Abstract
In fulfilling its biosynthetic roles in nuclear replication and in several types of repair, DNA polymerase δ (pol δ) is assisted by replication protein A (RPA), the single-stranded DNA-binding protein complex, and by the processivity clamp proliferating cell nuclear antigen (PCNA). Here we report the effects of these accessory proteins on the fidelity of DNA synthesis in vitro by yeast pol δ. We show that when RPA and PCNA are included in reactions containing pol δ, rates for single base errors are similar to those generated by pol δ alone, indicating that pol δ itself is by far the prime determinant of fidelity for single base errors. However, the rate of deleting multiple nucleotides between directly repeated sequences is reduced by ∼10-fold in the presence of either RPA or PCNA, and by ≥90-fold when both proteins are present. We suggest that PCNA and RPA suppress large deletion errors by preventing the primer terminus at a repeat from fraying and/or from relocating and annealing to a downstream repeat. Strong suppression of deletions by PCNA and RPA suggests that they may contribute to the high replication fidelity needed to stably maintain eukaryotic genomes that contain abundant repetitive sequences.
INTRODUCTION
DNA polymerase δ (pol δ) has a major and essential role in eukaryotic nuclear DNA replication (1). Pol δ also performs DNA synthesis during homologous recombination and fills DNA gaps during mismatch repair, long patch base excision repair of damaged bases and nucleotide excision repair of bulky DNA lesions [reviewed in (2)]. Because all these transactions influence eukaryotic genome stability, it is of interest to understand the fidelity of DNA synthesis conducted by pol δ. Previous studies (3–5) have shown that pol δ is a highly accurate enzyme whose fidelity derives from high nucleotide selectivity at the polymerase active site and from proofreading by its intrinsic 3′ exonuclease activity.
In performing its roles in replication and repair, pol δ is assisted by accessory proteins. The three-subunit replication protein A (RPA) complex (6) binds single-stranded DNA and coordinates the exchange of pol δ and other proteins at template–primer termini (7). In addition, the processivity of pol δ is enhanced by proliferating cell nuclear antigen (PCNA) (8), the sliding clamp that is loaded onto template–primers by the five-subunit RFC complex (9). There are several reasons to consider whether RPA or PCNA modulate the fidelity of DNA synthesis by pol δ. Genetic studies have identified mutations in the genes encoding the large subunit of RPA (10,11), RFC subunits (12,13) and PCNA (14,15) that elevate mutation rates. Among several possible explanations for these mutator effects, one is that they may result from reduced DNA synthesis fidelity by pol δ during replication (15), repair or recombination. Single-stranded DNA-binding proteins have been shown previously to affect the fidelity of other DNA polymerases [e.g. see (16,17) and references therein]. Proteins that enhance polymerase processivity promote the extension of mismatches (18–20), which could reduce base substitution fidelity by preventing partitioning of mismatches to the active sites of proofreading exonucleases (21). On the other hand, several studies [reviewed in (22)] have shown that the processivity of DNA polymerases correlates with their insertion/deletion (indel) fidelity in mononucleotide repeat sequences, such that proteins that increase processivity may improve indel fidelity.
Here we test these ideas by examining the fidelity of DNA synthesis by three-subunit yeast DNA polymerase δ alone and its fidelity in the presence of RPA alone, PCNA (plus its loader RFC) and all three accessory protein complexes. To evaluate the effects of these accessory proteins on both nucleotide selectivity and proofreading, we compare error rates of proofreading-proficient (wild-type) pol δ to those observed with two different proofreading-deficient derivatives. To obtain a comprehensive view of the effects of accessory proteins on pol δ fidelity, we use an assay (23) that scores a variety of base substitution and indel errors. The results indicate that fidelity for errors involving single base pairs is largely determined by pol δ itself. However, the accessory proteins strongly modulate the ability of pol δ to delete large numbers of nucleotides between directly repeated sequences. The results are discussed in relation to earlier studies (cited below) on the effects of accessory proteins, and in light of models for how accessory proteins may modulate fidelity.
MATERIALS AND METHODS
Materials
Yeast RPA, PCNA and RFC were purified from Escherichia coli overproducing strains as described elsewhere (24,25). All materials for the fidelity assay were from previously described sources (23,26).
Overexpression and purification of Pol δ
Plasmid pBL335 (bluescript, 2 µM ori, TRP1, M13 ori, GAL1-10 GST-POL3) contains the Schistosoma japanicum glutathione S-transferase gene (GST) fused to the N-terminus of the POL3 gene in vector pRS424-GALGSTPKA. The GST tag is separated from the POL3 gene by a recognition sequence for the human rhinoviral protease (LEVLFQ/GP), followed by a recognition site for the catalytic subunit of cAMP dependent protein kinase (27). After cleavage by the protease the N-terminal sequence of the Pol3 polypeptide is altered from MSEKRSLPM to GPEFRRASVGSM. Plasmid pBL341 (bluescript, 2 µM ori, URA3, M13 ori, GAL1-10, POL31, POL32) has the POL31 and POL32 genes placed divergently under control of the bidirectional GAL1-10 promoter into vector pRS426-GAL. Plasmids and sequences are available upon request from P.M.B. Plasmid pBL335-DV (as pBL335, but pol3-5DV=pol3D520V) was made by gap repair. pBL335 was cut with BglII and NdeI, releasing that portion of the POL3 gene containing the intended mutation, and the isolated large fragment was transformed into strain YH712 (MATα ade5-1 his7-2 leu2-3,112::lys2D5'-LEU2 lys2::InsHS-D trp1-289 ura3-52 pep4::KanMX pol3-5DV) (28). After plasmid recovery, the mutation was confirmed by sequencing.
Overexpression was in Saccharomyces cerevisiae strain BJ2168 (MATa, ura3-52, trp1-289, leu2-3, 112, prb1-1122, prc1-407, pep4-3) transformed with pBL341 and with either pBL335 or pBL335-DV. Growth and induction was as described elsewhere, and so was the preparation of cell lysates by blending with dry ice (29). Cells (60 g of packed cells resuspended in 20 ml of water) frozen previously in liquid nitrogen in the form of popcorn were blended with 40 ml of buffer 3A (buffer A, 30 mM HEPES–NaOH, pH 7.8, 10% glycerol, 2 mM EDTA, 1 mM EGTA, 0.02% Nonidet P-40, 2 mM DTT, 10 mM sodium bisulfite, 10 µM pepstatin A and 10 µM leupeptin). All further operations were carried out at 0–4°C. After thawing of the lysate, 1 mM phenylmethanesulfonyl fluoride (from a 100 mM stock in isopropanol) and 150 mM ammonium sulfate (from a 4 M stock) were stirred in, followed by 0.45% polymin P (from a 10% stock at pH 7.3). After stirring for 15 min, the lysate was cleared at 40 000 g for 40 min, and the supernatant precipitated with 0.28 g/ml of solid ammonium sulfate. The precipitate was collected at 40 000 g for 45 min, and redissolved in ∼75–125 ml of buffer A until the conductivity equals that of A250 (subscript denotes NaCl concentration). Batch binding to 2 ml of glutathione–Sepharose 4B beads (GE Healthcare), equilibrated previously in buffer A250, was accomplished by gentle rotation in the cold room for 2 h. The beads were collected at 1000 r.p.m. in a swinging bucket rotor, batch washed, by resuspension and spinning, with 3× 30 ml of buffer A250, transferred to a 10 ml column, and washed at 2 ml/min with 100 ml of A250. Bound chaperones, particularly Ssa1, were removed by a 30 ml wash with A250 containing 1 mM ATP and 5 mM Mg-acetate. After another 10 ml wash with A150 to remove residual nucleotide and decrease salt, the beads were resuspended in 2 ml A150 containing 20 mM glutathione (pH adjusted to 8.0). The capped column was incubated on ice for 10 min, and the eluant collected. This procedure was repeated four times. Most of the protein eluted in fractions 1–4. These fractions (∼0.5–1 mg protein) were incubated overnight at 4°C with 30 U of PreScission protease (GE Healthcare), and then directly loaded onto a 1 ml MonoS column as described elsewhere (30). Concentrated pure enzyme eluted at ∼350–400 mM NaCl.
Gap-filling DNA synthesis reactions and product analysis
Reactions (25 µl) contained 20 mM Tris–HCl (pH 7.7), 8 mM MgAc2, 75 mM NaCl, 0.5 mM ATP, 100 µM of each dNTP, 1 mM DTT, 100 mg/ml BSA and 40 fmol (1.6 nM) gapped M13mp2 DNA. When included, the amounts of the accessory proteins used were 500 fmol PCNA, 200 fmol RFC and 10 pmol RPA, an amount more than sufficient to coat the single-stranded DNA within the gap. Polymerization reactions were performed at 30°C. The amount of pol δ and reaction times were: pol δ alone, 2.0 pmol for naked DNA or 1.5 pmol with RPA-coated DNA, both 30 min; pol δ plus accessory proteins, 150 fmol pol δ for 5 min for naked DNA and for 2 min with RPA-coated DNA. These quantities of pol δ and incubation times were chosen such that, when DNA products were analyzed by agarose gel electrophoresis as described elsewhere (23), all reactions filled the 407 nt gap without obvious strand displacement [data not shown, but for typical result see Figure 3 in Ref. (23)]. Note that synthesis by pol δ alone is only moderately processive, such that complete gap filling likely involves multiple cycles of binding and dissociation. Importantly, reactions containing 150 fmol of pol δ alone failed to fill the gap, indicating that synthesis catalyzed by pol δ was indeed stimulated by the presence of the accessory proteins.
DNA products of gap-filling reactions were introduced into E.coli cells and plated as described elsewhere (23) to score blue M13 plaques (correct synthesis) and light blue and colorless plaques (containing errors). The types of errors were determined by sequencing the lacZ α-complementation gene in single-stranded DNA isolated from independent mutant M13 plaques, allowing calculation of error rates as described previously (5). The statistical significance of differences in error rates with and without accessory proteins was calculated using the Fisher's exact test (31). Because several such comparisons were made, the multiple comparisons method of Benjamini and Hochberg (32) was used to control the false discovery rate to no more than 0.05.
RESULTS AND DISCUSSION
Fidelity measurements and calculation of error rates
Pol δ fidelity with and without accessory proteins was determined for synthesis to fill a single-stranded gap in a circular duplex M13mp2 DNA substrate. This gap contains the lacZ α-complementation template sequence (Figure 1) that when copied correctly results in a blue M13 plaque phenotype. Polymerization errors are detected as light blue and colorless plaques. A total of 12 gap-filling reactions were conducted and the products analyzed for lacZ mutant frequencies. Four reactions contained wild-type pol δ (Table 1, Experiment 1), either alone, with RPA, with PCNA and RFC or with all three accessory protein complexes. The lacZ mutant frequencies for all reactions with wild-type pol δ were several-fold lower than for parallel reactions performed with exonuclease-deficient pol3-5DV pol δ (Experiment 2) or exonuclease-deficient pol3-01 pol δ (Experiment 3), reflecting the contribution of proofreading to the overall fidelity of the wild-type enzyme (see more below). DNA samples were prepared from independent lacZ mutants collected from each of the 12 reactions, and were sequenced to identify the types (Table 1) and locations (Figure 1) of sequence changes responsible for reduced plaque colors. As observed previously for three-subunit yeast pol δ alone (5), four main classes of sequence changes were observed: single base substitutions, single nucleotide deletions, single nucleotide insertions and deletions of larger numbers of nucleotides between direct repeat sequences (Table 1). The mutant frequency and sequence specificity information (Table1) was then used to calculate average rates (errors per detectable nucleotide polymerized) for single base errors (Table 2) and mutant frequencies for large deletions (Table 3).
Figure 1.
Spectra of point mutations made by exonuclease-deficient Pol δ ± accessory proteins. Errors generated by exonuclease-deficient Pol δ in the M13mp2 lacZ target sequence are shown, with base substitution mutations above the sequence, and single nucleotide deletions (Δ) and additions below. The lacZ (+)-strand is shown, with the transcriptional start site designated as position 1, and the first 53 codons displayed as triplets. The gapped region ends at position 191. (A) Pol δ alone. (B) Pol δ plus RPA, RFC and PCNA.
Table 1.
Mutant frequencies and sequences changes generated by Pol δ with and without accessory proteins
| Pol δ | Pol δ + RPA | Pol δ + RFC + PCNA | Pol δ + RPA + RFC + PCNA | |
|---|---|---|---|---|
| Experiment 1: wild-type Pol δ | ||||
| Total plaques | 32 997 | 43 891 | 43 955 | 37 302 |
| lacZ mutants | 125 | 44 | 79 | 55 |
| Mutant frequency | 0.0038 | 0.0010 | 0.0018 | 0.0015 |
| Base substitutions | 31 | 32 | 50 | 42 |
| One base deletions | 7 | 2 | 21 | 7 |
| One base additions | 0 | 0 | 0 | 0 |
| Larger deletions | 81 | 4 | 6 | 0 |
| Other changes | 4 | 4 | 3 | 5 |
| Experiment 2: exonuclease-deficient pol3-5DV Pol δ | ||||
| Total plaques | 8017 | 11 823 | 13 067 | 6303 |
| lacZ mutants | 101 | 84 | 111 | 46 |
| Mutant frequency | 0.0126 | 0.0071 | 0.0085 | 0.0073 |
| Base substitutions | 40 | 52 | 61 | 29 |
| One base deletions | 17 | 14 | 35 | 15 |
| One base additions | 10 | 15 | 3 | 3 |
| Larger deletions | 29 | 2 | 6 | 0 |
| Other changes | 5 | 1 | 5 | 0 |
| Experiment 3: exonuclease-deficient pol3-01 Pol δ | ||||
| Total plaques | 12 379 | 14 509 | 11 977 | 17 004 |
| lacZ mutants | 106 | 96 | 125 | 102 |
| Mutant frequency | 0.0086 | 0.0066 | 0.0104 | 0.0060 |
| Base substitutions | 55 | 63 | 71 | 72 |
| One base deletions | 15 | 17 | 43 | 27 |
| One base additions | 6 | 10 | 0 | 5 |
| Larger deletions | 26 | 4 | 3 | 0 |
| Other changes | 2 | 0 | 11 | 4 |
Larger deletions include loss of multiple nucleotides between direct repeats of two or more bases. Other changes include di- and tri-nucleotide deletions, additions of multiple nucleotides, tandem base substitution/deletions, complex deletions and (rare, spontaneous) deletions (40). The total number of specific mutations reported is sometimes different from the number of lacZ mutants due to the presence of two errors in single mutant (increasing the number of errors reported) and/or the occasional inability to obtain sequence data from a mutant (decreasing the number of errors reported).
Table 2.
Pol δ single base error rates ± accessory proteins
| Mutation type | Replication proteins | Error rate (× 10−5) | |
|---|---|---|---|
| WT | Exo− | ||
| Base substitutions | Pol δ only | 1.3 (0.45) | 6.3 |
| + RPA | 1.0 | 5.9 | |
| + RFC + PCNA | 1.5 | 7.0 | |
| + RPA + RFC + PCNA | 1.5 | 5.5 | |
| One nucleotide deletions | Pol δ only | 0.18 (0.033) | 1.3 |
| + RPA | 0.039 | 1.0 | |
| + RFC + PCNA | 0.39 | 2.6 | |
| + RPA + RFC + PCNA | 0.16 | 1.4 | |
| One nucleotide additions | Pol δ only | ≤0.026 (0.012) | 0.66 |
| + RPA | ≤0.020 | 0.80 | |
| + RFC + PCNA | ≤0.019 | 0.10 | |
| + RPA + RFC + PCNA | ≤0.022 | 0.27 | |
Background values for uncopied DNA are in parentheses.
Table 3.
Frequency of large deletions between direct repeats
| WT Pol δ | Exo− Pol δ | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Repeat + inverted repeat | Repeat only | Repeat + inverted repeat | Repeat only | ||||||
| Number | Frequencya | Number | Frequency | Number | Frequency | Number | Frequency | ||
| Pol δ only | 80 | 240 | 1 | 3.0 | 50 | 250 | 5 | 25 | |
| + RPA | 3 | 6.8 | 1 | 2.3 | 6 | 23 | 0 | ≤3.8 | |
| + PCNA + RFC | 4 | 9.1 | 2 | 4.6 | 4 | 16 | 5 | 20 | |
| + RPA + PCNA + RFC | 0 | ≤2.7 | 0 | ≤2.7 | 0 | ≤4.3 | 0 | ≤4.3 | |
These large deletions involve loss of multiple nucleotides between direct repeats of two or more bases. Those listed as involving an inverted repeat could theoretically be stabilized by a ‘stem’ of at least two correct base pairs, in rare cases allowing for one mismatch, e.g. a T–G mismatch.
aWhile error rates for single base events in Table 2 are expressed as ‘errors per nucleotide incorporated’, the values for deletion of multiple bases are expressed here simply as mutant frequencies (× 10−5).
In order to separate the effects of the accessory factors on polymerase insertion fidelity from those on proofreading efficiency, we will first discuss our results with the exonuclease-deficient pol δ, followed by those with the wild-type enzyme. Because similar results were obtained for each of the two exonuclease-deficient polymerases, those data were combined.
Effects on selectivity against base–base mismatches
The most common errors generated by pol δ were single base substitutions. The calculated average single base substitution error rate of exonuclease-deficient pol δ alone is 6.3 × 10−5 (Table 2), confirming an earlier report that yeast pol δ has high base substitution fidelity (5). Interestingly, similar base substitution error rates were obtained for pol δ reactions that contained RPA, PCNA plus RFC or all three accessory proteins. Detecting errors in this type of fidelity assay requires both nucleotide misinsertion and extension of the resulting mismatch. Thus, the similarities in average base substitution error rates suggest that the accessory proteins do not strongly influence the nucleotide selectivity of the pol δ active site or strongly alter discrimination for extension of matched versus mismatched primer termini.
The base substitution values in Table 2 are average error rates for numerous different mismatches in a variety of sequence contexts. From these average rates, it cannot be excluded that the accessory proteins have modest effects on nucleotide selectivity and/or mismatch extension for specific mismatches and/or in specific sequence contexts. For example, a previous kinetic study demonstrated that PCNA decreases the selectivity of exonuclease-deficient yeast pol δ for misinsertions opposite a specific template guanine by 2- to 4-fold (4). Any such effects here would be small compared to the >10 000-fold contribution to selectivity conferred by exonuclease-deficient pol δ alone. This conclusion is reinforced by studies of several other replicative polymerases (17–20,33–38) indicating that accessory proteins have only small effects that sometimes enhance and sometimes reduce discrimination against base substitution errors. Thus we conclude that pol δ itself is by far the primary determinant of selectivity against base substitution errors resulting from copying an undamaged DNA template.
Effects on proofreading of base–base mismatches
An estimate of the contribution of proofreading to base substitution fidelity in the absence and presence of the accessory proteins can be obtained by comparing error rates for exonuclease-deficient pol δ to those for wild-type pol δ (Table 2). We believe that these are minimal estimates because the base substitutions recovered from reactions by wild-type pol δ are thought to largely reflect background noise in the assay due to very low levels of spontaneous substitutions and cryptic damage (e.g. cytosine deamination) in the gapped DNA substrate. With this caveat in mind, the ratios of base substitution error rates are similar for all four reactions. This is interesting in light of studies indicating that proteins that increase processivity promote extension of base–base mismatches by T7 DNA polymerase (18), T4 DNA polymerase (19) and human DNA polymerase γ (20). The present results suggest that the accessory proteins neither strongly enhance nor strongly suppress proofreading by pol δ when copying an undamaged DNA template, as expected in order to maintain both high processivity and high fidelity during yeast chromosomal DNA replication.
Effects on single nucleotide deletion and addition errors
The second most common single base error made by pol δ is deletion of 1 nt (Table 2). The rates at which these errors are generated by pol δ in the absence or presence of RFC/PCNA and/or RPA either do not differ in a statistically significant manner, or they differ in a statistically significant manner but by <2-fold. Such effects are small relative to the 100 000-fold discrimination imposed by the polymerase alone, indicating that PCNA, RFC and RPA contribute very little to preventing single base deletion errors by pol δ. However, in all four reactions, single base deletion error rates are higher for exonuclease-deficient pol δ than for wild-type pol δ, suggesting that misaligned intermediates are proofread with or without the accessory proteins present.
Exonuclease-deficient pol δ also generates single base additions at a readily detectable overall average rate of 0.66 × 10−5 (Table 2). This rate is not appreciably influenced by RPA (0.80 × 10−5), but is reduced ∼7-fold by PCNA plus RFC (P-value for difference = 0.0007). Wild-type pol δ alone (error rate ≥0.026 × 10−5) is at least 25-fold more accurate for one base additions than exonuclease-deficient pol δ. This difference is larger than observed for single base deletions, supporting our earlier interpretation (5) that pol δ proofreads addition intermediates more efficiently than deletion intermediates. Just as for deletions, single base addition error rates in all four reactions are higher for exonuclease-deficient pol δ than for wild-type pol δ, suggesting that addition intermediates are proofread with or without accessory proteins present.
Effects on deletions between direct repeats
The effects of accessory proteins on single base error rates are modest in comparison with the dominant role of the polymerase itself in discriminating against single base errors. However, the situation is different for deletions of larger numbers of nucleotides located between direct repeat sequences. We found previously that pol δ alone is particularly prone to generating these types of deletions (5), and does so at frequencies that are similar for the wild-type and exonuclease-deficient enzymes, indicating that the misaligned intermediates are not efficiently proofread. The present study confirms those observations (Table 3, line 1). More importantly, the results show that the ability of pol δ to generate large deletions between direct repeats is strongly suppressed by RPA alone (line 2) or by PCNA plus RFC (line 3). Moreover, no deletions between direct repeats were generated by wild-type pol δ in the presence of all three accessory proteins, representing a ≥90-fold increase in fidelity for this class of errors.
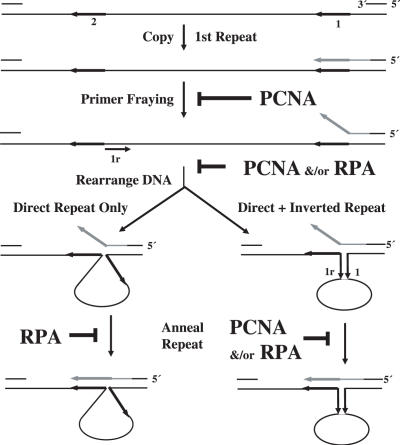
How RPA and PCNA may suppress formation of large deletions by pol δ can be considered in light of a model for deletions between direct repeats [Figure 2 (39)]. In this model, after the first repeat sequence encountered by the polymerase is copied, the primer frays and then relocates to the second repeat sequence. Hybridization would provide a duplex primer–template for continued polymerization, but in a misaligned intermediate that contains a loop of unpaired, template strand bases that are eventually deleted. In certain sequence environments, when an inverted repeat (designated 1r in Figure 2) is present at the deletion junction, the misaligned intermediate may be stabilized through formation of a stem at the base of the single-stranded loop (Figure 2, lower right). Within the context of this model, PCNA may suppress deletion formation by preventing fraying of the primer terminus at the first repeat, and/or by preventing relocation of the primer to the downstream repeat. RPA may suppress deletion formation by coating the single-stranded DNA to reduce the probability of DNA rearrangement and/or prevent the frayed primer terminus from annealing to the downstream repeat. The observation that all three accessory proteins together yield lower large deletion frequencies than either RPA alone or PCNA plus RFC (Table 3) is consistent with the idea that RPA and PCNA affect large deletion fidelity at least partly by different mechanisms. This possibility is further supported by the observation that RPA suppresses deletions generated by exonuclease-deficient pol δ that can be modeled by both pathways depicted in Figure 2 to about the same extent (∼10-fold, Table 3). However, while PCNA also strongly suppresses deletions between repeats that are potentially stabilized by a stem containing two or more correct base pairs (Table 3, compare 250 × 10−5 to 16 × 10−5, Figure 2, right), it has little effect on deletions that lack an obvious inverted repeat (Figure 2, left). By virtue of their ability to prevent wild-type pol δ from generating these types of replication errors, PCNA and RPA may have important roles in protecting eukaryotic genomes against the biological consequences of large deletions, and perhaps also in modulating the stability of large tracts of repetitive sequences whose instabilities are associated with hereditary degenerative diseases.
Figure 2.
Model for accessory protein suppression of large deletions between direct repeats. For descriptions see text.
Acknowledgments
We thank Stephanie Nick McElhinny and Zachary Pursell for critically reading this manuscript and for thoughtful suggestions. We thank Dinh Nguyen for expert assistance in DNA sequence analysis of lacZ mutants. This work was funded by the Division of Intramural Research, NIEHS, NIH, DHHS, and by extramural support from the National Institutes of Health (GM32431 to P.B.). Funding to pay the Open Access publication charges for this article was provided by the Division of Intramural Research of the NIEHS, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Garg P., Burgers P. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K., Kunkel T.A. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 3.Thomas D.C., Roberts J.D., Sabatino R.D., Myers T.W., Tan C.K., Downey K.M., So A.G., Bambara R.A., Kunkel T.A. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto K., Shimizu K., Nakashima N., Sugino A. Fidelity of DNA polymerase delta holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42:14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 5.Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A. Saccharomyces cerevisiae DNA polymerase {delta}: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 6.Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 7.Yuzhakov A., Kelman Z., Hurwitz J., O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan C.K., Castillo C., So A.G., Downey K.M. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. J. Biol. Chem. 1986;261:12310–12316. [PubMed] [Google Scholar]
- 9.Tsurimoto T., Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc. Natl Acad. Sci. USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Kolodner R.D. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nature Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Putnam C.D., Kane M.F., Zhang W., Edelmann L., Russell R., Carrion D.V., Chin L., Kucherlapati R., Kolodner R.D., et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nature Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 12.McAlear M.A., Tuffo K.M., Holm C. The large subunit of replication factor C (Rfc1p/Cdc44p) is required for DNA replication and DNA repair in Saccharomyces cerevisiae. Genetics. 1996;142:65–78. doi: 10.1093/genetics/142.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amin N.S., Tuffo K.M., Holm C. Dominant mutations in three different subunits of replication factor C suppress replication defects in yeast PCNA mutants. Genetics. 1999;153:1617–1628. doi: 10.1093/genetics/153.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eissenberg J.C., Ayyagari R., Gomes X.V., Burgers P.M. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol. Cell. Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C., Merrill B.J., Lau P.J., Holm C., Kolodner R.D. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 1999;19:7801–7815. doi: 10.1128/mcb.19.11.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts J.D., Kunkel T.A. Fidelity of DNA replication. In: De Pamphilis M., editor. DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. Cold Spring Harbor: Cold Spring Harbor Laboratories; 1996. pp. 217–247. [Google Scholar]
- 17.Bebenek A., Carver G.T., Kadyrov F.A., Kissling G.E., Drake J.W. Processivity clamp gp45 and ssDNA-binding-protein gp32 modulate the fidelity of bacteriophage RB69 DNA polymerase in a sequence-specific manner, sometimes enhancing and sometimes compromising accuracy. Genetics. 2005;169:1815–1824. doi: 10.1534/genetics.104.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel T.A., Patel S.S., Johnson K.A. Error-prone replication of repeated DNA sequences by T7 DNA polymerase in the absence of its processivity subunit. Proc. Natl Acad. Sci. USA. 1994;91:6830–6834. doi: 10.1073/pnas.91.15.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroutil L.C., Frey M.W., Kaboord B.F., Kunkel T.A., Benkovic S.J. Effect of accessory proteins on T4 DNA polymerase replication fidelity. J. Mol. Biol. 1998;278:135–146. doi: 10.1006/jmbi.1998.1676. [DOI] [PubMed] [Google Scholar]
- 20.Longley M.J., Nguyen D., Kunkel T.A., Copeland W.C. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 22.Bebenek K., Kunkel T.A. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb. Symp. Quant. Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- 23.Bebenek K., Kunkel T.A. Analyzing the fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 24.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 25.Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 26.Shcherbakova P.V., Pavlov Y.I., Chilkova O., Rogozin I.B., Johansson E., Kunkel T.A. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 27.Cordingley M.G., Callahan P.L., Sardana V.V., Garsky V.M., Colonno R.J. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J. Biol. Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 28.Jin Y.H., Obert R., Burgers P.M., Kunkel T.A., Resnick M.A., Gordenin D.A. The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl Acad. Sci. USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgers P.M. Overexpression of multisubunit replication factors in yeast. Methods. 1999;18:349–355. doi: 10.1006/meth.1999.0796. [DOI] [PubMed] [Google Scholar]
- 30.Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 31.Fleiss J.L. Statistical Methods For Rates and Proportions. NY: John Wiley and Sons; 1973. [Google Scholar]
- 32.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 33.Mo J.Y., Schaaper R.M. Fidelity and error specificity of the alpha catalytic subunit of Escherichia coli DNA polymerase III. J. Biol. Chem. 1996;271:18947–18953. doi: 10.1074/jbc.271.31.18947. [DOI] [PubMed] [Google Scholar]
- 34.Mozzherin D.J., McConnell M., Jasko M.V., Krayevsky A.A., Tan C.K., Downey K.M., Fisher P.A. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase delta. J. Biol. Chem. 1996;271:31711–31717. doi: 10.1074/jbc.271.49.31711. [DOI] [PubMed] [Google Scholar]
- 35.Bloom L.B., Chen X., Fygenson D.K., Turner J., O'Donnell M., Goodman M.F. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of beta, gamma complex processivity proteins and epsilon proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- 36.Pham P.T., Olson M.W., McHenry C.S., Schaaper R.M. The base substitution and frameshift fidelity of Escherichia coli DNA polymerase III holoenzyme in vitro. J. Biol. Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- 37.Johnson A.A., Johnson K.A. Fidelity of nucleotide incorporation by human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:38090–38096. doi: 10.1074/jbc.M106045200. [DOI] [PubMed] [Google Scholar]
- 38.Bebenek A., Carver G.T., Dressman H.K., Kadyrov F.A., Haseman J.K., Petrov V., Konigsberg W.H., Karam J.D., Drake J.W. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162:1003–1018. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunkel T.A., Soni A. Mutagenesis by transient misalignment. J. Biol. Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 40.Kunkel T.A. The mutational specificity of DNA polymerase-ß during in vitro DNA synthesis: Production of frameshift, base substitution, and deletion mutations. J. Biol. Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]