Abstract
DGCR8/Pasha is an essential cofactor for Drosha, a nuclear RNase III that cleaves the local hairpin structures embedded in long primary microRNA transcripts (pri-miRNAs) in eukaryotes. Although our knowledge of pri-miRNA processing has significantly advanced in recent years, the precise role of DGCR8 in this pathway remains unclear. In our present study, we dissect the domains in DGCR8 that contribute to the processing of pri-miRNAs and the subcellular localization of DGCR8. Drosha is stabilized through an interaction between its middle domain and the conserved C-terminal domain of DGCR8. Furthermore, DGCR8, but not Drosha, can directly and stably interact with pri-miRNAs, and the tandem dsRNA-binding domains (dsRBDs) in DGCR8 are responsible for this recognition. Moreover, the DGCR8 N-terminal region upstream of its dsRBDs is unnecessary for pri-miRNA processing but is critical for nuclear localization. Our study thus provides further insights into the mechanism of action of the Drosha–DGCR8 complex in pri-miRNA processing.
INTRODUCTION
MicroRNAs (miRNAs) constitute an abundant class of key regulatory molecules that control diverse cellular functions in eukaryotes, such as differentiation, development and antiviral defense (1,2). These molecules are single-stranded RNAs (ssRNAs) of ∼22 nt, which anneal to their target mRNA molecules and induce specific degradation and translational repression. Both computational and biological studies indicate that each miRNA can target a number of different mRNAs, and that ∼20–30% of human genes may be regulated by these factors (3,4). Interestingly, the expression profiles of the miRNAs often show a strong correlation with the disease status of a cell [reviewed by Croce and Calin, (5)]. Furthermore, at least some of these miRNAs are likely to be involved in tumorigenesis, as indicated in a recent study showing that the over-expression of the miR-17 cluster, when co-expressed with c-Myc, induces B cell lymphoma in mice (6). The tight control of both the temporal and spatial expression of miRNAs thus seems to be crucial for the maintenance of cellular integrity. MiRNA expression appears to be regulated at multiple stages during the biogenesis of these molecules, although, it remains to be determined how this control is achieved. The characterization of biogenesis factors will therefore be of critical importance towards our increased understanding of the miRNA-guided gene regulatory network.
MiRNA biogenesis is initiated by transcription with RNA polymerase II (7–9) and their primary transcripts (pri-miRNAs) harbor a local hairpin structure that is then cropped by a nuclear RNase III, Drosha, into ∼70 nt precursor-miRNAs (pre-miRNAs) (10,11). Drosha functions in a complex known as Microprocessor that also contains a dsRNA-binding protein, DGCR8 (DiGeorge syndrome chromosomal region 8; also known as Pasha in Caenorhabditis elegans and Drosophila) (12–15). Pre-miRNAs then exit the nucleus via the action of exportin-5 (Exp5) (16–18). Dicer, a cytoplasmic RNase III nuclease, then removes the terminal loop of the pre-miRNAs to generate the ∼22 nt miRNA duplex (19–23). One strand of the duplex is then loaded onto the RNA induced silencing complex (RISC) (24,25).
DsRNA-binding proteins that contain dsRNA-binding domains (dsRBDs) serve diverse roles during RNA metabolism. A recently emerging theme in RNA silencing pathways is that multidomain RNase III proteins, such as Drosha and Dicer, necessitate dsRNA-binding proteins for miRNA processing and/or RISC formation (26–31). For example, Dcr-1 in Drosophila interacts with Loquacious/R3D1 that contains three dsRBDs and is required for miRNA processing, and possibly also for RISC assembly (28–30,32). Furthermore, TRBP and PACT, the human homologues of Loquacious/R3D1, interact with human Dicer and thereby assist in the assembly of RISC (31,33,34). However, the biochemical functions of these proteins remain largely unknown.
DGCR8/Pasha has been identified as a Drosha-interacting protein in Drosophila by both yeast two hybrid screening (13,15,35) and immunopurification from human cells (12,14). The human DGCR8 gene is located on chromosome 22q11 and is expressed ubiquitously from fetus to adult (36). Monoallelic deletion of this genomic region is associated with several clinical defects, most notably including DiGeorge syndrome/conotruncal anomaly face syndrome/velocardiofacial syndrome (37). In experiments in which DGCR8 was depleted by RNAi, pri-miRNAs were found to accumulate whereas pre-miRNA and mature miRNA levels decreased. In addition, neither recombinant DGCR8 nor Drosha alone was observed to be active during pri-miRNA cleavage, whereas a combination of these factors restores activity, indicating that DGCR8 is an essential cofactor for Drosha (12,14). The addition of recombinant DGCR8 also slightly reduced non-specific cleavage by Drosha (14). Because DGCR8/Pasha contains two dsRBDs at the C-terminus, it is thought that this cofactor facilitates the interaction of Drosha with its substrate RNA. DGCR8 also contains a putative WW domain (also termed Rsp5/wwp) in its middle region, which contains two highly conserved tryptophan residues separated by ∼20 amino acids (38). Because the WW domain is known to interact with both proline-rich motifs (39) and Drosha is proline-rich at its N-terminus, it has been proposed that DGCR8 may interact with Drosha through its WW domain.
In our current study, we examine how the various DGCR8 domains contribute to Drosha interaction, and to RNA recognition and subcellular localization. Our findings further our understanding of the mechanism of action of the Drosha–DGCR8 complex during pri-miRNA processing.
MATERIALS AND METHODS
Ultraviolet (UV)-crosslinking
A total of 20–50 ng of purified FLAG-DGCR8 and radiolabeled RNAs of (1 × 106 c.p.m.; 50–100 fmol) were mixed in 15 µl of binding buffer [10 mM Tris (pH 7.5), 50 mM KCl, 0.5 mM DTT, 1 U of RNase inhibitor, TAKARA] in 96-well plates, and then incubated at 4°C for 30 min. For competition assays, cold transcripts prepared by in vitro transcription were added to the reaction mixtures. The 96-well plates containing the reaction mixture were then placed in a UV-crosslinker (CL-1000 UV-crosslinker, UVP) for 5 min. After treatment with an RNase A/T1 mixture, the reaction mixtures were loaded onto 7.5% SDS–polyacrylamide gels.
In vitro protein binding assay
Immunopurified Drosha-FLAG proteins (∼2 µg) were immobilized on 15 µl of anti-FLAG M2 agarose mouse affinity gel (Sigma) and incubated with 106 c.p.m. of in vitro translated DGCR8 protein in 1 ml of buffer D-K′250 [20 mM Tris (pH 8.0), 250 mM KCl, 0.2 mM EDTA, 0.2 mM phenylmethlysulfonyl fluoride (PMSF), 0.1% Triton X-100]. After incubation for 90 min at 4°C, the resin was washed six times with 1 ml of buffer D-K′250 and the bound fraction was eluted by boiling in 40 µl of SDS–PAGE sample buffer and resolved by SDS–PAGE.
Purification of recombinant DGCR8 proteins
DGCR8 proteins fused to glutathione S-transferase (GST) at the N-terminus were purified as described previously (12). The GST tag was then removed from by incubation with 10 U/ml of thrombin at 16°C overnight. FLAG-DGCR8 proteins were prepared as follows: four 10 cm dishes of HEK293T cells were transfected with the FLAG-DGCR8 expression vector. Two days after transfection, the cells were harvested and sonicated in 1.5 ml of ice-cold buffer D-K′200 [20 mM Tris (pH 8.0), 200 mM KCl, 0.2 mM EDTA and 0.2 mM PMSF]. After centrifugation at 13 200 r.p.m. at 4°C for 15 min, the supernatant was treated with 50 µg/ml of RNase A at 4°C for 30 min. This extract was then incubated with 30 µl of anti-FLAG antibody that had been conjugated to agarose beads (anti-FLAG M2 agarose mouse affinity gel, Sigma) with constant rotation for 120 min at 4°C. The beads were then washed four times in buffer D-Na′2500 [20 mM Tris (pH 8.0), 2.5 M NaCl, 0.2 mM EDTA, 0.2 mM PMSF and 1% Triton X-100) to remove DGCR8-interacting proteins from the FLAG-DGCR8 protein. After three additional washings with FLAG-elution buffer [50 mM Tris (pH 7.4) and 150 mM NaCl], FLAG-DGCR8 proteins were eluted three times by incubating the beads with FLAG-elution buffer containing 400 µg/ml of 3× FLAG peptide (Sigma) at 4°C for 60 min. The eluant was then concentrated at 20 ng/µl using Centricon YM-30 tubes (Millipore).
Preparation and labeling of substrate RNA
Pri-let-7a-1, pri-miR-16-1 and pri-miR-30a molecules were prepared by in vitro transcription as described previously (10,12). The labeling of pri-miR-16-1 at the 5′ end was also carried out as described previously (12).
In vitro processing of pri-miRNAs
In vitro processing of pri-miRNAs using either whole cell extracts or FLAG-immunoprecipitates was carried out as described previously (10,11). Briefly, reactions were carried out in 30 µl volumes containing either 15 µl of whole cell extract or immunoprecipitation beads, and also comprising 6.4 mM MgCl2, 1 U/µl of Ribonuclease Inhibitor (TAKARA) and labeled transcripts at 1 × 104–1 × 105 c.p.m.. The reaction mixture was incubated at 37°C for 90 min, and RNA was extracted by phenol and analyzed on a 12.5% denaturing polyacrylamide gel.
Mutagenesis of DGCR8
PCR products of the DGCR8 deletion mutants were subcloned into the EcoRI and NotI sites of the FLAG-pCK or V5-pcDNA3 vectors for expression in human cells. Point mutagenesis of DGCR8 was carried out using a site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The primer sequences used for mutagenesis are shown in Supplementary Table.
Cell culture and transfection
HeLa cells and HEK293T cells were cultured in DMEM (WelGENE) supplemented with 10% FBS (WelGENE). Transfections were carried using the calcium-phosphate method.
Gel retardation assay
A total of 50 ng of recombinant DGCR8 proteins purified from Escherichia coli cells was mixed with radiolabeled RNAs (5 × 105 c.p.m.) in 10 µl of reaction buffer [15 mM HEPES (pH 7.4), 100 mM KOAc, 2 mM Mg(OAc)2, 5 mM DTT, 0.2 % BSA, 1 U of RNase Inhibitor, TAKARA] and incubated at 4°C for 20 min. Samples were loaded onto 4% non-denaturing polyacrylamide gel (acrylamide: bis-acrylamide = 39:1) and electrophoresed at 250 V at 4°C.
Immunofluorescence
HeLa cells were grown on glass coverslips in DMEM, supplemented with 10% FBS. Cell transfections were carried out using the calcium-phosphate method. Two days after transfection, the cells were fixed using 2% formaldehyde in phosphate-buffered saline (PBS) for 30 min, washed with PBS and then permeabilized in PBS containing 0.1% Triton X-100 for 15 min. After several washes, the cells were saturated with 4% BSA in PBS for 30 min and immunostained for 2 h at RT with rabbit anti-V5-tag serum (Sigma) in PBS containing 4% BSA as the blocking agent. Subsequently, the cells were washed with PBS and incubated for 1 h at room temperature with Alexa Fluor 594 secondary antibody (Molecular Probes). The cells were analyzed using an Axioplan2 (Carl Zeiss) fluorescent microscope.
RESULTS
DGCR8 interacts with Drosha through its conserved C-terminal region
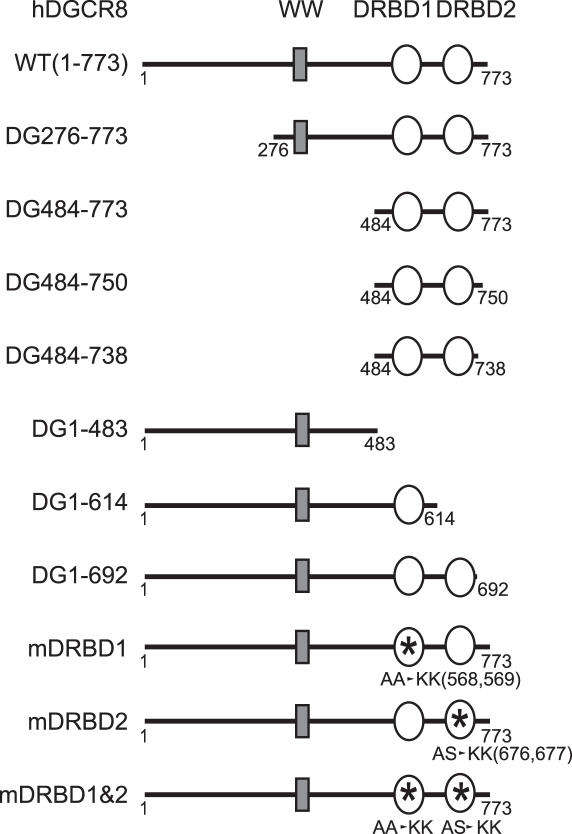
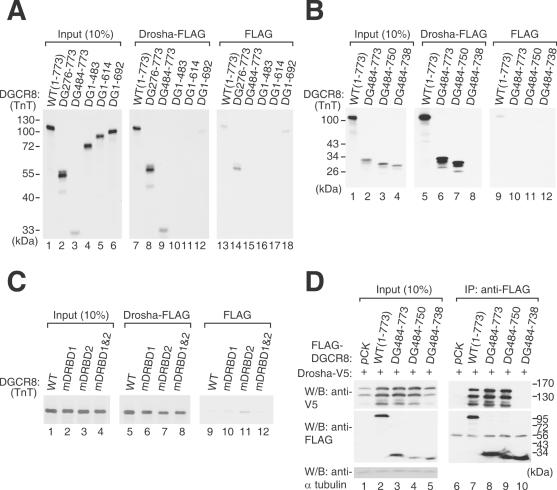
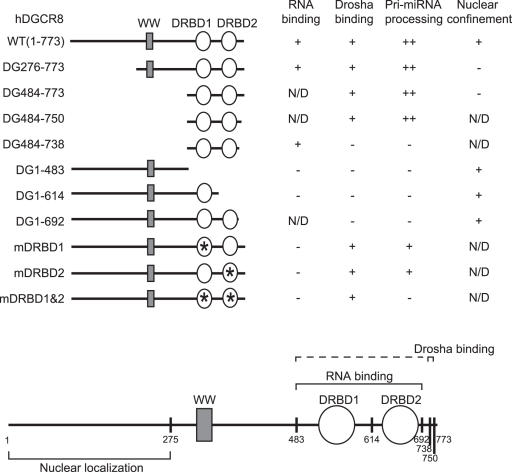
In order to determine the region required for the interaction of DGCR8 with Drosha, five deletion mutants of DGCR8 (DG276–773, DG484–773, DG1–483, DG1–614 and DG1–692) were tested (Figure 1). These truncation mutants were prepared as 35S-methionine labeled proteins by TnT (in vitro transcription and translation) reactions and incubated with Drosha-FLAG proteins that had been immobilized on anti-FLAG antibody-conjugated beads (Figure 2A, lanes 7–12). The expression plasmid encoding only the FLAG epitope was also transfected as a control to determine the background binding activities (Figure 2A, lanes 13–18). These pull-down experiments showed that two N-terminal deletion mutants of DGCR8 (DG276–773 and DG484–773), in addition to the wild-type protein (WT), efficiently bind Drosha-FLAG (Figure 2A). In contrast, the C-terminal deletion mutants of DGCR8 (DG1–483, DG1–614 and DG1–692) do not interact with Drosha-FLAG (Figure 2A). Therefore, the C-terminal region of DGCR8 is important for the interaction between DGCR8 and Drosha, whereas the N-terminal region, containing the putative WW domain, is dispensable for this binding. Previously, the WW domain of DGCR8/Pasha has been predicted to interact with the proline-rich region of Drosha (39). However, taken together with our previous finding that the proline-rich region of human Drosha is dispensable for DGCR8 binding (12), our current findings demonstrate that the C-terminus of DGCR8, and not the WW domain, is responsible for the association with Drosha.
Figure 1.
DGCR8 mutants used in this study. Asterisks represent the sites of point mutagenesis.
Figure 2.
Identification of the Drosha-interacting domain. (A–C) In vitro binding assay. DGCR8 wild-type and the indicated mutant proteins were synthesized by TnT (in vitro transcription and translation, Invitrogen) reactions and incubated with the Drosha-FLAG protein immobilized on anti-FLAG antibody that is conjugated on agarose beads. The left panels show 10% of the input proteins used in the binding reactions, whereas the middle panels visualize the proteins interacting with Drosha-FLAG. As a control (right panels, pull-down with FLAG), an empty vector expressing FLAG tag only was transfected and the cell lysates were used for immunoprecipitation. (D) Immunoprecipitation followed by western blot analysis. FLAG-DGCR8 mutant constructs were co-expressed with Drosha-V5. Immunoprecipitation was performed with anti-FLAG antibody and western blotting was carried out using either anti-V5 antibody (upper panels) or anti-FLAG antibody (lower panels).
To elucidate the Drosha-binding domain of DGCR8 more precisely, we generated two additional deletion mutants, DG484–750 and DG484–738, which contain smaller deletions at the C-terminus of this protein (Figure 1). These truncation mutants were prepared in TnT reaction and used for in vitro binding assays (Figure 2B). DG484–750 was found to interact with Drosha, whilst DG484–738, which is shorter by an additional 12 amino acids, failed to interact with Drosha, indicating that the C-terminal residues 739–750 of DGCR8 are critical for Drosha-binding (Figure 2B, lanes 7 and 8). It is noteworthy that this region is highly conserved among the DGCR8/Pasha homologues of various species (Supplementary Figure S1).
We performed additional pull-down experiments to test the Drosha-binding ability of three point mutants of DGCR8; mDRBD1, mDRBD2 and mDRBD1&2 (Figure 1). Point mutagenesis was carried out to substitute the highly conserved alanines (568 and 569) in dsRBD1 to lysine residues (mDRBD1). Similarly, the conserved alanine and serine residues at positions 676 and 677 of dsRBD2 were also converted to lysine residues (mDRBD2) (Figure 1). Each of these mutant proteins were capable of binding Drosha, although the dsRBD2 mutations (mDRBD2 and mDRBD1&2) slightly affected the binding activity (Figure 2C, lanes 7 and 8).
These results were confirmed by co-immunoprecipitated (co-IP) experiment followed by western blot analysis (Figure 2D). For this, the DGCR8 mutants were expressed as FLAG-tagged proteins together with V5-tagged Drosha protein in HEK293T cells. The mutants, DG484-773 and DG484-750 co-precipitated Drosha-V5 efficiently whereas the mutant DG484–738 failed to precipitate Drosha-V5, demonstrating that the C-terminal residues of DGCR8 are important for the Drosha–DGCR8 interaction (Figure 2D).
It is noted that when DGCR8 was overexpressed, Drosha protein level was also increased (Figure 2D, compare lanes 1 and 2). Moreover, the DGCR8 mutants that retain the Drosha-binding activity had the same effect on Drosha protein level (Figure 2D, lanes 3 and 4). This result suggests that DGCR8 may stabilize Drosha through protein–protein interaction.
The two dsRBDs of DGCR8 are responsible for pri-miRNA binding
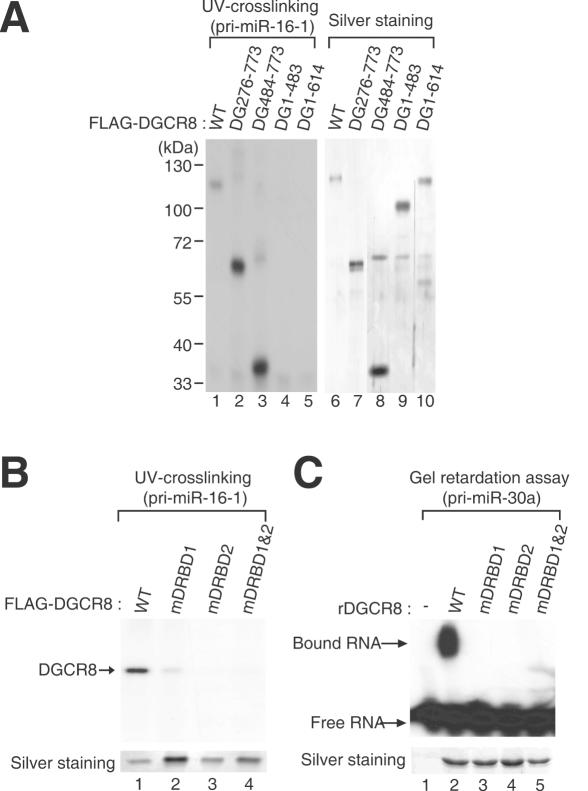
To determine the exact region of the DGCR8 protein that is required for pri-miRNA binding, we utilized four deletion mutants (DG276–773, DG484–773, DG1–483 and DG1–614) and three point mutants (mDRBD1, mDRBD2 and mDRBD1&2) in UV-crosslinking experiments (Figure 3A and B). These proteins were transiently expressed and immunopurified from HEK293T cells by extensive washing with high salt buffer containing 2.5 M NaCl to remove Drosha, as described previously (12). The immunopurified mutant proteins were then UV-crosslinked to radiolabeled pri-miR-16-1 transcripts. Both the N-terminal deletion mutants (DG276–773 and DG484–773), which retain two intact dsRBDs, bound the pri-miRNAs as efficiently as the wild-type protein, whereas the mutants, DG1–483 and DG1–614, lacking the dsRBDs, were deficient in this binding (Figure 3A). This result indicates that the C-terminal region of DGCR8 containing the two dsRBDs is required for pri-miRNA binding. The dsRBD point mutants (mDRBD1, mDRBD2 and mDRBD1&2) had also lost RNA binding activity, as indicated by both UV-crosslinking analysis (Figure 3B) and gel retardation assays (Figure 3C), further confirming that both dsRBDs in DGCR8 are required for pri-miRNA binding.
Figure 3.
DGCR8 domains responsible for pri-miRNA binding. (A) UV-crosslinking experiments between a series of DGCR8 deletion mutants and 32P-UTP radiolabeled pri-miR-16-1. After UV-crosslinking, residual RNA was removed by treatment with an RNase A/T1 mix. The left panel visualizes the radioactivity associated with recombinant FLAG-DGCR8 proteins, whereas the right panel indicates the amounts of protein used in this experiment as visualized by silver staining. (B) UV-crosslinking between a series of DGCR8 point mutants and 32P-UTP radiolabeled pri-miR-16-1. The upper panel displays the radioactivity crosslinked to the immunopurified FLAG-DGCR8 proteins and the lower panel indicates the amount of protein used in this experiment as determined by silver staining. (C) Gel retardation assay of pri-miR-30a binding to DGCR8 point mutants. Recombinant DGCR8 proteins prepared from E.coli were pre-treated with thrombin to separate the GST tag prior to the assay. The lower panel shows silver stained gel of recombinant DGCR8 protein used in this experiment.
The DGCR8 domains required for pri-miRNA processing in vitro
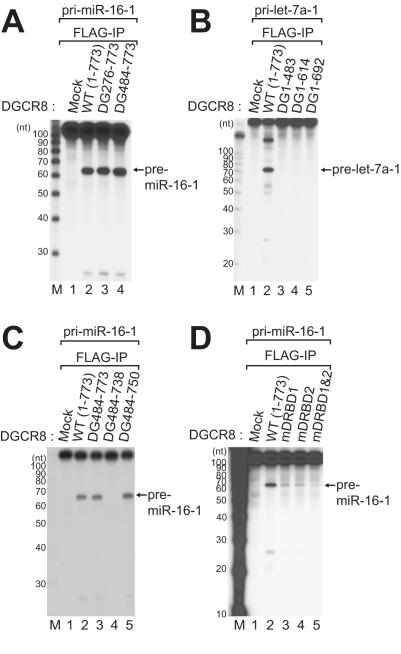
To investigate how the various mutations that we introduced into the DGCR8 protein would affect the pri-miRNA processing activity of the Microprocessor complex, an in vitro processing assay was carried out using DGCR8 proteins immunoprecipitated with low salt buffer (buffer D-K′200 containing 200 mM KCl). This was done to ensure that the endogenous Drosha protein would be co-IP (12). First, the deletion mutants were incubated with either pri-let-7a-1 or pri-miR-16-1 substrates to allow pri-miRNA processing by the Microprocessor complex (Figure 4). As expected, four DGCR8 mutants (DG1–483, DG1–614, DG1–692 and DG484–738) that lack the Drosha-binding domain were deficient in pri-miRNA cleavage activity (Figure 4B, lanes 3–5 and C, lane 4). In contrast, both the N-terminal deletion mutants and one C-terminal deletion mutant of DGCR8 (DG276–773, DG484–773 and DG484–750) efficiently cleaved pri-miRNA into pre-miRNA, indicating that the N-terminal residues 1–483 and C-terminal residues 751–773 are dispensable for pri-miRNA processing in vitro (Figure 4A, lanes 3 and 4 and C, lane 5). We next tested our three dsRBD point mutants in these assays as they bind poorly to pri-miRNA, and found that the alanine point mutation in each of the dsRBDs (mDRBD1 or mDRBD2) significantly reduced the pri-miRNA processing activity of DGCR8 (Figure 4D, lanes 3 and 4). In addition, a combination of these substitutions (mDRBD1&2) almost completely abolished the processing activity (Figure 4D, lane 5). Hence, the RNA binding activity of DGCR8 may be required to enable Microprocessor to recognize and process pri-miRNAs.
Figure 4.
DGCR8 domains required for pri-miRNA processing. In vitro pri-miRNA processing assay of DGCR8 mutants. FLAG-DGCR8 was immunoprecipitated using anti-FLAG antibody-conjugated agarose beads in low salt buffer (buffer D-K′200), and subsequently subjected to an in vitro pri-miRNA processing assay. Pri-miR-16-1 or pri-let-7a-1, labeled internally with 32P-UTP, was used as the substrate. Decade markers (Ambion) were used as the size markers.
Nuclear localization of DGCR8
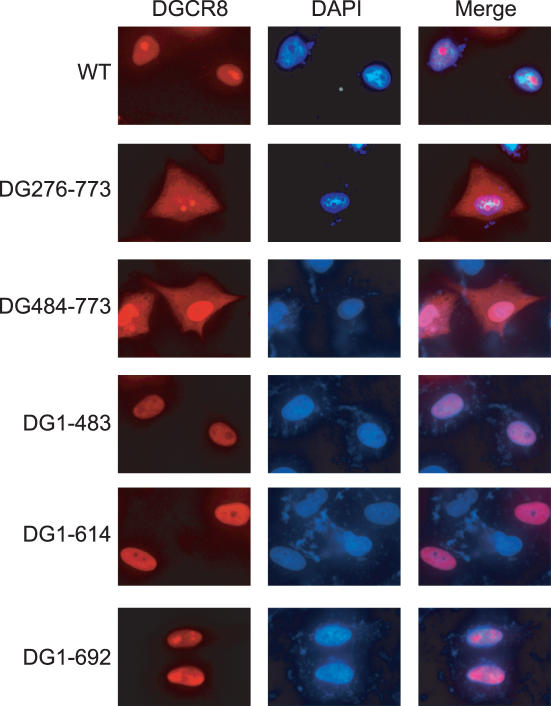
To examine the subcellular localization of DGCR8, we performed immunofluorescence studies using ectopically expressed, V5-tagged DGCR8 wild-type and mutant proteins (Figure 5). Wild-type DGCR8 was found to be restricted in the nucleus of HeLa cells (Figure 5, WT). We subsequently analyzed the localization pattern of our DGCR8 deletion mutants to determine the regions of the protein that were important for this nuclear localization. The N-terminal truncation mutant, DG276-773, was detectable throughout the cell (Figure 5), indicating that the nuclear localization signal and/or nuclear retention signal for DGCR8 is contained within its N-terminal 275 residues. A similar localization pattern was observed for another N-terminal deletion mutant, DG484–773 (Figure 5). Consistently, three C-terminal deletion mutants, DG1–483, DG1–614 and DG1–692 were detected in the nucleus (Figure 5, DG1–692), further indicating that the nuclear localization/retention signal of DGCR8 may be confined in its N-terminal region.
Figure 5.
Subcellular localization of DGCR8 and its deletion mutants. HeLa cells were transiently transfected with V5-tagged constructs, including WT DGCR8, and the DG276–773, DG484–773, DG1–483, DG1–614 and DG1–692 mutants. The left panels show DGCR8 protein signals visualized with rabbit anti-V5 serum (1:500) and Alexa Flour 594 goat anti-rabbit IgG (1:400). The middle panels show DAPI staining of the nucleus and the right panels display an overlay of these two signals.
DISCUSSION
A summary of the results of our current DGCR8 mutagenesis study is shown in Figure 6. A relatively small region of the DGCR8 protein (residues 484 through 750) was found to be sufficient to facilitate pre-miRNA processing in vitro. This minimal region of DGCR8 contains both the dsRBDs and the C-terminal Drosha-binding domain. The N-terminal region was not critical for processing but was shown to be important for nuclear localization of DGCR8.
Figure 6.
Summary of the DGCR8 mutagenesis findings. Our DGCR8 mutants can be characterized into four different groups; pri-miRNA binding, Drosha-binding, pri-miRNA processing and nuclear confinement activity. N/D, not determined.
DGCR8 is capable of direct interaction with pri-miRNA. On the contrary, we could not detect any significant pri-miRNA binding activity for Drosha in any of our assays (including gel mobility shift assays, UV-crosslinking, immunoprecipitations and GST pull-downs) (data not shown). Thus, DGCR8 may be the primary factor that recognizes the pri-miRNA structure, whereas Drosha interacts only with the RNA substrates transiently during catalysis. In addition, DGCR8 appears to stabilize the Drosha protein through its C-terminal Drosha-binding domain. It will be of great interest in the future to resolve the 3D structure of pri-miRNA and the Drosha–DGCR8 complex to fully decipher the associations between the substrate and the enzyme complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank the members of our laboratory and Dr Yunje Cho and Soyoung Sohn for helpful discussion. Supported by the Basic Research Program (R02-2004-000-10173-0) from the Ministry of Education and Human Resources Development and by the National Research Laboratory Program (M1050000010905J000010910) and the Molecular and Cellular BioDiscovery Research Program (2005-00518) from the Ministry of Science and Technology. K.-Y., Y.L., J.H. and M.R.S. are supported by a BK21 Research Studentship. Funding to pay the Open Access publication charges for this article was provided by the Basic Research Program (R02-2004-000-10173-0).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kim V.N. Small RNAs: Classification, Biogenesis, and Function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 3.Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Croce C.M., Calin G.A. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 6.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev. Mol. Cell. Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 14.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 15.Landthaler M., Yalcin A., Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 20.Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 21.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 22.Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight S.W., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 25.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 26.Tabara H., Yigit E., Siomi H., Mello C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 28.Leuschner P.J., Obernosterer G., Martinez J. MicroRNAs: loquacious speaks out. Curr. Biol. 2005;15:R603–R605. doi: 10.1016/j.cub.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 29.Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito K., Ishizuka A., Siomi H., Siomi M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase A.D., Jaskiewicz L., Zhang H., Laine S., Sack R., Gatignol A., Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 Complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 35.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 36.Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 2003;304:184–190. doi: 10.1016/s0006-291x(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 37.Goldberg R., Motzkin B., Marion R., Scambler P.J., Shprintzen R.J. Velo-cardio-facial syndrome: a review of 120 patients. Am. J. Med. Genet. 1993;45:313–319. doi: 10.1002/ajmg.1320450307. [DOI] [PubMed] [Google Scholar]
- 38.Sudol M. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 39.Macias M.J., Wiesner S., Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.