshold level of UspA2 protein necessary to evade host defenses. The lack of a relevant animal model for disease caused by M. catarrhalis (28) precludes direct testing of this hypothesis at the experimental level.
Exactly how these AGAT repeats control the levels of uspA2 mRNA remains to be established definitively. However, examination of mRNA stability in the present study revealed that transcripts from a uspA2 gene with no AGAT repeats had a significantly shorter half-life than did those that were transcribed from a uspA2 gene with 18 AGAT repeats (Fig. 9). The predicted secondary structure of the 5′-UTR of mRNA from uspA2 genes with smaller numbers of repeats appeared to be different from that with larger numbers of repeats (Fig. 10). This predicted structural difference affected the relative conformation of the RNA region containing the predicted ribosome-binding site (Fig. 10), which could consequently affect the efficiency of binding of ribosomes and therefore mRNA susceptibility to endonucleolytic activity (16). It is also possible that a change in the secondary structure unrelated to potential masking of the ribosome-binding site is responsible for the difference in mRNA stability. For example, we cannot exclude the possibility that another M. catarrhalis gene product might interact with the 5′-UTR of the uspA2 mRNA and thereby affect the stability of this message.
The identification in the present study of naturally occurring variants of M. catarrhalis strain O12E with different numbers of AGAT repeats (i.e., 18, 19, and 23) in their uspA2 genes provides ample evidence that this repeat motif can undergo spontaneous loss and gain of these tetranucleotide units. However, in screening many different strains of M. catarrhalis with UspA2-reactive antibodies, we and others (39) have only rarely encountered M. catarrhalis strains that either do not express UspA2 or express a reduced amount of this protein. Even after many passages in vitro over a 15-year period, M. catarrhalis strain O35E still expresses its UspA2 protein, suggesting that there may exist some selective advantage to UspA2 protein expression even outside of the nasopharynx.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI36344 to E.J.H.
We thank John Nelson, Anthony Campagnari, Steven Berk, Merja Helminen, and Frederick Henderson for providing the wild-type isolates of M. catarrhalis used in this study. We thank Thomas Rosche for the isolation and characterization of the spontaneous uspA2 variants of M. catarrhalis O12E and Wade Winkler and John (Trey) Fondon for very helpful discussions concerning the RNA secondary structure.
Footnotes
Published ahead of print on 8 September 2006.
REFERENCES
- 1.Aebi, C., L. D. Cope, J. L. Latimer, S. E. Thomas, C. A. Slaughter, G. H. McCracken, Jr., and E. J. Hansen. 1998. Mapping of a protective epitope of the CopB outer membrane protein of Moraxella catarrhalis. Infect. Immun. 66:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. R. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 74:1597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayliss, C. D., K. M. Dixon, and E. R. Moxon. 2004. Simple sequence repeats (microsatellites): mutational mechanisms and contributions to bacterial pathogenesis: a meeting review. FEMS Immunol. Med. Microbiol. 40:11-19. [DOI] [PubMed] [Google Scholar]
- 7.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayliss, C. D., W. A. Sweetman, and E. R. Moxon. 2005. Destabilization of tetranucleotide repeats in Haemophilus influenzae mutants lacking RnaseHI or the Klenow domain of PolI. Nucleic Acids Res. 33:400-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bootsma, H. J., H. G. Der Heide, P. S. van De, L. M. Schouls, and F. R. Mooi. 2000. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J. Infect. Dis. 181:1376-1387. [DOI] [PubMed] [Google Scholar]
- 10.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson, S. D., B. Stone, M. Beucher, J. Fu, and P. F. Sparling. 2000. Phase variation of the gonococcal siderophore receptor FetA. Mol. Microbiol. 36:585-593. [DOI] [PubMed] [Google Scholar]
- 12.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, D., V. Barniak, K. R. VanDerMeid, and J. C. McMichael. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun. 67:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, and G. H. McCracken, Jr. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cripps, A. W., D. C. Otczyk, and J. M. Kyd. 2005. Bacterial otitis media: a vaccine preventable disease? Vaccine 23:2304-2310. [DOI] [PubMed] [Google Scholar]
- 16.Deana, A., and J. G. Belasco. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19:2526-2533. [DOI] [PubMed] [Google Scholar]
- 17.Faden, H. 2001. The microbiologic and immunologic basis for recurrent otitis media in children. Eur. J. Pediatr. 160:407-413. [DOI] [PubMed] [Google Scholar]
- 18.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, R. C. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 19.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 20.Hays, J. P., C. van der Schee, A. Loogman, K. Eadie, C. Verduin, H. Faden, H. Verbrugh, and A. Van Belkum. 2003. Total genome polymorphism and low frequency of intra-genomic variation in the uspA1 and uspA2 genes of Moraxella catarrhalis in otitis prone and non-prone children up to 2 years of age: consequences for vaccine design? Vaccine 21:1118-1124. [DOI] [PubMed] [Google Scholar]
- 21.Helminen, M. E., I. Maciver, J. L. Latimer, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution, and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches: the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 24.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)beta Gal. Mol. Microbiol. 9:1275-1282. [DOI] [PubMed] [Google Scholar]
- 25.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 27.Jennings, M. P., D. W. Hood, I. R. Peak, M. Virji, and E. R. Moxon. 1995. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol. Microbiol. 18:729-740. [DOI] [PubMed] [Google Scholar]
- 28.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes. Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, L., V. S. Panangala, and K. Dybvig. 2002. Trinucleotide GAA repeats dictate pMGA gene expression in Mycoplasma gallisepticum by affecting spacing between flanking regions. J. Bacteriol. 184:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke, N. R., A. J. Howlett, J. Shao, and A. A. Campagnari. 2004. Expression of type IV pili by Moraxella catarrhalis is essential for natural competence and is affected by iron limitation. Infect. Immun. 72:6262-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, P., K. Makepeace, S. A. Hill, D. W. Hood, and E. R. Moxon. 2005. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA 102:3800-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, P., L. Sun, D. W. Hood, and E. R. Moxon. 2004. Involvement of genes of genome maintenance in the regulation of phase variation frequencies in Neisseria meningitidis. Microbiology 150:3001-3012. [DOI] [PubMed] [Google Scholar]
- 35.Martin, P., D. van, V., N. Mouchel, A. C. Jeffries, D. W. Hood, and E. R. Moxon. 2003. Experimentally revised repertoire of putative contingency loci in Neisseria meningitidis strain MC58: evidence for a novel mechanism of phase variation. Mol. Microbiol. 50:245-257. [DOI] [PubMed] [Google Scholar]
- 36.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 37.Meier, P. S., S. Freiburghaus, A. Martin, N. Heiniger, R. Troller, and C. Aebi. 2003. Mucosal immune response to specific outer membrane proteins of Moraxella catarrhalis in young children. Pediatr. Infect. Dis. J. 22:256-262. [DOI] [PubMed] [Google Scholar]
- 38.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 39.Meier, P. S., R. Troller, N. Heiniger, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2005. Moraxella catarrhalis strains with reduced expression of the UspA outer membrane proteins belong to a distinct subpopulation. Vaccine 23:2000-2008. [DOI] [PubMed] [Google Scholar]
- 40.Mollenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin d-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 185:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, G. L., T. D. Connell, D. S. Barritt, M. Koomey, and J. G. Cannon. 1989. Phase variation of gonococcal protein II: regulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell 56:539-547. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, T. F. 2000. Bacterial otitis media: pathogenetic considerations. Pediatr. Infect. Dis. J. 19:S9-S15. [DOI] [PubMed] [Google Scholar]
- 43.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Antigenic specificity of the mucosal antibody response to Moraxella catarrhalis in chronic obstructive pulmonary disease. Infect. Immun. 73:8161-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, T. F., A. L. Brauer, C. Aebi, and S. Sethi. 2005. Identification of surface antigens of Moraxella catarrhalis as targets of human serum antibody responses in chronic obstructive pulmonary disease. Infect. Immun. 73:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 47.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. W. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 50.Seib, K. L., I. R. A. Peak, and M. P. Jennings. 2002. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol. Med. Microbiol. 32:159-165. [DOI] [PubMed] [Google Scholar]
- 51.Stutzmann, M. P., N. Heiniger, R. Troller, and C. Aebi. 2003. Salivary antibodies directed against outer membrane proteins of Moraxella catarrhalis in healthy adults. Infect. Immun. 71:6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 53.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. Van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiser, J. N., J. M. Love, and E. R. Moxon. 1989. The molecular mechanism of phase variation of Haemophilus influenzae lipopolysaccharide. Cell 59:657-665. [DOI] [PubMed] [Google Scholar]
- 55.Weiser, J. N., D. J. Maskell, P. D. Butler, A. A. Lindberg, and E. R. Moxon. 1990. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J. Bacteriol. 172:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]