Abstract
A synapomorphy is a phylogenetic character that provides evidence of shared descent. Ideally a synapomorphy is ubiquitous within the clade of related organisms and nonexistent outside the clade, implying that it arose after divergence from other extant species and before the last common ancestor of the clade. With the recent proliferation of genetic sequence data, molecular synapomorphies have assumed great importance, yet there is no convenient means to search for them over entire genomes. We have developed a new program called Conserv, which can rapidly assemble orthologous sequences and rank them by various metrics, such as degree of conservation or divergence from out-group orthologs. We have used Conserv to conduct a largescale search for molecular synapomorphies for bacterial clades. The search discovered sequences unique to clades, such as Actinobacteria, Firmicutes and γ-Proteobacteria, and shed light on several open questions, such as whether Symbiobacterium thermophilum belongs with Actinobacteria or Firmicutes. We conclude that Conserv can quickly marshall evidence relevant to evolutionary questions that would be much harder to assemble with other tools.
INTRODUCTION
Biology textbooks typically use phenotypic characters to describe clades, e.g. milk and hair for mammals. Not only do these synapomorphies aid in phylogenetic inference, but they also record key innovations in the history of life, as exemplified by such famous clades as Amniota and Eutheria (placental mammals). A number of papers have used molecular synapomorphies to weigh in on phylogenetic debates. A convincing molecular synapomorphy can often resolve a phylogeny that cannot be unambiguously determined by more continuously varying characters (1). Moreover, characteristic proteins (2) or regulatory sequences (3)—i.e. sequences restricted to hypothesized clades—may represent landmark evolutionary events, such as the divergence of metazoans (2) or the origin of the bilaterian body plan (4). Characteristic proteins are currently found, with some effort, by local alignment searching each gene in each genome of interest against all other genomes (2), or by the use of predefined ortholog collections, such as the COGs database (5–7). More subtle synapomorphies, such as insertions or deletions are found serendipitously by researchers studying specific genes (8,9), or more systematically by manual examination of multiple alignments (10–13). As more sequence becomes available, there is a need and opportunity to further automate the search for molecular synapomorphies.
In this paper, we report on a synapomorphy search tool, called Conserv, that takes as input two sets of genomes: those for the putative clade, or in-group, and those for an out-group. The types of molecular synapomorphies we consider are as follows: (i) signature genes ubiquitous and unique to the clade, (ii) large insertions or deletions (indels) present only within the clade and (iii) sequence motifs well conserved within the clade but quite different outside the clade. Type (i) is generally the rarest and type (iii) the most common, so these types are roughly ordered from strongest to weakest phylogenetic evidence. Each type includes both strong and weak examples, however, and sequence alone cannot distinguish orthologs with novel function or structure, so we somewhat arbitrarily set the boundary between types (i) and (iii) using BLAST score thresholds that varied with the probe sequence length. We do not consider other types of synapomorphies, such as gene fusions (14,15) or changes in gene order (16,17). No matter the type, synapomorphies possess the same allure. They represent rare—possibly even unique—events that can potentially overcome the ‘ratio problem’ illustrated in Figure 1: clock-like evolutionary models are inherently limited in their ability to resolve a short internal branch followed by long branches to leaves (18). Sequence characteristics with an extremely large number of character states, however, as is the case with signature genes or long indels, can theoretically still retain information (19).
Figure 1.
A short, ancient, internal branch such as the one marked * is not easy to resolve with clock-like sequence evolution, but a rare event such as a signature gene may resolve the clade. The depicted phylogenetic tree is a consensus of those given in three recent studies (36,37,42). The enigmatic organisms and the placements considered here are shown with wiggly branches, with the solid wiggly lines indicating the placements best supported by our synapomorphy search.
Conceptually, we can think of Conserv as performing three steps. First it performs an all-against-all local alignment search, probing each protein-coding gene in each genome against every other genome. Second, it processes the resulting sets of hits to find the orthologous families most conserved over the in-group genomes. Third, it ranks the families by ‘synaptitude’, which measures in-group pairwise similarity scores relative to in-to-out similarity scores. All three types of molecular synapomorphies, (i–iii) above, show up near the top of the ranked list. Evaluation of the significance of the discovered synapomorphies remains a manual (and poorly understood) process, but this step can be facilitated by existing bioinformatics tools, such as local alignment search and multiple alignment programs (20,21).
We emphasize that Conserv is a search tool, and not a complete tool for inferring a phylogenetic tree or network. Conserv's candidate synapomorphies can be used in conjunction with methods, such as parsimony (5,22) or Dollo parsimony (22) to reconstruct a tree; however, because conserved genes and indels that occur in only a single putative clade are rare, Conserv is unlikely to find enough synapomorphies to reconstruct a large tree. In this case, the program can provide confirmatory evidence and help evaluate trees suggested by other means. Notice that phylogeny by synapomorphies and parsimony is quite distinct from phylogeny by gene content (24,25), as a single gene with the right distribution pattern may decide a branch, whereas such a gene counts no more heavily than one with a scattered distribution in gene-content methods. Finally, it is worth reiterating that Conserv is a relatively simple tool, optimized for speed. Because Conserv considers only highly conserved proteins and obvious homology (at least 25% identity), and performs only pairwise alignments, it has no need for sophisticated sequence modeling techniques, such as hidden Markov models (HMMs) (25,26).
Conserv is currently most useful for prokaryotic genomes. When run on a putative eukaryotic clade, e.g. Ecdysozoa, Conserv will return voluminous results that are hard to evaluate, due to the sparse and uneven taxon sampling of eukaryotes. Thus, to demonstrate the utility of Conserv, we ran the program over bacterial genomes in GenBank (27) for about 30 choices of in-groups and out-groups, both putative clades and other sets of genomes. In this test, we discovered possible synapomorphies for higher-level clades uniting Planctomycetes with Chlamydiales and Chloroflexi with Cyanobacteria, as in Figure 1. We also discovered strong evidence for placing Symbiobacterium in Firmicutes, and weaker evidence for placing the endosymbionts Buchnera and Wigglesworthia in Enterobacteria. The placement of Symbiobacterium with Firmicutes contradicts the current GenBank taxonomy, which places it in Actinobacteria, yet the discovered synapomorphies seem incontrovertible. We discovered signature genes for a number of clades, including Actinobacteria and Firmicutes. We also used Conserv to explore surprising similarities between two groups that do not together form a clade: ɛ-Proteobacteria and Spirochaetes. Finally, we used the tool to answer an intriguing peripheral question: what is the most conserved protein?
MATERIALS AND METHODS
Given a set of n genomes—n = 30 is typical—and a sequence ‘window’ length k, Conserv returns a list of families of orthologous protein sequences of the desired length (k amino acid residues). The number of families in the list can be specified by the user with a typical value being 1000, but if Conserv determines that there are not 1000 sufficiently conserved proteins in the set of genomes (e.g. if the set of organisms includes reduced genomes or both eubacteria and archaea), then Conserv will return a shorter list. The list is initially ranked from ‘most conserved’ to ‘least conserved’ over the first m genomes in the set, where the user supplies m ≤ n and thus defines the in-group and out-group. In order to find synapomorphies we further process the list as follows: (i) we rerank the list by synaptitude; (ii) from each ortholog family, we remove each sequence of very low pairwise similarity with all the in-group sequences and (iii) we use MUSCLE (21) to compute multiple alignments. Step (ii) is necessary because, in the case of a genome without a close ortholog, Conserv will return a distant homolog or a completely unrelated sequence that could corrupt the multiple alignment. To remove sequences, we use a log odds threshold that corresponds to about 20–25% identity, Conserv reports the gene distribution, with presence (*) or absence (−) in each genome, as shown in the example in the Supplementary Data. It also reports a P-value, the likelihood that this distribution, or a better one, would arise by chance in a random model in which each genome independently chooses the gene with probability equal to its frequency over all n genomes. For example, if n = 10 and a gene appears in three of the five in-group members and just one of the five out-group members, then the chance that it appears in any given genome is (4/10) = 0.4 and the chance that it does not appear is 0.6. The P-value is the probability that at least three in-group members, and at most one out-group member, contain the gene, or
If the gene distribution indicates a signature gene, or if manual inspection of the multiple alignment shows an indel synapomorphy, we add another step: (iv) a BLAST search [PSI-BLAST (28) with default settings] against all prokaryotic genomes in GenBank, possibly followed by another multiple alignment, to check whether we have indeed discovered a synapomorphy of types (i) or (ii).
Rather than find its own orthologs, Conserv could, at least in principle, process the orthologs found by BLAST or an ortholog assembler (29), or use predefined ortholog databases such as COGs (6,7,30) as in (5). There are two reasons why Conserv does its own ortholog assembly. The first reason is simply speed. Conserv is much faster than using BLAST; e.g. Conserv can find the 1000 most conserved proteins for a window size of 90 in 12 bacterial genomes in 15 min on a SunFire V440, compared to 7 h for BLAST-searching each gene in each of the 12 genomes against each other genome. The second reason is quality of results. Reciprocal best BLAST searching may not find the best representative from a set of paralogs (31). Similarly, the COGs database draws a line at a certain level of homology, and does not try to separate paralogs even in cases where they can be distinguished fairly reliably, e.g. annotating both ClpA and ClpB genes as 0542. (Another, larger, drawback of a predefined database is that it overlooks rare proteins, such as the one annotated simply ‘putative protein’ that appears in only Aquifex and Thermotoga). Conserv attempts to find the best representatives from sets of paralogs by simultaneously minimizing all pairwise distances within each ortholog family. Conserv rarely mixes up paralogs that are separately annotated in GenBank. With k = 60, it sometimes confused peptide chain release factors RF-1 and RF-2, but with k ≥ 90 it correctly separates these two highly homologous proteins.
In order to explain how Conserv works, we begin by defining a conservation score for a set of orthologous protein sequences. Let and be orthologous proteins from two different organisms. Let denote the subsequence of k amino acid residues starting at residue i in protein . We obtain the alignment score S by aligning and using the standard dynamic programming algorithm (32), scored with the BLOSUM-50 matrix with a gap penalty of 8. Higher scores represent more similar sequences, but the conservation score is more easily understood if we describe it in terms of a distance function, such as . Then for a highly conserved family of proteins , each protein will have a sequence of length k, such that all the distances will be small. In other words, the set of sequences will form a tight cluster, whose maximum pairwise distance (diameter) is small. Thus a reasonable definition of conservation score over all n organisms is the diameter:
where a smaller score means greater conservation. Finding the minimizing choices of is a computationally hard problem; below we describe a heuristic algorithm that works well in practice.
In practice, however, the definition of conservation score just given is too brittle. If an organism t has no homolog of a protein, then the algorithm described below will select a nonhomologous protein (the one with the smallest distance) and will be large for all r. Thus instead of using the maximum pairwise distance, we use a more robust rank statistic. We sort the set of distances , and select the quartile distance Q so that 1/4 of the distances are larger than Q and 3/4 are smaller.
If m < n, that is, if we are considering both an in-group and and out-group, we look for minimizing choices of over all n organisms, and record these choices (which define the ortholog family), but we record the conservation score as .
In its internal computations, Conserv uses a data structure we call a conservation table. This is a table with a row for each family of orthologous proteins and a column for each organism. If there are 1000 rows in the conservation table, Conserv's goal is to fill in the table so that the rows represent the 1000 most conserved proteins over the m genomes of the in-group. We now explain how Conserv arrives at the set of proteins for each row and how it finds approximately minimizing choices of . As mentioned above, computing the minimizing choices of is likely to be an inherently hard problem, like most multiple alignment problems, so Conserv uses a heuristic algorithm that works well in practice. The heuristic finds a family of orthologous proteins P (i.e. builds a row of the conservation table) at the same time that it computes an approximation of the row's conservation score.
The heuristic first picks two genomes O1 and O2 at random among the first m genomes. For each protein in O1 it finds the most similar protein (more precisely, the protein with a most similar k-long segment) in O2. This gives two columns (any many rows) of the conservation table. Then for each row (nascent orthologous family) it finds the best k-long homologs in the n − 2 other organisms. This results in a conservation table with a candidate set of rows and a tentative score for each row. Then the whole process is repeated for another random pair of organisms, and the two conservation tables are merged. When merging, if the two tables each have a row with at least two proteins in common, only the row with the better score is retained.
To construct the first two columns of the table, we use the standard technique [used in both BLAST and FASTA (33,34)] of building a table of 4mers (contiguous four-residue sequences), only considering potential homologs if they have a 4mer in common. However, the completed row (orthologous family) is not required to have a single 4mer common to all members of that row. Along with the ‘mer table’ of 4mers, we use another temporary data structure called a ‘match table’, a list of protein pairs sharing common subsequences at least four long. The algorithm in more detail:
Build a table of 4mers. We start by picking two organisms, O1 and O2 from the in-group of organisms under study. We place each 4mer from each protein in the two organisms into a mer table of size 204 = 160 000. Each entry in the mer table will have a list of the mer's occurrences in O1 and a list of its occurrences in O2.
Build a match table. For each 4mer in the mer table, look at each pair of occurrences, one from O1 and the other from O2. This defines a potential match, that is, an alignment matching the common 4mers. Check to see if the alignment extends to a neighborhood of the 4mer. If it does, put it into a match table. If the number of elements in the match table is too small or large, change the definition of ‘extends to a neighborhood’ (that is, reduce or enlarge the neighborhood and the number of allowed misses) until the match table is a reasonable size.
Compute scores for match table entries. For each item in the match table (a pair of proteins with a common 4mer), compute the score of each k-long alignment that extends from the common 4mer. The score of the item is the maximum of these scores.
Fill in the first two columns of conservation table. Iterate through the proteins in O1. For each protein P in O1 find all entries in the match table that contain a subsequence from P. Select the entry (P,Q) with the highest score. Then place P in column 1, and Q in column 2.
Build a new mer table. Each entry in the just constructed first two columns of the conservation table is a protein together with an k-long sequence with a good alignment. Take each 4mer that occurs in such a sequence and place it into a new mer table. Do this for each protein in columns 1 and 2.
Fill in the rest of the columns of the conservation table. Pick a new column (organism O) and iterate through its proteins. For each protein P in O, examine all 4mers from P. For each one that matches a 4mer in the mer table, constructed in the previous step, compute the score between P and all proteins with that mer from the mer table. Suppose the highest score obtained is s and is with protein Q (from either column 1 or 2). If the entry for the row containing protein Q and column k is empty, place P into that slot. If the entry is already occupied, replace it if the score s is higher than the score of the current entry. Finally we sort the resulting table and only retain the rows of sufficiently high degree of conservation.
Iterate over other starting pairs O1, O2. Repeat steps (i–vi) for a different choice of starting organisms O1 and O2. Merge the resulting conservation tables. Whenever there is a row from each table sharing at least two proteins in common, use the row with the highest score.
We performed several tests to validate the robustness of Conserv's algorithm for finding and ranking orthologous sequences by conservation level. In one test, we varied the number of iterations in step (vii), i.e. the number of new choices of starting pair, and determined that most subsequences and ranks stabilize after 2–3 iterations, and all of the top 50 proteins stabilized after at most 10 iterations. In another test, we ran Conserv (with m = n ) on 10 different sets of bacteria. Each set was chosen randomly, but with the constraint that it include one bacterium from each of 19 different phyla represented in GenBank at the time. For each of the top 50 proteins, we computed its relative range, i.e. the difference between the highest and lowest ranks it achieved on the 10 sets, divided by its median rank. For over half of the proteins, the relative range was less than 0.5, showing that the list of most conserved proteins is fairly stable over varying taxon samples.
Finally we mention a further practical speedup used in Conserv. Since we are examining only highly similar sequences, we can speed up the computation of the similarity score of two sequences by only filling in the dynamic programming table in a narrow band around the diagonal. The band width sets an upper limit on the length of indels considered. We typically set the band width to be ⌊k/60⌋ + 10 for a window size of k; we empirically found that larger widths did not turn up any more synapomorphies. Our use of mer and match tables means that overall running time is not very dependent on genome sizes; it depends more heavily on the number of organisms. It bears repeating that because Conserv is intended to find only the most conserved proteins, at least 25% identity over each pair of genomes, it does not need sensitive techniques for detecting remote homology, such as profiles and HMMs. Thus Conserv is specialized in the opposite direction from most recent bioinformatics tools. Rather than being slower but more sophisticated than BLAST (33), it is faster but simpler.
RESULTS
Conserv takes as input a set of n genomes and a sequence ‘window’ length k. The program returns ortholog families, with one amino acid sequence of length k from each genome. Conserv uses genome annotations to identify protein-coding genes, but does not use the annotations to identify orthologs. The use of a fixed window length distinguishes Conserv from other alignment search tools, such as BLAST (28,33). The fixed window enables Conserv to make a fair comparison of degree of conservation for genes of different lengths, and more importantly, a search over a range of window lengths can uncover synapomorphies of all three types. For example, a search with k = 30 will find short motifs and small indels that would make negligible contributions to whole-gene similarity scores. At k = 100 the search can uncover homologous domains, and at k = 300 it can find whole-gene synapomorphies.
Molecular synapomorphies
We first report on using Conserv to find molecular synapomorphies for accepted and hypothesized bacterial clades. For this experiment, m genomes form a putative clade, the in-group, and n − m genomes putatively lie outside that clade and form the out-group. Conserv finds sequences that are highly conserved in the in-group and quite different in the out-group. More specifically, it ranks ortholog families by a metric we call synaptitude. We calculate the synaptitude of an ortholog family by first computing all pairwise similarity scores from the putative clade, sorting them, and taking the median. Then we compute all m(n − m) pairwise distances between orthologous in-group and out-group sequences, and take the three-quartile score (the -th largest, where larger scores are less similar). Synaptitude is the median score minus the three-quartile score. Thus for synaptitude to be positive, a typical inside-inside score must exceed a good inside-to-outside score. All three types of molecular synapomorphies, i.e. (i) signature genes, (ii) conserved indels and (iii) characteristic sequence motifs, should have high synaptitude for some choice of k. We typically ran Conserv for k = 30,60,80,100,200,300. Conserv reports the discovered sequences in three groups: those for which the minimum inside similarity score exceeds the maximum in-out score; those for which the median inside score exceeds the maximum in-out score; and finally those for which the median inside score exceeds the three-quartile in-out score, i.e. the remaining proteins of positive synaptitude. (See the Supplementary Data for example results.)
A large number of high-synaptitude ortholog families offers some evidence that the hypothesized clade is real, but manual curation is important to further evaluate the evidence and determine synapomorphy type. As usual in phylogenetic inference, taxon sampling is an important issue (35). A gene could appear to be a strong synapomorphy for a clade relative to a given out-group, but then fail as a synapomorphy relative to a slightly larger out-group. Rather than use an enormous out-group and slow down the search, we found it best to use a more modest out-group (say 20 diverse genomes) and then doublecheck sequences of high synaptitude by BLAST searching against all available out-group genomes. The most common situation handled by manual curation is a gene with an otherwise-good synapomorphy missing from one or more of the clade organisms. Such a lack does not necessarily disqualify a sequence as a synapomorphy. For example, a gene common to all Proteobacteria except Buchnera would still be a good synapomorphy, because Buchnera is a highly reduced genome, safely interior to the clade. On the other hand, a gene missing from all ɛ-Proteobacteria would not be a good synapomorphy for Proteobacteria, as ɛ-Proteobacteria are the first class of Proteobacteria to branch off (36,37).
We ran Conserv for various choices of in-groups and out-groups, including all eubacterial phyla with more than one sequenced genome in GenBank, and a number of hypothesized higher-level clades such as Chlamydiales/Planctomycetes. We also ran a number of control experiments with in-groups formed by randomly swapping organisms between an accepted or putative clade and a diverse out-group, in order to test the background level of chance synapomorphies. Control experiments with more than two randomly chosen organisms in the in-group gave no synapomorphies, but two-organism in-groups gave signature genes 15 out of 200 times. Bacteroides seems especially prone to forming pairs, sharing unique genes with Treponema (a hexokinase containing ividaggtnfrsclvrf), Pirellula (a glycosyltransferase containing sgwksdilavngfdermqyggqdre) and Helicobacter (lwqidmihirkgsrydgyfekvaeri).
Table 1 gives signature genes for several accepted clades, both phyla and classes. As mentioned above, the sequences returned by Conserv were doublechecked by BLAST searching against all the prokaryotic genomes in GenBank. Except as noted, the proteins in Table 1 occur only within the named clades—the short probe sequences given do not return any hits outside the clades—and hence are strong molecular synapomorphies. Such signature genes can arise through gene duplication and subsequent rapid evolution, or through horizontal transfer into the ancestral genome from some genome, possibly a bacteriophage genome (38), now lost or unsequenced. In either case, the signature gene supports the validity of the putative clade.
Table 1.
This table gives some synapomorphies of type (1), that is, ‘signature genes’ ubiquitous within the clade and nonexistent outside, for various clades. The sequence given is from one organism in the clade, but is sufficient for BLAST searching. None of the signature genes for Actinobacteria have orthologs in Symbiobacterium, but both of the signature genes for Firimicutes do. This finding is evidence that Symbiobacterium belongs with Firmicutes.
| Clade | Proteins (Annotations) | Sequence | Occurs outside clade? |
|---|---|---|---|
| Actinobacteria (all except Symbiobacterium) | Proteasome element | QRADHVWEGVSSATTRSRPII | |
| Transcriptional regulator | NRLANFDDANLRRSARAAVAA | ||
| Nuclease of recB family | LAEHIETLGDGYSLVRREYPT | Homolog in Archaea? | |
| Phosphoserine aminotransferase | AETTPYVTDPAKRSLVVGTID | ||
| (From a total of 15) | WhiB-type transcript'l regulator | EQALCAQTDPEAFFPEKGGST | |
| Transcriptional regulator | GYRVPIACQVAGITYRQLDYW | ||
| WhiB-type transcript'l regulator | HEAVCRDEDPELFFPVGNSGP | ||
| Firmicutes | yvcL | LRFEDIRIVRDMRNSVNRLVN | In Symbiobact'm, also distant homologs |
| (From a total of 2) | purine operon repressor | RKDNKVTEGPTVSINYVSGSS | In Symbiobact'm, distant homologs in Archaea |
| Cyanobacteria | PsaA & PsaB chlorophyll apoprotein | SRLIPDKANLGFRFPCDGPGR | Homolog in Chlorobium |
| (From a total of 63) | PsbD & PsbB Photosystem II proteins | QSQGWFDALDDWLKKDRFVFI | |
| PsbA Photosystem II proteins | DEWLYNGGPYQLVVFHFLIGI | ||
| Probable glycosyl transferase | LYGWEDLELGVRLKNLGLQLI | ||
| Hypothetical protein | QALIKLGNNYGKFDREGKKLF | ||
| γ-Proteobacteria | ygdH, predicted Rossman fold | IGMTEPSIIAAEPPNPLVNEL | |
| (From a total of 9) | glycerol-3-phosphate acyltransferase | GHEIVYVPCHRSHMDYLLLSY | |
| ɛ-Proteobacteria | hdhA dehydrogenase | GFAPFMRLKPRGLNNIYTATV | Many distant homologs |
| (From a total of 45) | OorD oxidoreductase | PQNVPVWVDETRCKACDVCVS | Homolog of Ferredoxin |
| Hypothetical protein | KVENIVFDYNGRNPERFYHKA | ||
| Hypothetical protein | AMDLMQYQDIHRQKIERVINV |
Not all accepted clades have ‘perfect’ signature genes, ubiquitous and conserved within the clade and nonexistent outside the clade. For Enterobacteria the closest such gene is one containing pwyhvvmedddgqpvhtylaeaql, which is missing from Buchnera within the clade and has a distant homolog in Idiomarina outside the clade. The rarity of signature genes contrasts with the relatively large number of clade-specific ORFan genes (85 for Enterobacteria) found by Daubin and Ochman (38). The difference is that we require a gene to be nearly ubiquitous within the clade, whereas Daubin and Ochman require only that the gene be confined within the clade and not within one of a small number of subsumed clades, in particular, a gene specific to Enterobacteria need not appear in the reduced genomes Buchnera and Wigglesworthia.
For Proteobacteria, a large, diverse, and relatively well-sequenced phylum, we found only a few signature genes, none of them perfect: e.g. ubiquinol-cytochrome c reductase containing psakakaagapvevniskvepgq, phospholipid and glycerol acyltransferase containing wvlkkellripffgwglamtspiaidr, and succinyl-diaminopimelate desuccinylase containing iallitsdeegpavdgt, all of which are missing from a small number of proteobacteria. We did, however, find a few indel and many motif synapomorphies for Proteobacteria. We found no signature genes or clean indels, and only a few small motifs, for the putative clade marked by * in Figure 1.
The signature genes in Table 1 place Symbiobacterium with Firmicutes, because horizontal transfer would be unlikely to produce such a consistent pattern. BLAST searches of key proteins (39) and maximum-likelihood phylogeny using representative proteins (37) (see the Supplementary Data) also argue for this placement, but GC-content and 16S rRNA initially placed Symbiobacterium with Actinobacteria (40).
Figures 2–4 give example synapomorphies of types (ii) and (iii), i.e. indels and motifs. Figure 2 gives several indels in one gene and illustrates the current importance of manual evaluation of evidence. The qvr insertion in columns 9–11 and the lr insertion in columns 114–115 separate Chlamydiales and Pirellula [the only sequenced representative of the enigmatic phylum Planctomycetes (41)], an hypothesized clade (37), from all the other genomes. Especially important is the separation from Leptospira, a spirochaete, as Spirochaetes is the most probable sister taxon for Chlamydiales (36,37,42). (Other sequenced members of Spirochaetes do not have close homologs of this gene.) The qvr insertion, however, is somewhat compromised by the poor conservation of the flanking sequences and the existence of a similarly positioned, but quite different, insertion in Chlorobi/Bacteroides and Geobacter. The evaluation of flanking sequences could be automated, but this step would not be part of Conserv, as it would necessarily take place after multiple alignment. Incidentally, the close similarity of the sequences for Chlorobi/Bacteroides and Geobacter in Figure 2 must be a case of horizontal transfer, as most genes clearly place Geobacter with δ-Proteobacteria (represented here by Desulfovibrio). Examples such as this one caution against using single-gene synapomorphies, no matter how compelling, for prokaryotic phylogenetic inference.
Figure 2.
This MUSCLE (21) alignment of a 125-aa subsequence of succinate dehydrogenase, flavoprotein subunit (COG1053), provides evidence that Pirellula and Chlamydiales, the first five rows, form a clade. The first five rows share overall sequence similarity, along with the qvr insertion in columns 9–11 and the lr insertion in columns 114–115. The alignment also uncovers an apparent horizontal transfer from Chlorobi/Bacteroides to Geobacter, evidenced by great sequence similarity and the insertion at columns 114–119. The top and bottom rows give the consensus sequence of Chlamydiales and Pirellula. A dot indicates agreement with a consensus residue, and a blank indicates agreement with a consensus gap. The columns with indels are marked by *.
Because Pirellula shares several signature genes with Bacteroides, we also searched for synapomorphies supporting a Pirellula/Bacteroides/Chlorobi clade. The best synapomorphy found was glucosamine-6-phosphate isomerase containing the sequence revqphqiyaagdlsphgthrtcld, present in Pirellula, Porphyromonas and Bacteroides, but missing from Chlorobium. Due to the tendency of Bacteroides to form pairs, we prefer to place Pirellula with Chlamydiales.
Dehalococcoides ethenogenes (43) is currently the only fully sequenced representative of the enigmatic phylum Chloroflexi. The position of this phylum has implications for the evolutionary history of photosynthesis (11,44–46), specifically whether reaction center RC-1 (in Cyanobacteria, Chlorobium and Heliobacteria (within Firmicutes)), preceded RC-2 (in Cyanobacteria, Chloroflexi and Proteobacteria), or vice versa, or whether both evolved together. Using maximum-likelihood phylogeny and a draft genome of Chloroflexus aurantiacus from the DOE Joint Genome Institute, we previously placed (37) Chloroflexi next to Cyanobacteria as the first two phyla to diverge from an hypothesized clade containing Chloroflexi, Cyanobacteria, Deinococcus/Thermus and Actinobacteria. The same phylogeny method using the complete Dehalococcoides genome places Chloroflexi near Fusobacterium on a path to Firmicutes, but with very poor bootstrap support. (See the Supplementary Data).
We turned Conserv to the problem of placing Chloroflexi. We found no synapomorphies supporting the hypothesized clade containing Chloroflexi, Cyanobacteria, Deinococcus and Actinobacteria. Although there are a number of genes (e.g. polyribonucleotide nucleotidyltransferase) that are well-conserved over this in-group, some out-group members (typically members of Firmicutes) also have sequences similar to those of the in-group. We also found no synapomorphies supporting the hypothesized clade of Chloroflexi, Fusobacterium and Firmicutes. We did, however, find a few synapomorphies to support a Cyanobacteria/Chloroflexi clade: a motif sequence (sidfglcvfcglcvescpt in Dehalococcoides) in NADH dehydrogenase subunit I, and two characteristic indels in IMP dehydrogenase, shown in Figure 3. An in-group of Dehalococcoides, Deinococcus and Thermus gave no synapomorphies. An in-group of Dehalococcoides, Chlorobium, Bacteroides and Pirellula, i.e. a putative clade containing both green sulfur and green nonsulfur bacteria, also gave no synapomorphies, even though such a clade appears in gene trees of photosynthesis genes (46). Both searches, however, found several genes that appear in only the in-group genomes and a few out-group members, again generally members of Firmicutes, such as Clostridium and Listeria. Listeria's sequence in Figure 3 shows a related anomaly: it is sufficiently similar to the other sequences that the multiple alignment is probably correct, and not so different from the sequences of the other Firmicutes (Lactococcus and Streptococcus) as to constitute a clear case of horizontal transfer, yet it includes the insertion at columns 60–65 characteristic of the hypothesized Cyanobacteria/Chloroflexi clade. PSI-BLAST searching reveals that this insertion—not so well-conserved—also occurs in two members of Bacteroidetes (Bacillus fragilis and Porphyromonas gingivalis) and a spirochaete (Treponema denticola). Possible explanations for this pattern include repeated horizontal transfers and/or a tendency for insertion in this sequence.
Figure 3.
This MUSCLE alignment of a 129-aa subsequence of IMP dehydrogenase (COG0516) provides evidence that Chloroflexi (represented by Dehalococcoides) and Cyanobacteria, the first eight rows, form a clade. The first eight rows share overall sequence similarity, along with an insertion in columns 60–65 and a deletion in columns 111–120. The top and bottom rows give the consensus sequence of the first eight rows. In this figure, | indicates a column with complete conservation over all rows, and : indicates nearly complete conservation.
Both our placement of Chloroflexi made with synapomorphies, on the path to Cyanobacteria, and our weakly supported maximum-likelihood placement of Chloroflexi on a path to Firmicutes, are consistent with an evolutionary model (44,46) in which RC-2 (bacteriochlorophyll) precedes RC-1 (chlorophyll). Our finding that a number of genes appear only in Dehalococcoides, Chlorobium and its relatives, and members of Firmicutes close to Heliobacterium, suggests ancient horizontal transfer into Firmicutes as a plausible explanation for the appearance of RC-1 in this early-diverging phylum.
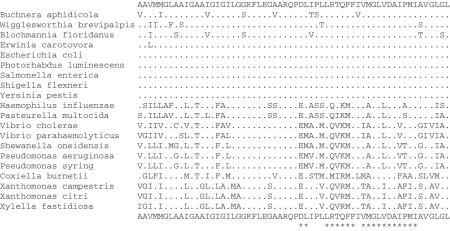
Figure 4 studies the position of the endosymbionts Buchnera and Wigglesworthia with respect to other γ-Proteobacteria, showing the strongest synapomorphy—a motif synapomorphy—for the inclusion of Buchnera and Wigglesworthia with Enterobacteria, as in (47,48) and the current GenBank taxonomy. An almost equally good motif synapomorphy occurs in cold shock protein CspC. An alternative taxonomy (36,37) that places Buchnera/Wigglesworthia outside an Enterobacteria/Pasteurellales clade had less persuasive motif synapomorphies. Neither placement yielded signature gene synapomorphies, but the alternative placement had four genes rather than two with median in-in scores exceeding maximum in-out scores. In this case, even if the alternative placement had found signature genes, we might favor the motif synapomorphies, because Buchnera and Wigglesworthia are both reduced genomes that could easily have lost signature genes characteristic of the Enterobacteria/Pasteurellales clade. Due to long branches and reduced genomes, the placement of Buchnera and Wigglesworthia is not easily made with either distance- or sequence-based phylogenetic methods (36,37), and even the fact that these two organisms form a clade is not obvious (47).
Figure 4.
This MUSCLE alignment of a highly conserved 60-aa sequence in ATP synthase, subunit C, gives a motif synapomorphy arguing for the inclusion of the endosymbionts Buchnera and Wigglesworthia with Enterobacteria. Notice the complete conservation in Enterobacteria of the columns marked *. The first nine organisms are Enterobacteria, the next two (Haemophilus and Pasteurella) are Pasteurellales and the last nine are other γ-Proteobacteria, roughly in order of distance from Enterobacteria.
We also looked at the position of Aquifex: does it belong with Thermotoga near the base of the bacterial tree (36,37,42) or did it diverge later, close to Proteobacteria (13)? Along with many motif synapomorphies, we found two signature-gene synapomorphies for the hypothesized Aquifex/Thermotoga clade, a protein containing the well-conserved sequence qkrawneyrmllipgfevtnn and another containing khgirditatgvkaititkre. Are these proteins the smoking guns proving that Aquifex and Thermotoga diverged together, or are they more recent horizontal transfers from one to the other? Although we found no signature-gene synapomorphies when we tried alternative placements for Aquifex, we consider this question still open.
We are not restricted to giving Conserv putative clades. For example, we can give it a set of organisms sharing some biochemical capability, in order to find genes related to that capability. We have used Conserv in this way to study photosynthetic bacteria and endoparasites/endosymbionts (Mycoplasma, Buchnera, Wigglesworthia). Such computational experiments have been performed previously using all-against-all BLAST; for example, Raymond et al. (49) find genes related to photosynthesis in this way. Conserv is faster than all-against-all BLAST, and also ranks its output by conservation level or synaptitude so that a user can decide where to cut. ‘Phylogenetic profiles’ (50) cluster predetermined genes by which genomes contain them, thereby discovering possible functional relationships. Conserv enables a reverse search: find sequences (not necessarily full genes) with predetermined phylogenetic profiles.
Here we report on using Conserv to study a false clade not obviously related by a shared capability. The ɛ-Proteobacteria are generally accepted as true Proteobacteria, yet molecular phylogeny sometimes places them with Spirochaetes (37,42). Table 2 gives the annotated proteins with top synaptitude for this (presumably) false clade. The out-group for this experiment included several members of Chlamydiales, the likely sister clade of Spirochaetes (36,42). Evidently, the flagellar proteins of ɛ-Proteobacteria resemble those of Spirochaetes, although none of the proteins are signature-gene synapomorphies as in Table 1. It is conceivable that a block of interacting genes were horizontally transferred to an organism ancestral to one of these taxa.
Table 2.
By giving Conserv a false clade (ɛ-Proteobacteria and Spirochaetes), we can study what two taxa have in common. This list of indel and motif synapomorphies tells a consistent ‘tail’.
| Annotation | Gene | COG |
|---|---|---|
| flagellar hook protein | FlgE | 1749 |
| KH/HDIG domain protein | – | 1418 |
| flagellar motor switch protein | fliM | 1868 |
| histidyl-tRNA synthetase | hisS | 0124 |
| cell division protein | FtsK | 1674 |
| flagellar motor rotation protein A | MotA | 1291 |
| flagellar motor switch protein | FliG-2 | 1536 |
| flagellar motor switch protein | FliN | 1886 |
| flagellar synthesis regulator? | ylxH-1, FleN | 0455 |
| flagellar basal-body rod protein | FlgG | 4786 |
| K+ transport protein | ntpJ | 0168 |
| M23/M37 peptidase domain protein | – | 0739 |
| flagellar GTP-binding protein | flhF | 1419 |
| ribosomal protein S2 | rpsB | 0052 |
| valyl-tRNA synthetase | valS | 0525 |
| flagellar filament 41 kDa core protein | flaB | 1344 |
What's the most conserved protein?
Biologists frequently describe proteins as ‘very highly conserved’ or ‘one of the most highly conserved’ proteins. In this section, we use Conserv to make this quantitative, ranking proteins by degree of conservation over all n input genomes. We considered both eubacteria and archaea in this experiment.
To answer the question for eubacteria, we used 28 representative bacteria from 11 phyla; results vary slightly with other, equivalently diverse, taxon samples. Figure 5 and Table 3 summarize the output of Conserv over window sizes ranging from about 50 to 500 amino acids. In order to measure the conservation of an orthologous family, we computed all pairwise similarity scores (BLOSUM-50 with a gap penalty of 8) between orthologous sequences, and took the conservation score to be the score such that one-fourth of the pairs of orthologs score worse (less similarity) and three-fourths score better. The vertical axis represents score per residue and shows that normalized score depends irregularly on window size.
Figure 5.
The plot shows conservation level, measured by quartile similarity score, as a function of window size for the seven most conserved proteins over a sample of 28 diverse eubacteria, representing 11 phyla. The similarity score per residue generally decreases, but sometimes increases when a less-conserved stretch is flanked by two well-conserved subsequences. Because EF-Tu claims top rank at a variety of window lengths, it is the most natural candidate for the most conserved protein over all Bacteria.
Table 3.
The 20 most conserved eubacterial proteins by quartile similarity score for the diverse set of 28 bacteria from 11 phyla. Rank (Rk) is the protein's highest rank; Window gives a window size (or range) for which the protein achieves its highest rank; Score/Res (S/R) is the quartile score divided by the window size; Length (Len) is the number of amino acid residues for a representative bacterium; COG and Class (Cls) are from (7). Class J is translation, ribosomal structure and biogenesis; K is transcription; L is replication, recombination and repair; and O is post-translational modification, protein turnover, and chaperones.
| Rk | Window | S/R | Gene | Cls | COG | Len | Name |
|---|---|---|---|---|---|---|---|
| 1 | 60–90, 330–390 | 5.7 | tufB | JE | 0050 | 405 | elongation factor TU |
| 1 | 120 | 5.2 | hflB | O | 0465 | 618 | cell division protein FtsH |
| 1 | 150–300 | 4.7 | rpoC | K | 0086 | 1546 | DNA-dir'd RNA polymerase, β′ |
| 1 | 420–480 | 4.0 | uvrA | L | 0178 | 1016 | excinuclease ABC, subunit A |
| 1 | 510 | 3.8 | fusA | J | 0480 | 698 | elongation factor G |
| 2 | 60 | 5.8 | rpoB | K | 0085 | 1179 | DNA-dir'd RNA polymerase, β |
| 2 | 420 | 4.0 | groL | O | 0459 | 548 | Chaperonin GroEL (HSP60) |
| 2 | 450 | 3.8 | dnaK | O | 0443 | 628 | Hsp70 chaperone |
| 3 | 330 | 4.2 | atpD | C | 0055 | 478 | ATP synthase F1 β subunit |
| 4 | 60–120 | 5.1 | clpB | O | 0542 | 875 | ATP-dependent Clp protease |
| 4 | 150 | 4.9 | clpX | O | 1219 | 403 | ATP-dependent Clp protease |
| 4 | 180 | 4.7 | recA | L | 0468 | 363 | recombination protein |
| 4 | 510 | 3.7 | uvrB | L | 0556 | 730 | excinuclease ABC, subunit B |
| 5 | 60 | 5.5 | rpoD | K | 0568 | 360 | RNA polymerase σ-70 factor |
| 6 | 60–90 | 5.2 | rpsL | J | 0048 | 131 | ribosomal protein S12 |
| 6 | 510 | 3.5 | lepA | N | 0481 | 606 | GTP-binding elongation factor |
| 7 | 60 | 5.4 | aspS | J | 0173 | 577 | aspartyl-tRNA synthetase |
| 8 | 360 | 3.7 | gyrA | L | 0188 | 812 | DNA gyrase, subunit A |
| 9 | 450 | 3.4 | atpA | C | 0056 | 503 | ATP synthase F1 α subunit |
| 9 | 480–510 | 3.1 | infB | J | 0532 | 803 | translation initiation factor IF-2 |
There is a natural way to combine different window sizes and obtain an answer to the question of ‘What's the most conserved protein?’. For each window size k, with k = 60, 90, 120, … , 510, we ranked the proteins. Then for each protein, we found the k for which it had the highest rank and assigned this best rank to the protein. Five different proteins (EF-Tu, FtsH, etc.) achieve the top rank for some choice of k, but because EF-Tu is most conserved for both small and large windows, we considered it first among the proteins that achieve rank 1. Table 3 shows some interesting trends among the most conserved proteins. The only ribosomal protein to make the list is S12, which is physically central to the ribosome and is suspected to interact directly with EF-Tu (personal communication with Irina Gabashvili). There are four heat shock proteins (GroEL, DnaK, ClpB, ClpX), but only one metabolic protein (two subunits of ATP synthase F1). For a more systematic study of the connection between function and conservation, we use the COG functional categories (6,7). Of the top 20 proteins, 60% are from categories J, K and L, the information storage and processing categories. This pattern holds up for larger lists. Of the top 100 proteins, 56 turn out to be in categories L (DNA replication, recombination and repair) and J (translation, ribosomal structure and biogenesis). Along with functional category (51), conservation level has also been correlated with protein length (52), expression level (53), essential proteins (54–56) and number of interaction partners (53,57–59). By rapidly assembling and ranking orthologs by conservation, Conserv enables systematic study of the phenomenon of sequence conservation.
For archaea, we used 11 representative genomes. Four genes achieved top rank for some choice of k: Cdc48 (cell division control protein), AtpA and AtpB (H+-transporting ATP synthase, subunits A and B), and RpoB1 (DNA-directed RNA polymerase, subunit β′). We also ran Conserv to find what protein is most conserved over all prokaryotes (archaea and eubacteria). Now three metabolic proteins, CTP synthase, enolase and GMP synthase, which do not even make the top 20 for eubacteria, leap to the top of the list; this finding is further corroboration of the relative similarity of eubacterial and archaeal metabolic genes compared to transcriptional and translational genes (51). It seems likely that the genes for these most conserved proteins were horizontally transferred sometime after the last common ancestor of Archaea and Bacteria.
DISCUSSION
Molecular synapomorphies are potentially very valuable phylogenetic characters, because rare discontinuous events—a large insertion or deletion, or the ‘sudden’ appearance of a novel, highly conserved gene—are not easily erased by subsequent point mutations. Moreover, molecular synapomorphies are complementary to popular sequence-based methods, such as maximum likelihood, which do not ordinarily take into account non-ubiquitous characters, such as insertions and deletions (‘gap columns') and proteins unique to a clade. To date, however, molecular synapomorphies have been used on an ad hoc basis, with phylogenies inferred from a handful of manually discovered synapomorphies. Nevertheless, the prokaryotic phylogenies computed in the 1990s by Gupta et al. (10,11) from hand-picked indels in selected genes show broad agreement with recent phylogenies computed using complete genomes and the latest tools (36,37,42). We do not expect molecular synapomorphies to replace modern tree-building methods, but we can imagine hybrid methods akin to those devised for gene trees (60) and routine use of synapomorphies to rescore a small number of alternative trees. Until now there has been, to our knowledge, no effort or means to automatically gather ‘all’ synapomorphies bearing on a phylogenetic question. Hence we believe that Conserv, simple as it is, can play a role in phylogenetic inference, as well as in data mining for unexpected nuggets, such as the similarity of the flagellar proteins of ɛ-Proteobacteria and Spirochaetes.
The sensitivity and specificity of Conserv are hard to assess at this point, as we do not have a test set of agreed-upon synapomorphies or a validated mathematical model. Conserv appears to be fairly effective at finding signature-gene and motif synapomorphies, which are easy to recognize from pairwise alignments, but indel synapomorphies remain somewhat problematic. Conserv found most of the indels manually discovered by Gupta et al. (10–12) from multiple alignments, but missed some of the less obvious (and more arguable) one- and two-residue indels. The great difference in the numbers of signature genes found by our study and that of Daubin and Ochman reveals that Conserv's sensitivity could be improved by a more flexible definition of synaptitude that allows signature genes to be missing from organisms deemed safely interior to the in-group.
Future research should assess the strengths and weaknesses of signature genes, indels, and motifs as phylogenetic characters. The Listeria anomaly in Figure 3 highlights the fact that the evolutionary mechanism of insertion and deletion is not well understood. Until more is known about this process, the evidentiary strength of indels will be open to question, and such characters would be better used not as absolute evidence as in the work of Gupta et al. but simply as relative evidence to decide among competing hypotheses. Another type of discrete character—gene order—has been judged relatively weak for deep bacterial phylogeny (61), yet valuable for eukaryotic phylogeny based on mitochondrial genomes (16,17). Other basic questions also remain to be explored. Is our classification of synapomorphy types adequate, or would a more detailed and exhaustive classification be useful? Will the discovery of new molecular synapomorphies elucidate epochal events [e.g. colonization of land (36)] in the history of prokaryotic life?
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Palo Alto Research Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rokas A., Holland P.W.H. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 2.Müller W.E., Schröder H.C., Skorokhod A., Bünz C., Müller I.M., Grebenjuk V.A. Contribution of sponge genes to unravel the genome of the hypothetical ancestor of Metazoa (Urmetazoa) Gene. 2001;276:161–173. doi: 10.1016/s0378-1119(01)00669-2. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli A.E., McCoy A., Jimenez E., Salo E., Ruvkun G., Martindale M.Q., Baguna J. Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution? Evol. Dev. 2003;5:372–378. doi: 10.1046/j.1525-142x.2003.03044.x. [DOI] [PubMed] [Google Scholar]
- 4.Bruce A.E., Shankland M. Expression of the head gene Lox22-Otx in the leech Helobdella and the origin of the bilaterian body plan. Dev. Biol. 1998;201:101–112. doi: 10.1006/dbio.1998.8968. [DOI] [PubMed] [Google Scholar]
- 5.Mirkin B.G., Fenner T.I., Galperin M.Y., Koonin E.V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov R.L., Koonin E.V., Lipman D. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 7.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusche K., Burmester T. Diplopod hemocyanin sequence and the phylogenetic position of the myriapoda. Mol. Biol. Evol. 2001;18:1566–1573. doi: 10.1093/oxfordjournals.molbev.a003943. [DOI] [PubMed] [Google Scholar]
- 9.Culligan K.M., Meyer-Gauen G., Lyons-Weiler J., Hays J.G. Evolutionary origin, diversification and specialization of eukaryotic MutS homolog mismatch repair proteins. Nucleic Acids Res. 2000;28:463–471. doi: 10.1093/nar/28.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R.S. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among Archaeabacteria, Eubacteria, and Eukaryotes. Microbiol. Mol. Biol. Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R.S., Mukhtar T., Singh B. Evolutionary relationships among photosynthetic prokaryotes (Heliobacterium chlorum, Chloroflexus aurantiacus, cyanobacteria, Chlorobium tepidum, and proteobacteria): implications regarding the origin of photosynthesis. Mol. Microbiol. 1999;32:893–906. doi: 10.1046/j.1365-2958.1999.01417.x. [DOI] [PubMed] [Google Scholar]
- 12.Gupta R.S. The phylogeny and signature sequences characteristic of Fibrobacteres, Chlorobi, and Bacteroidetes. Crit. Rev. Microbiol. 2004;30:123–143. doi: 10.1080/10408410490435133. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths E., Gupta R.S. Signature sequences in diverse proteins provide evidence for the late divergence of the order Aquificales. Int. Microbiol. 2004;7:41–52. [PubMed] [Google Scholar]
- 14.Stechman A., Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- 15.Yanai I., Wolf Y.I., Koonin E.V. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 2002;3:research0024.1–0024.13. doi: 10.1186/gb-2002-3-5-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boore J.L., Collins T.M., Stanton D., Daehler L.L., Brown W.M. Deducing the patterns of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 18.Mossel E., Steel M. How much can evolved characters tell us about the tree that generated them? In: Gascuel O., editor. Mathematics of Evolution and Phylogeny. Oxford: Oxford University Press; 2005. pp. 384–412. [Google Scholar]
- 19.Steel M.A., Penny D. Maximum parsimony and the phylogenetic information in multistate characters. In: Albert V., editor. Parsimony, Phylogeny, and Genomics. Oxford: Oxford University Press; 2005. pp. 163–178. [Google Scholar]
- 20.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swofford D.L., Olsen G.J., Waddell P.J., Hillis D.M. Phylogenetic inference. In: Hillis D.M., Moritz C., Mable B.K., editors. Molecular Systematics. Massachusetts: Sinauer Associates Sunderland; 1996. pp. 407–514. [Google Scholar]
- 23.Snel B., Bork P., Huynen M.A. Genome phylogeny based on gene content. Nat. Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 24.Kunin V., Ahren D., Goldovsky L., Janssen P., Ouzonis C.A. Measuring genome conservation across taxa: divided strains and United Kingdoms. Nucleic Acids Res. 2005;33:616–621. doi: 10.1093/nar/gki181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin R., Eddy S., Krogh A., Mitchison G. Biological Sequence Analysis. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 26.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 27.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remm M., Strom C.E.V., Sonnhammer E.L.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 30.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koski L.B., Golding G.B. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 32.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S.G., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillis D.M., Pollock D.D., McGuire J.A., Zwickl D.J. Is sparse taxon sampling a problem for phylogenetic inference? Syst. Biol. 2003;52:124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battistuzzi F.U., Feijao A., Hedges S.B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bern M., Goldberg D. Automatic selection of representative proteins for bacterial phylogeny. BMC Evol. Biol. 2005;5:34. doi: 10.1186/1471-2148-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daubin V., Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda K., Yamashita A., Ishikawa J., Shimada M., Watsuji T.O., Morimura K., Ikeda H., Hattori M., Beppu T. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 2004;32:4937–4944. doi: 10.1093/nar/gkh830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno M., Shiratori H., Park M.J., Saitoh Y., Kumon Y., Yamashita N., Hirata A., Nishida H., Ueda K., Beppu T. Symbiobacterium thermophilum gen. nov., sp. nov., a symbiotic thermophile that depends on co-culture with a Bacillus strain for growth. Int. J. Syst. Evol. Microbiol. 2000;50:1829–1832. doi: 10.1099/00207713-50-5-1829. [DOI] [PubMed] [Google Scholar]
- 41.Chistoserdova L., Jenkins C., Kalyuzhnaya M.G., Marx C.J., Lapidus A., Vorholt J.A., Staley J.T., Lidstrom M.E. The enigmatic Planctomycetes may hold a key to the origin of methanogenesis and methylotrophy. Mol. Biol. Evol. 2004;21:1234–1241. doi: 10.1093/molbev/msh113. [DOI] [PubMed] [Google Scholar]
- 42.Wolf Y.I., Rogozin I.B., Grishin N.V., Tatusov R.L., Koonin E.V. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seshadri R., Adrian L., Fouts D.E., Eisen J.A., Phillippy A.M., Methe B.A., Ward N.L., Nelson W.C., Deboy R.T., Khouri H.M., et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- 44.Burke D.H., Hearst J.E., Sidow A. Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc. Natl Acad. Sci. USA. 1993;90:134–138. doi: 10.1073/pnas.90.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baymann F., Brugna M., Mühlenhoff U., Nitschke W. Daddy, where did (PS)I come from? Biochim. Biophys. Acta. 2001;1507:291–310. doi: 10.1016/s0005-2728(01)00209-2. [DOI] [PubMed] [Google Scholar]
- 46.Xiong J., Fischer W.M., Inoue K., Nakahara M., Bauer C.E. Molecular evidence for the early evolution of photosynthesis. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 47.Lerat E., Daubin V., Moran N.A. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-proteobacteria. PLoS Biol. 2003;1:1–9. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moran N.A., Mira A. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2001;2:research0054.1–0054.12. doi: 10.1186/gb-2001-2-12-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raymond J., Zhaxybayeva O., Gogarten J.P., Gerdes S.Y., Blankenship R.E. Whole-genome analysis of phytosynthetic prokaryotes. Science. 2002;298:1616–1620. doi: 10.1126/science.1075558. [DOI] [PubMed] [Google Scholar]
- 50.Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivera M.C., Jain R., Moore J.E., Lake J.A. Genomic evidence for two functionally distinct gene classes. Proc. Natl Acad. Sci. USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lipman D.J., Souvorov A., Koonin E.V., Panchenko A.R., Tatusova T.A. The relationship of protein conservation and sequence length. BMC Evol. Biol. 2002;2:20. doi: 10.1186/1471-2148-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraser H.B., Hirsh A.E. Evolutionary rate depends on number of protein–protein interactions independently of gene expression level. BMC Evol. Biol. 2004;4:13. doi: 10.1186/1471-2148-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirsh A.E., Fraser H.B. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- 55.Jordan I.K., Gorozin I.B., Wolf Y.I., Koonin E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pal C., Papp B., Hurst L.D. Rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. [DOI] [PubMed] [Google Scholar]
- 57.Bloom J.D., Adami C. Apparent dependence of protein evolutionary rate on number of interactions is linked to biases in protein–protein interactions data sets. BMC Evol. Biol. 2003;3:21. doi: 10.1186/1471-2148-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraser H.B., Wall D.P., Hirsh A.E. A simple dependence between protein evolution rate and the number of protein–protein interactions. BMC Evol. Biol. 2003;3:11. doi: 10.1186/1471-2148-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jordan I.K., Wolf Y.I., Koonin E.V. No simple dependence between protein evolution rate and the number of protein–protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol. Biol. 2003;3:1. doi: 10.1186/1471-2148-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durand D., Halldórsson B., Vernot B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. Proceedings of the Research in Computational Molecular Biology (RECOMB), LNBI 3500; Springer-Verlag: 2005. pp. 250–264. [DOI] [PubMed] [Google Scholar]
- 61.Mushegian A.R., Koonin E.V. Gene order is not conserved in bacterial evolution. Trends Genet. 1996;12:289–290. doi: 10.1016/0168-9525(96)20006-x. [DOI] [PubMed] [Google Scholar]