Abstract
The frequency distribution of mutation-induced aberrant 3′ splice sites (3′ss) in exons and introns is more complex than for 5′ splice sites, largely owing to sequence constraints upstream of intron/exon boundaries. As a result, prediction of their localization remains a challenging task. Here, nucleotide sequences of previously reported 218 aberrant 3′ss activated by disease-causing mutations in 131 human genes were compared with their authentic counterparts using currently available splice site prediction tools. Each tested algorithm distinguished authentic 3′ss from cryptic sites more effectively than from de novo sites. The best discrimination between aberrant and authentic 3′ss was achieved by the maximum entropy model. Almost one half of aberrant 3′ss was activated by AG-creating mutations and ∼95% of the newly created AGs were selected in vivo. The overall nucleotide structure upstream of aberrant 3′ss was characterized by higher purine content than for authentic sites, particularly in position −3, that may be compensated by more stringent requirements for positive and negative nucleotide signatures centred around position −11. A newly developed online database of aberrant 3′ss will facilitate identification of splicing mutations in a gene or phenotype of interest and future optimization of splice site prediction tools.
INTRODUCTION
Mutations that affect pre-mRNA splicing have been shown to account for up to a half of disease-causing gene alterations (1,2), potentially representing the most frequent cause of hereditary disorders (3). The most common consequence of splicing mutations is skipping of one or more exons, followed by the activation of aberrant 5′ (donor) splice sites (5′ss), 3′ (acceptor) splice sites (3′ss) and retention of full introns in mRNA (4,5). Each of these four events may have a dramatic impact on the structure or outcome of mature transcripts, function of their translation products and phenotypic manifestations. However, gene mutations or variants can also have more subtle effects at the level of splicing by altering the expression of pre-existing alternatively spliced mRNA isoforms, which can considerably modify not only phenotypic severity of both Mendelian and complex traits, but also their population prevalence (6–9).
Mutation-induced aberrant splice sites have been classified into two categories (10): (i) cryptic splice sites, which are only used when a mutation disrupts use of the authentic site, and (ii) de novo splice sites, which are induced by mutations elsewhere in introns or exons and increase the match to a splice site consensus. However, distinction between the two categories may be ambiguous in some cases since disruption of the authentic site may create a new splice site consensus, and is less obvious for 3′ss than 5′ss because accurate recognition of acceptor sites requires additional signal sequences in introns (11). The splicing signals of acceptor sites, namely the branch point sequence (BPS), polypyrimidine tract (PPT), and 3′AG, are recognized by RNA–protein interactions involving splicing factor 1 (SF1) and 65 and 35 kDa subunits of the U2 small nuclear RNP auxiliary factor (U2AF65 and U2AF35), respectively (12–17). The overall strength of 3′ss is defined by optimal sequences for interaction with each cognate factor as well as their distances from each other (18,19).
Cryptic 5′ss have a similar frequency distribution in exons and introns and their number decreases with an increasing distance from authentic 5′ss (10). In contrast, the localization of cryptic 3′ss is biased towards exons, whereas de novo 3′ss usually reside in introns, particularly within the PPT of authentic 3′ss (11). The distribution bias and a lower prevalence of aberrant 3′ss than 5′ss in vivo is most likely due to sequence constraints near intron/exon boundaries, including depletion of AG dinucleotides and the presence of PPT and BPS upstream of 3′ss (11). In addition, the multifaceted distribution of aberrant 3′ss would be predicted to reflect variable distances between the 3′ss signal sequence from intron to intron (18–21), including the presence of putative ‘distant BPS’ that are not located within an optimal distance of 18–40 nt 5′ of 3′ss, but may reside up to several hundred nucleotides further upstream (22). Despite a growing number of reported splicing mutations and associated phenotypes, the localization of the resulting aberrant 3′ss and their effect on gene expression remain difficult to predict.
Currently available computational tools that estimate the splice site strength have been based on a variety of methods, including nucleotide frequency matrices (23,24), machine learning approaches (25), neural networks (26), information theory (27) and interdependence between adjacent (the first-order Markov model) or more distant (the maximum entropy model) positions of the splicing consensus sequences (28). Gene prediction algorithms that take into account protein coding information have been shown to perform better than those that rely only on signals present in the splice sites (29). However, the strength of mutation-induced aberrant splice acceptor sites has not been systematically analyzed, and it is unknown at present which models best predict the localization of cryptic or de novo 3′ss activated in vivo.
Here, nucleotide sequences of aberrant 3′ss that were reported previously in human disease genes have been compiled and made available to the public through an online retrieval tool. Comparison of the splice site strength using current prediction algorithms showed that the maximum entropy model allowed the best discrimination between authentic and mutation-induced aberrant 3′ss, validating this model as the most sensitive instrument. In addition, this study provides a detailed characterization of the underlying mutation pattern and comparison of nucleotide composition upstream of aberrant and corresponding authentic 3′ss.
MATERIALS AND METHODS
Compilation of mutation-induced aberrant 3′ss in human disease genes
Published reports of cryptic and de novo 3′ss were identified by searching PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) and home pages of peer-reviewed journals. A subset of case reports were identified by searching locus-specific mutation databases (http://archive.uwcm.ac.uk/uwcm/mg/docs/oth_mut.html). The search was restricted to human genes with sequence-verified aberrant RNA products published before May 2006 that resulted from disease-associated mutations or variants. Nine cases in which no patient RNA was available but aberrant RNA products of wild-type and mutated alleles were characterized in minigene splicing reporter assays were also included. Aberrant 3′ss were manually validated by mapping the information in the literature to sequences in the Human Genome Project databases. Nucleotide sequences of authentic, mutated and aberrant 3′ss are available at http://www.dbass3.soton.ac.uk/ in the first online database of aberrant 3′ss termed DBASS3.
Comparison of computational methods to predict aberrant 3′ss
Validated sequences of aberrant and corresponding authentic 3′ss were used as input files for several splice site prediction algorithms. The Shapiro and Senapathy (S&S) matrix is based on nucleotide frequencies at each position of the 3′ss consensus sequence (23,24). The S&S matrix scores were computed using an online tool available at http://ast.bioinfo.tau.ac.il/. The information theory-based server (27) available at https://splice.cmh.edu/ was used to obtain the information content (Ri) of 3′ss in bits. To accommodate dependencies between adjacent and non-adjacent positions, the compiled sequences were analyzed using the first-order Markov (MM) and the maximum entropy (ME) models (28). The former method considers dependencies between adjacent positions, whereas the latter approximates short sequence motif distributions with the ME distribution and may include dependencies between non-adjacent as well as adjacent positions. The MM and ME scores (28) were derived for each 3′ss using online tools available at http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html. The Wilcoxon Mann–Whitney rank test (Stat-200, v. 2.01, Biosoft, UK) was employed to test the significance of score differences between authentic, mutated and aberrant 3′ss in each category.
DBASS3 construction
DBASS3 is an online retrieval and submission tool for mutation-induced aberrant 3′ss available at http://www.dbass3.soton.ac.uk/. The web application was created using the ASP server technology (Microsoft), and SQL database software (http://www.sql.org). In addition to aberrant 3′ss induced by germ-line and somatic mutations, DBASS3 contains naturally occurring variants common in the population if they have been convincingly shown to modify both alternative pre-mRNA splicing and disease phenotypes, such as FECH IVS3-48T/C in protoporphyria (8). Genetic polymorphisms that may influence utilization of tandemly arranged ‘NAGNAG’ 3′ss (30) and exert putative functional effects have been reported elsewhere (31) and were not included in DBASS3, nor were the mutations leading to exon skipping or complete intron retention.
RESULTS
Mutations that activate aberrant 3′ss
An exhaustive search for previously published aberrant 3′ss identified 218 unique aberrant acceptors in 131 genes (Table 1). They were generated by a total of 16 deletions/insertions (32–46) and 211 point mutations (Table 2). Single-nucleotide substitutions of purine residues were much more frequent than those of pyrimidines (165 versus 46, P < 10−16). This overrepresentation was not attributable solely to substitutions at 3′YAG (102 versus 8), but was also observed for de novo 3′ss (63 versus 38, P = 0.004). The most frequently introduced base in each of the four categories of aberrant 3′ss was guanine (G), accounting for ∼42% (89/211) of all point mutations (Table 2).
Table 1.
Summary of aberrant 3′ss
| Location of cryptic or de novo 3′ss | |||||
|---|---|---|---|---|---|
| Exon | Intron | Both | |||
| Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | All mutations | |
| Number of genes | 54 | 25 | 23 | 56 | 131 |
| Number of cryptic and de novo 3′ ss (per cent) | 88 (39) | 32 (14) | 29 (13) | 78 (34) | 227 (100) |
| Number of unique 3′ss (per cent) | 83 (38) | 29 (13) | 28 (13) | 78 (36) | 218 (100) |
| Number of aberrant 3′ss affecting terminal exons (per cent) | 11 (13) | 4 (14) | 8 (29) | 4 (5) | 27 (12) |
| Median distance (nucleotide) between authentic and aberrant 3′ss | 12 | 55 | −44 | −14 | 1 |
| Change in the reading frame for unique aberrant 3′ss | |||||
| 0 | 29 | 10 | 8 | 27 | 74 |
| +1 | 38 | 7 | 13 | 26 | 84 |
| +2 | 21 | 15 | 8 | 25 | 69 |
Table 2.
Summary of mutations leading to aberrant 3′ss
| Location of cryptic or de novo 3′ss | |||||
|---|---|---|---|---|---|
| Exon | Intron | Both | |||
| Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | All mutations | |
| Number of deletions/insertions | 7 | 3 | 0 | 6 | 16 |
| Number of single-nucleotide substitutions | 81 | 29 | 29 | 72 | 211 |
| Wild-type nucleotide | |||||
| A | 36 | 3 | 13 | 29 | 81 |
| C | 2 | 9 | 5 | 9 | 25 |
| G | 43 | 13 | 10 | 18 | 84 |
| T | 0 | 4 | 1 | 16 | 21 |
| Mutated nucleotide | |||||
| A | 23 | 8 | 5 | 28 | 64 |
| C | 21 | 3 | 5 | 2 | 31 |
| G | 25 | 9 | 14 | 41 | 89 |
| T | 12 | 9 | 5 | 1 | 27 |
| Number of AG-creating mutations | |||||
| Total number (%) | 16 (7) | 12 (5) | 8 (4) | 62 (27) | 98 (43) |
| Not used as aberrant 3′ss (%) | 0 | 5 | 2 | 4 | 11 (5) |
As expected, point mutations were most common in highly conserved positions −1 (53/211; 25%) and −2 (48/211; 23%) relative to natural intron/exon junctions (Table 3). Position −3 was mutated in nine cases (∼4%). As noted in the initial analysis of all splice site mutations for position −2 (47), G-to-Y (in position −1; Y is pyrimidine) and A-to-Y (position −2) transversions were under-represented as compared with G-to-A and A-to-G transitions, respectively (P < 0.01 and P < 0.00001, assuming that substitutions to the remaining nucleotides were equally probable; Table 3). Since transitions are in significant excess in humans compared with the expected frequency of 33% (47), the expected numbers were calculated for each substitution using previously published single-nucleotide mutability rates in disease genes (Table 3). However, the observed number of G−1-to-T−1 mutations was too low to be explained by chance, suggesting that primary transcripts carrying the A−2T−1 acceptors generate on average more canonical mRNAs as compared with 3′AG mutated to other dinucleotides, leading to a detection bias against less severe phenotypes. This notion is supported by similar frequencies of G>T/C>A and G>C/C>T alterations among disease-causing point mutations (48) and by the presence of residual amounts of natural transcripts in some 5′G+1T+2–3′A−2T−1 introns both in Saccharomyces cerevisiae (49) and humans (50). However, comparison of the observed and expected distributions derived from di-nucleotide mutability rates that allow for the influence of neighbouring nucleotides (48) failed to confirm any bias for both intron positions (Table 3). Thus, although small effects of ‘leaky’ dinucleotides on the observed distribution cannot be excluded, these data are consistent with dramatic consequences for splicing of any point mutation in the highly conserved 3′AG and with indistinguishable defects of the second splicing step previously observed in vitro both for intron position −1 (51) and −2 (52).
Table 3.
Number of single-nucleotide substitutions in 3′YAG that resulted in cryptic 3′ss
| Observed | Expected | ||||
|---|---|---|---|---|---|
| Location of cryptic 3′ splice site | Exon | Intron | Both | Mono-a | Di-b |
| Point mutations in position IVS-1 | 43 | 10 | 53 | — | — |
| −1G>A | 23 | 4 | 27 | 25.2c | 31.5d |
| −1G>C | 14 | 3 | 17 | 10.9 | 13.2 |
| −1G>T | 6 | 3 | 9 | 16.9 | 8.3 |
| Point mutations in position IVS-2 | 36 | 12 | 48 | — | — |
| −2A>C | 7 | 2 | 9 | 9.3e | 8.8f |
| −2A>G | 23 | 8 | 31 | 30.6 | 35.5 |
| −2A>T | 6 | 2 | 8 | 8.1 | 3.7 |
| Point mutations in position IVS-3 | 2 | 7 | 9 | — | — |
| −3C>N | 2 | 5 | 7 | — | — |
| −3T>N | 0 | 1 | 1 | — | — |
| −3A>N | 0 | 1 | 1 | — | — |
Expected numbers of both exonic and intronic cryptic 3′ss were calculated as a weighted average of relative mono-a (47) and di-b (48) nucleotide mutability rates in the sense and antisense DNA strands that were published previously for a large number of point mutations in human disease genes. Relative substitution rates at the di-nucleotide level allow for the nearest-neighbour effects as previously described (48). cχ2=6.2, P = 0.046, dχ2=1.3, P > 0.05, eχ2=0.02, P > 0.05; fχ2=4.4, P > 0.05.
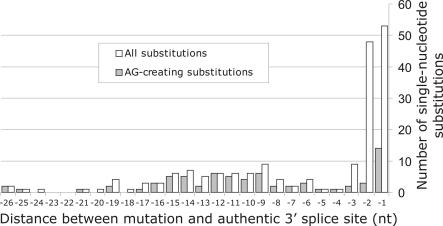
Interestingly, as many as 14/53 (26%) point mutations in position −1 (IVS-1G>A if the first exon nucleotide was G) (34,53–64), 3/48 (6%) substitutions in position −2 (IVS-2A>G) (65,66) and 2/9 (22%) point mutations in position −3 [IVS-3T>G (67) and IVS-3A >G(68)] created new 3′AG sites that were used in vivo (Figure 1). The proportion of AG-creating mutations in position −1 was higher than in position −2 (P = 0.01, Fisher's exact test), which may have contributed to the higher number of substitutions observed in position −1 than −2 (Table 3). In contrast to mutations in the 3′YAG consensus, the majority of substitutions in the PPT were AG-creating mutations. For example, in positions −5 to −26 relative to natural intron/exon junctions as many as 61/73 (84%) point mutations mutations created new AGs (Figure 1). The overall proportion of AG-creating mutations that resulted in aberrant 3′ss was 43%, and ∼95% of the newly introduced 3′AGs were used in vivo (Table 2).
Figure 1.
Frequency distribution of 184 intronic point mutations that activated aberrant 3′ss
Purine transitions, which accounted for ∼54% (113/211) of all aberrant 3′ss and dominated the mutation pattern of cryptic 3′ss, were also the most frequent point mutations leading to de novo 3′ss (54/101; 53%). De novo sites in introns resulted from purine transitions more often than de novo sites in exons (45/72 versus 9/29, χ2 = 7.0, P = 0.008). Intronic de novo 3′ss were most frequently induced by substitutions of A (29/72, 40%), whereas exonic de novo 3′ss were most commonly activated by point mutations of G (13/29, 45%; Table 2).
Comparison of computational tools to predict mutation-induced aberrant 3′ss in vivo
The predicted strength of aberrant, mutated and corresponding authentic 3′ss was analyzed using publicly available computational tools shown in Table 4. Each of the tested models distinguished authentic, mutated and aberrant 3′ss, with authentic sites giving, on average, the highest scores or information bits, followed by aberrant and then by mutated 3′ss (Table 5). However, this was not the case for each category of aberrant acceptors.
Table 4.
Comparison of the strength of authentic, mutated and aberrant 3′ss
| Location of aberrant 3′ss | |||||
|---|---|---|---|---|---|
| Exon | Intron | Both | |||
| Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | All mutations | |
| Shapiro and Senapathy matrix score | |||||
| A (SD) | 84.5. (6.5) | 79.3 (9.8) | 85.0 (6.1) | 81.0 (7.8) | 82.6 (7.7) |
| M (SD) | 67.5 (8.0) | 78.9 (9.4) | 70.2 (6.6) | 79.4 (8.7) | 73.5 (10.0) |
| CR (SD) | 74.5 (9.2) | 78.6 (10.4) | 82.7 (8.5) | 77.3 (9.0) | 77.1 (9.5) |
| A–M | 17.1 | 0.5 | 14.8 | 1.6 | 9.2 |
| M–CR | 7.1 | −0.3 | 12.4 | −2.1 | 3.6 |
| A–CR | 10.0 | 0.7 | 2.4 | 3.6 | 5.5 |
| Maximum entropy model | |||||
| A (SD) | 8.7 (3.5) | 6.7 (4.0) | 8.6 (3.0) | 7.3 (2.9) | 7.9 (3.4) |
| M (SD) | −0.3 (4.5) | 5.8 (5.1) | −0.4 (3.6) | 3.8 (4.1) | 2.0 (5.0) |
| CR (SD) | 4.1 (3.9) | 5.5 (5.6) | 6.4 (3.6) | 5.2 (4.5) | 5.0 (4.4) |
| A–M | 8.4 | 0.5 | 9.0 | 3.2 | 5.6 |
| M–CR | 4.1 | −0.4 | 6.8 | 1.8 | 3.0 |
| A–CR | 4.4 | 0.9 | 2.2 | 1.4 | 2.6 |
| First-order Markov model | |||||
| A (SD) | 9.1 (3.0) | 7.1 (4.1) | 8.9 (2.8) | 7.5 (3.0) | 8.2 (3.2) |
| M (SD) | 0.1 (3.9) | 6.2 (5.1) | 0.3 (3.0) | 4.0 (4.1) | 2.3 (4.7) |
| CR (SD) | 4.4 (3.9) | 5.3 (5.9) | 7.1 (3.2) | 5.4 (4.8) | 5.2 (4.6) |
| A–M | 8.9 | 0.8 | 8.5 | 3.5 | 5.9 |
| M–CR | 4.3 | −0.9 | 6.7 | 1.4 | 2.9 |
| A–CR | 4.7 | 1.7 | 1.8 | 2.1 | 3.0 |
| Weight matrix model | |||||
| A (SD) | 9.9 (3.8) | 7.3 (4.3) | 9.6 (3.0) | 7.5 (4.1) | 8.7 (4.0) |
| M (SD) | 1.1 (4.3) | 7.1 (4.2) | 1.4 (3.1) | 6.6 (4.0) | 3.9 (4.9) |
| CR (SD) | 4.6 (4.8) | 6.2 (5.3) | 8.1 (3.5) | 6.2 (4.8) | 5.8 (4.8) |
| A–M | 8.8 | 0.3 | 8.2 | 0.9 | 4.8 |
| M–CR | 3.6 | −0.9 | 6.8 | −0.4 | 2.0 |
| A–CR | 5.3 | 1.2 | 1.5 | 1.3 | 2.8 |
| Information content | |||||
| A (SD) | 9.9 (3.9) | 8.1 (3.9) | 9.6 (3.1) | 7.9 (3.9) | 8.9 (3.9) |
| M (SD) | 2.1 (3.8) | 7.6 (3.8) | 2.4 (3.3) | 7.0 (3.7) | 4.6 (4.5) |
| CR (SD) | 5.9 (3.7) | 7.2 (3.3) | 8.0 (3.5) | 7.5 (3.6) | 6.9 (3.7) |
| A–M | 7.8 | 0.5 | 7.2 | 0.8 | 4.3 |
| M–CR | 3.5 | −0.7 | 5.5 | 0.1 | 2.2 |
| A–CR | 4.1 | 1.0 | 1.7 | 0.7 | 2.2 |
Means and SD of splice prediction scores (S&S, ME, MM, weight matrix model) or bits (information content) for authentic (A), mutated authentic (M) and aberrant (CR) 3′ss. The length of input sequences was 15 (−14 to +1 relative to authentic 3′ss), 23 (−20 to +3), 23 (−20 to +3), 23 (−20 to +3) and 28 (−26 to +2) nt, respectively. The information content algorithm failed to recognize 7 authentic, 12 mutated and 28 aberrant 3′ss used in vivo. The missing values (47/654, 7%) were treated as a group mean.
Table 5.
Discrimination of computational tools between authentic, mutated and cryptic/de novo 3′ss
| Location of aberrant 3′splice sites | |||||
|---|---|---|---|---|---|
| Exon | Intron | Both | |||
| Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | Mutation in 3′YAG (cryptic) | Mutation outside 3′YAG (‘de novo’) | All mutations | |
| Shapiro and Senapathy matrix score | |||||
| A–M | 7.2 × 10−26 | 0.4 | 1.3 × 10−9 | 0.16 | 2.1 × 10−23 |
| CR–M | 5.2 × 10−8 | 0.5 | 4.1 × 10−7 | 0.09 | 2.9 × 10−5 |
| A–CR | 1.2 × 10−13 | 0.4 | 0.13 | 0.01 | 9.5 × 10−11 |
| Maximum entropy model | |||||
| A–M | 2.4 × 10−28 | 0.3 | 1.6 × 10−10 | 8.4 × 10−9 | <10−32 |
| CR–M | 1.3 × 10−11 | 0.4 | 2.2 × 10−8 | 8.4 × 10−3 | 6.9 × 10−13 |
| A–CR | 1.5 × 10−15 | 0.2 | 4.5 × 10−3 | 1.9 × 10−3 | 1.8 × 10−15 |
| First-order Markov model | |||||
| A–M | 2.3 × 10−28 | 0.3 | 1.2 × 10−10 | 1.5 × 10−8 | <10−32 |
| CR–M | 1.0 × 10−11 | 0.2 | 5.2 × 10−9 | 9.3 × 10−3 | 1.7 × 10−12 |
| A–CR | 5.1 × 10−16 | 0.1 | 0.01 | 6.3 × 10−3 | 1.4 × 10−14 |
| Weight matrix model | |||||
| A–M | 1.9 × 10−24 | 0.4 | 4.1 × 10−10 | 0.09 | 2.5 × 10−24 |
| CR–M | 3.2 × 10−7 | 0.2 | 1.4 × 10−8 | 0.4 | 8.1 × 10−6 |
| A–CR | 2.7 × 10−13 | 0.2 | 0.04 | 0.07 | 1.8 × 10−10 |
| Information contents | |||||
| A–M | 9.2 × 10−23 | 0.3 | 4.8 × 10−9 | 0.08 | 1.9 × 10−20 |
| CR–M | 1.8 × 10−10 | 0.3 | 2.9 × 10−7 | 0.2 | 5.7 × 10−9 |
| A–CR | 6.8 × 10−11 | 0.2 | 0.02 | 0.2 | 3.2 × 10−9 |
Table cells contain P-values of Wilcoxon Mann–Whitney rank tests comparing authentic (A), mutated (M) and cryptic/de novo (CR) 3′ss. P-values < 0.05 are in bold.
First, each computational tool was more effective in discriminating authentic and aberrant 3′ss that resulted from mutations in the 3′YAG consensus than from mutations elsewhere (Table 5). This was owing to significantly higher scores for authentic 3′ss that corresponded to cryptic 3′ss than for authentic counterparts of de novo sites. For example, the S&S scores for authentic counterparts of de novo and cryptic 3′ss were 80.5 ± 8.4 (±SD) and 84.6 ± 6.4 (P < 10−7, Wilcoxon Mann–Whitney rank test), respectively. Similarly, the ME scores were 7.2 ± 3.2 and 8.6 ± 3.3, respectively (P < 10−7). In contrast, the score differences between cryptic and de novo 3′ss were not statistically significant (means of the S&S matrix scores were 76.5 versus 77.7, P = 0.3; means of the ME scores were 4.7 versus 5.3, P = 0.4, respectively). Scores or information bits for each category of aberrant acceptors are shown in Table 4. These results indicate that authentic counterparts of de novo 3′ss are intrinsically weak and can be outcompeted by newly created splicing consensus elements. They also suggest that mutations or genetic variants flanking weak splice sites are more likely to play a role in regulated splicing than those near well-defined sites, consistent with weakening of splicing signals in evolution from virtually invariable sequences in yeasts to highly degenerate in humans and a need for more sophisticated regulation in complex organisms at the level of alternative splicing.
Second, each algorithm could distinguish cryptic and authentic 3′ss in exons, whereas matrix-based scores struggled to differentiate between authentic and cryptic 3′ss in introns where the ME and MM were the only models that showed P-values of 0.01 or lower (Table 5).
Third, de novo 3′ss could not be discriminated from authentic sites by any algorithm if located in exons. Although this could be partly attributed to a smaller sample size of exonic than intronic de novo sites (Table 1), a similar sample of intronic cryptic 3′ss did show statistically significant differences for a subset of algorithms (Table 5). Finally, the difference between intronic de novo sites and their authentic counterparts was statistically significant with the ME and MM models but not with the remaining algorithms, except for the S&S matrix scores.
Taken together, these results indicated that the value of computational tools to predict aberrant 3′ss depended on their localization in introns and exons as well as on the underlying mutation, and that the ME was the best model discriminating mutation-induced aberrant 3′ss in vivo from corresponding authentic 3′ss. They also suggested that the failure to distinguish exonic de novo 3′ss from authentic counterparts may be due to our as yet incomplete understanding of the role of exonic splicing silencers or enhancer elements in 3′ss selection.
Single-nucleotide composition upstream of aberrant 3′ss
Comparison of the nucleotide structure upstream of aberrant and authentic 3′ss revealed a significantly higher proportion of purines in aberrant 3′ss. For example, in intronic positions −3 through −26 aberrant 3′ss had 1760 purines as opposed to 1526 purines in authentic 3′ss (χ2 = 23.7, P < 0.00001; Supplementary Figure 1). Overall, this was attributable to a higher number of As (χ2 = 13.5, P < 0.001) rather than Gs (χ2 = 6.4, P = 0.01; Supplementary Figure 1A). The increase of purine residues was almost exclusively at the expense of uridines for aberrant 3′ss in exons (Supplementary Figure 1B and C). In contrast, aberrant 3′ss in introns showed only a borderline increase of purine residues (χ2 = 3.2, P = 0.07), largely owing to cytosine depletion (Supplementary Figure 1D and E). De novo 3′ss in exons had a smaller number of Gs as compared with authentic 3′ss, but the difference was not statistically significant (Supplementary Figure 1C).
The increase of purines in aberrant 3′ss was the highest in position −3 where As were 6× more frequent than in authentic 3′ss (Supplementary Figure 2A, χ2 = 26.5, P < 0.000001). The number of aberrant 3′ss with G in position −3 was also higher (7 versus 2) in aberrant (65,66,69–73) than in corresponding authentic (70,74) 3′ss. Positive associations between −3C and upstream Cs in the PPT and between −3T and upstream Ts, which were described previously for authentic 3′ss (75), were observed also for aberrant 3′ss (Supplementary Table 2). Although the influence of −3C on the relative usage of C versus T in the PPT may be attributed to autocorrelation due to compositional similarities of local genomic regions (75), sequence constraints resulting from cooperative interactions at the 3′ss could not be excluded. Indeed, non-random distributions at −3 observed for positions −11, −12, −17 and −19 of aberrant 3′ss (Supplementary Table 2) may be explained by inefficient binding of U2AF to RNAs carrying 3′TAG as compared to 3′CAG (76) and a need for functional compensation of the former by stronger interaction of U2AF65 (or other PPT-binding proteins) with uridines at positions −11 and −12 rather than cytosines. Associations further upstream may involve similar compensation by more optimal BPS interactions with the RS domain of U2AF (77–79) and/or, possibly, other BPS-interacting factors, including K- and Quaking-homology 2 domains of SF1 (12,80,81) or U2 small nuclear RNA (82,83). Similar associations at −3 with upstream intron positions were seen also for authentic counterparts of aberrant 3′ss (data not shown), confirming previous findings with a larger dataset (75).
Although most of the analyzed positions upstream of aberrant 3′ss showed uridine depletion as compared to authentic sites (e.g. 565 versus 659 Ts in positions −5 to −10; χ2 = 12.8, P < 0.001; Supplementary Figure 2A), their numbers were similar further upstream between positions −11 and −13 (311 versus 319, P = 0.7). Cs were slightly under-represented between positions −11 and −13 in aberrant 3′ss (168 versus. 202, χ2 = 4.0, P = 0.04). The T-to-C ratio in aberrant 3′ss was the highest in position −11 (2.53 versus 1.70 in authentic), while the average (±SD) ratios between positions −4 and −26 in aberrant and authentic 3′ss were similar (1.52 ± 0.34 and 1.55 ± 0.28, respectively). Aberrant 3′ss with purine at −3 had higher T-to-C ratios between −11 and −13 than aberrant 3′ss with pyrimidine at −3 (2.54 versus 1.69). The number of Gs in this region was significantly higher in aberrant than authentic 3′ss (153 versus 92 in positions −9 to −12, χ2 = 17.0, P < 0.0001), particularly in cryptic sites, whereas the number of As in these positions was not different (125 versus 115, χ2 = 0.4, P = 0.5).
Di-nucleotide composition upstream of aberrant 3′ss
The number of AG dinucleotides, which are depleted in ‘AG exclusion zones’ upstream of authentic 3′ss (24,75,84), was significantly higher in aberrant than corresponding authentic 3′ss (Supplementary Figure 2B). In a 17 nt sequence upstream of 3′ss where the AG depletion in natural 3′ss is the most pronounced (75), the numbers of authentic and aberrant 3′ss with a non-3′ss (‘intervening’) AG were 15 and 36, respectively (χ2 = 8.8, P = 0.003), while the number of AGs in the two groups was 15 and 40 (binomial test, P = 0.0003). The observed frequency of authentic 3′ss with non-3′ss AGs in this region (∼16%) was similar to those previously reported for constitutively (14%) and alternatively (17%) spliced introns that contained intervening AGs downstream of predicted BPS (21). Between positions −3 and −26, there were 53 versus 80 AG-containing 3′ss (χ2 = 7.2, P = 0.007) and 64 versus 95 intervening AGs (binomial test, P = 0.003), respectively. No AG dinucleotides were found in positions −10 and −11 of aberrant 3′ss. Although the number of intervening AGs was low, putative differences of these and other purine dinucleotides between aberrant and authentic in intron positions −25, −24, −22, −20 or −19 upstream of 3′ss are consistent with a distinct average distance of the BPS from aberrant versus authentic 3′ss. Peak frequencies of the GA and AA dinucleotides that may signify the presence of branchpoint in the mammalian BPS consensus YNYURAY were shifted several nucleotides upstream in aberrant 3′ss (Supplementary Figure 2B).
The remaining purine dinucleotides were also more common in aberrant than in authentic sites. The increase of AA dinucleotides (253 versus 185 in positions −26 to −3, P = 0.001), which were found in excess upstream of authentic 3′ss as compared to pseudo-sites (75), was largely attributable to position −3 due to the excess of −3As in aberrant 3′ss (Supplementary Figure 2A, B). The GG dinucleotides (186 in aberrant versus 118 in authentic sites in the same region, P < 0.0001) also clustered in some positions, such as −17 to −21 (56 versus 19, χ2 = 17.9, P < 0.0001) and −8 to −12 (49 versus 26, χ2 = 6.7, P < 0.01, respectively).
A region upstream of 3′ss in vertebrates (75) and Arabidopsis thaliana (85) contains a higher number of TG dinucleotides as compared to pseudo-splice sites, suggesting that they are important for correct 3′ss recognition. Although the total number of TGs in positions −3 to −26 was similar in aberrant and authentic 3′ss (430 versus 428), there were 94 and 60 TGs in positions −10 to −13 in aberrant and corresponding authentic sites, respectively (χ2 = 7.7, P = 0.005). The number of GTs in the same region was also higher in aberrant sites (56 versus 34; χ2 = 5.2, P = 0.02). In contrast, the number of TTs in the same region was similar (235 versus 200, P > 0.05) both in cryptic and de novo sites, whereas aberrant 3′ss showed TT depletion for most of the remaining positions. The number of CC dinucleotides between position −10 and −13 was lower in aberrant 3′ss (71 versus 99, χ2 = 4.7, P = 0.03), but this difference was limited to de novo sites (χ2 = 10.7, P = 0.001). The TT-to-CC ratio in aberrant 3′ss was the highest in position −12 (8.14 versus 2.76 in authentic), whereas the average (±SD) between positions −5 to −26 was 2.26 (±1.43), with 2.21 ± 0.56 in authentic counterparts.
Position −11 shows peak uridine frequencies in vertebrate PPTs (86), most probably due to highly conserved interactions with the second RNA recognition motif (RRM2) of U2AF65, a central organizing force for 3′ss recognition in higher eukaryotes, or with competing pyrimidine-binding proteins (14,87,88). The same position was efficiently crosslinked to RRM2 of U2AF65 in several PPTs (87) and substitutions of T−11 generated lower levels of spliced products and prespliceosomal complexes than identical mutations of T−8 or T−14 (89), suggesting that the observed single- and di-nucleotide imbalances between aberrant and authentic 3′ss centred around this position have functional significance. Higher T-to-C and TT-to-CC ratios in aberrant 3′ss in this area are proposed to improve these interactions and functionally compensate their less favourable sequence context (Supplementary Figure 2A and Tables 4 and 5). The difference in the number of C−12C−11 between aberrant and authentic 3′ss (7 versus 21, χ2 = 6.4, P = 0.01) suggests that this di-nucleotide does not sufficiently promote U2AF binding and that at least one uridine is required in either position for the productive interaction since the numbers of T−12C−11 or C−12T−11 were not significantly different in aberrant and authentic 3′ss (Supplementary Figure 2B). This notion is in agreement with ∼80- to 100-fold inhibition of U2AF65 binding following chemical modification of the uridine N3 and O4 atoms, the only positions that differ between the two nucleosides (90). However, the CC dinucleotides in positions −11 to −13 were over-represented in authentic counterparts of de novo sites (53 versus 21, χ2 = 13.7, P < 0.001) but not cryptic sites (19 versus 26, P > 0.05), suggesting that they signify natural 3′ss that compete poorly with and may be susceptible to mutation-induced 3′ss.
In contrast to cytosines, both de novo and cryptic 3′ss showed an increase of TGs/GTs between positions −10 and −13 (64 versus 40, χ2 = 5.4, P < 0.05 and 86 versus 54, χ2 = 7.4, P < 0.01, respectively) as compared to authentic counterparts. A relative lack of G−12T−11/T−11G−10 in authentic sites suggests that such 3′ss may compete relatively well with newly introduced 3′ss, consistent with an earlier observation that GU tracts can substitute for pyrimidine tracts (91), probably as a result of flexible side chain rearrangements of U2AF65 and/or relocation of bound water molecules (92).
Depletion of aberrant 3′ss upstream and downstream of authentic 3′ss
Distribution of the distances between aberrant and authentic 3′ss with the updated sample confirmed a previously reported (11) bias of cryptic 3′ss towards exons and de novo sites towards introns (Supplementary Figure 3A and B). Major frequency peaks for cryptic and de novo 3′ss were 8 and −10 nt from authentic 3′ss, respectively (median distances in each category of aberrant 3′ss are in Table 1). In addition, a relative depletion of both in cryptic and de novo 3′ss emerged further upstream and downstream. A lack of cryptic 3′ss upstream is apparently due to AG depletion (11), although cryptic 3′ss activation may also be prevented by spliceosomal complexes assembled around the branch site. The latter explanation is likely to account for the observed depletion of de novo 3′ss, which is more upstream as compared to cryptic sites (∼50 nt, Supplementary Figure 3B).
Smaller areas of depletion for cryptic 3′ss 30–40 nt downstream of authentic 3′ss and ∼20 nt downstream for de novo sites was followed by a second peak at 50–60 nt. The exonic depletion may be explained by a lack of suitable alternative BP adenosines within an optimal distance from de novo 3′ss, cross-exon interactions, selection against codons carrying AGs or a combination of these factors. In contrast to asymmetric distribution of cryptic and de novo 3′ss, the frequency plot of all aberrant 3′ss was virtually symmetric, with a median distance of just 1 nt from authentic 3′ss (Table 1 and data not shown). Finally, the observed frequency distribution suggests that aberrant 3′ss retaining the BPS and PPT of their authentic counterparts may be more frequent than those that use a new BPS-PPT-3′AG unit.
DBASS3: a database of aberrant 3′ss
Nucleotide sequences of all aberrant 3′ss were compiled in a new online resource available at http://www.dbass3.soton.ac.uk/. The DBASS3 web interface provides access to the database through the ‘search’ option. The user can search DBASS3 by phenotype, gene designation, mutation, location of aberrant 3′ss and their distance from authentic 3′ss. Aberrant 3′ss generated in terminal exons can also be easily retrieved. In cases in which a search identifies more than one database entry, the results page displays the gene, phenotype and location of aberrant 3′ss for all corresponding hits. The user can then choose details pages that show nucleotide sequences flanking the authentic and cryptic 3′ss, literature references with PubMed links and the estimated strength of each splice site for the tested algorithms. In addition, the details page shows how aberrant 3′ss change the reading frame of each transcript (0, +1 and +2 nt). DBASS3 visitors can also submit published data to the corresponding author and receive regular updates by email. Potential applications of DBASS3 include the optimization of computational tools for prediction of aberrant splice sites, detection of introns or exons that are frequently involved in aberrant splicing, identification of splicing mutations and aberrant 3′ss in a gene or phenotype of interest, and investigating basic mechanisms of 3′ss selection.
DISCUSSION
A high proportion of AG-creating mutations activating aberrant 3′ss
This study is the first to provide a detailed survey of mutations leading to aberrant 3′ss. It showed that the distribution of single-nucleotide substitutions roughly reflected the degree of conservation of consensus sequences that define 3′ss (Figure 1) and revealed a high proportion of mutations creating the 3′AG consensus (Table 1). The observed frequency of AG-creating mutations (42%) was considerably higher than the estimated ∼13% in the initial analysis of splicing mutations (47). Only ∼5% (n = 11, Table 2) of these mutations failed to activate de novo 3′ss in situ and instead induced one or more aberrant 3′ss upstream (36,70,93–96) or downstream (62,97,98) of the newly introduced AGs. These mutations were in position −3 (36,93), −9 (62,98), −10 (96), −14 (70), −15 (97), −17 (95) and −24 (94) relative to authentic 3′ss (Supplementary Table 1). Mutations in positions −3 and −24 directly inactivated 3′YAG and BPS, respectively, but the remaining AG-creating mutations were all in ‘AG-exclusion zones’ downstream of the BPS. The distance between predicted BP adenosine and new 3′AG/ was 9–20 nt (Supplementary Table 1). Aberrant 3′ss with the ‘BP-new AG’ distances between 9 and 16 nt were either in exons or upstream of the BPS, and new AGs were never selected as 3′ss, consistent with protein complexes bound to ∼19 nt region downstream of BP (99). In the FALDH gene (70), this distance was 20 nt and normally silent AG located 9 nt downstream of the BPS was activated by the newly created AG further 11 nt downstream. However, this putative exception can be explained by inefficient recognition of new 3′AG, which was preceded by G, unlike the remaining aberrant 3′ss (Supplementary Table 1). Alternatively, selection of aberrant 3′ss in this FALDH intron can be explained by almost identical BPS sequences arranged in tandem, with the upstream BP in the optimal distance (18 nt) from aberrant 3′ss. In contrast, wild-type AGs 6 and 7 nt downstream of the predicted BP were not selected (36,98). Although the location of AG exclusion zones is likely to be substrate-dependent, these data suggest that the average zone is between ∼7 and ∼19 nt downstream of the BP adenosine, consistent with previous studies of intervening AGs (11,19,21,99).
Selection of cryptic 3′ss upstream of BPS
If 3′ss are selected by unidirectional scanning for 3′YAG downstream of the BPS (91), why are so many cryptic 3′ss upstream of the predicted BPS used in vivo? Inspection of downstream exonic sequences in 29 cases of intronic cryptic 3′ss (Table 2) showed that eight were in terminal introns (67,100–106) (Table 1), which was significantly more frequent (χ2 = 5.6, P = 0.018) than for the remaining categories of aberrant 3′ss (Table 1), one was activated in a downstream intron (107) and two were associated with cryptic 3′ss in the following exon (108). Of the remaining sites, 13 cases either completely lacked exonic 3′YAG consensus in the context of four or more upstream pyrimidines or contained this consensus only in the last 20 nt of the exon (2,65,66,72,93,109–116) These 3′YAGs are unlikely to be used as 3′ss given inefficient inclusion of very small exons in mRNA (117) and a typical recognition site of RRM of ∼4–7 nt [(87) and references therein]. This strongly suggests that the choice of upstream 3′ss is influenced by the availability of 3′YAGs in the downstream exon and their distance from the exon end, and is consistent with unidirectional scanning that is inefficient in terminal exons. It is therefore possible that a new, competing BPS-PPT-3′AG unit is selected after the initial scanning of the downstream exon for AGs is completed. However, there has been no obvious reason for using upstream 3′ss in at least some of the remaining introns (36,118,119). These rare cases and similar examples identified in the future might provide interesting insights into cellular mechanisms that discriminate between authentic 3′ss and pseudo-acceptors.
Random distribution of the reading frames in transcripts that use aberrant 3′ss
Aberrant splicing often results in transcripts containing premature termination codons (PTCs). Such transcripts are downregulated by nonsense mediated RNA decay (NMD), which degrades PTC-containing mRNAs whose translation may be deleterious for the cell (120). Whereas EST databases over-represent alternative splicing events that maintain the reading frame (121), neither cryptic 5′ss (10) nor aberrant 3′ss (Table 1, χ2 = 8.2, 6 d.f., P = 0.2) (11) showed any bias against splice sites involving a frameshift with respect to the authentic sites, even though many mRNAs frameshifted by +1 and +2 nt would be expected to trigger NMD. These results can be explained by a great reduction of RNA downregulation in response to a PTC in transcripts containing PPT Y-to-R mutations that reduced splicing (122). In addition, NMD usually does not completely eliminate RNAs with PTCs and the activated cryptic sites that result in frameshifts can still be detected with RT–PCR, a method used by the authors of most DBASS3 records.
The maximum entropy model as a method of choice for predicting aberrant 3′ss
This study demonstrated that the ability of current computational tools to predict utilization of aberrant 3′ss is influenced by their localization and the underlying mutation. The best overall model discriminating authentic and aberrant 3′ ss was the ME model, validating previous predictions based on comparisons of genuine 3′ss and pseudo-acceptors (28). The ME model outperformed the remaining algorithms for each category of aberrant 3′ss and, together with the MM model, was the only method that could separate authentic from de novo 3′ss in introns at a significance level <0.01. Since none of the tested tools discriminated between de novo 3′ss in exons and their authentic counterparts (Table 5), these aberrant 3′ss were tested with additional algorithms, including NetGene2 (25,123) available at http://genome.cbs.dtu.dk/services/NetGene2/ and ASSP (alternative splice site predictor; http://es.embnet.org/~mwang/assp.html) method (124). NetGene2 considers more distant features that include global coding information and distances between potential splice sites, whereas ASSP is based on two neural networks pre-processed by position specific matrix scores. However, neither method revealed a difference for this category of aberrant 3′ss.
Although this study is the first to focus on 3′ss utilized in vivo as opposed to previous comparisons with pseudo-sites, there are limitations of this approach. First, even though each aberrant 3′ss was confirmed by sequencing, aberrant splicing was reliably and accurately quantified only in a subset of case reports and was highly variable from mutation to mutation, ranging from a few to hundred per cent utilization. This could be improved in future case reports and, as DBASS3 submissions permit inclusion of this information in future database records, taken into account in subsequent analyses. Second, despite the cell-specific nature of alternative splicing, measurements of aberrant and authentic RNA products have been obtained largely for blood leukocytes and only rarely for other cell types. Even with these limitations, future updates of DBASS3 may provide valuable insights into nucleotide dependencies between individual positions and distribution of trinucleotides that were significantly favoured or avoided upstream of authentic 3′ss as compared to pseudo-sites (75), as well as other motifs.
CONCLUSIONS
This work showed that (i) almost one half of aberrant 3′ss resulted from AG-creating mutations and from the introduction of guanosine, a virtually invariant nucleotide in both terminal positions of U2-dependent introns; (ii) the higher frequency of transitions over transversions observed for both positions of 3′AG can be attributed to relative di-nucleotide mutability rates rather than a detection bias resulting from a differential splicing efficiency of mutated 3′AGs; (iii) purine transitions leading to de novo sites in introns were more frequent than for de novo sites in exons; (iv) the maximum entropy model was the best model discriminating authentic and mutation-induced aberrant 3′ss used in vivo; (v) authentic counterparts of de novo 3′ss were intrinsically weak; (vi) the nucleotide sequence upstream of aberrant 3′ss had a higher purine content than corresponding authentic sites, particularly in position −3; (vii) as with authentic sites, aberrant 3′ss showed positive associations at −3 with upstream positions that may result from functional compensation of weaker interactions of U2AF with 3′TAG by stronger interactions with PPT uridines around position −11 and with more optimal BPS; (viii) the extreme rarity of AGs between positions −6 and −15 in authentic 3′ss (75,84) was violated in aberrant 3′ss, particularly 5–9 nt upstream of new intron/exon junctions; (ix) although uridines were generally under-represented upstream of aberrant 3′ss, they maintained their high numbers at position −11 and flanking nt for predicted interaction with U2AF65 or other PPT-binding proteins; (x) in this region, aberrant 3′ss had higher T-to-C and TT-to-CC ratios, required a complete lack of AGs, but tolerated more guanosines and UG/GU dinucleotides than authentic sites. Finally, the development and maintenance of DBASS3 will facilitate prediction of cryptic or de novo 3′ss in mutated disease genes, identification of introns or exons that are frequently involved in aberrant splicing, structural dissection of interactions leading to selection of 3′ss in vivo, and refinement of computational methods that estimate the splice site strength.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The author thanks Dr Christopher Smith, University of Cambridge, and Dr Ellen Copson, University of Southampton, for critical reading of the manuscript and helpful comments. The author also thanks Martin Chivers and Raj Sood for excellent technical help. This study was supported by the Juvenile Diabetes Research Foundation International (1-2006-263). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal Ö., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 3.Lopez-Bigas N., Audit B., Ouzounis C., Parra G., Guigo R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900–1903. doi: 10.1016/j.febslet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 4.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 5.Nakai K., Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 6.Cooper T.A., Mattox W. The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nissim-Rafinia M., Kerem B. Splicing regulation as a potential genetic modifier. Trends Genet. 2002;18:123–127. doi: 10.1016/s0168-9525(01)02619-1. [DOI] [PubMed] [Google Scholar]
- 8.Gouya L., Puy H., Robreau A.M., Bourgeois M., Lamoril J., Da Silva V., Grandchamp B., Deybach J.C. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nature Genet. 2002;30:27–28. doi: 10.1038/ng809. [DOI] [PubMed] [Google Scholar]
- 9.Královičová J., Gaunt T.R., Rodriguez S., Wood P.J., Day I.N.M., Vořechovský I. Variants in the human insulin gene that affect pre-mRNA splicing: is-23HphI a functional single nucleotide polymorphism at IDDM2? Diabetes. 2006;55:260–264. doi: 10.2337/diabetes.55.01.06.db05-0773. [DOI] [PubMed] [Google Scholar]
- 10.Roca X., Sachidanandam R., Krainer A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Královičová J., Christensen M.B., Vořechovský I. Biased exon/intron distribution of cryptic and de novo 3′ splice sites. Nucleic Acids Res. 2005;33:4882–4898. doi: 10.1093/nar/gki811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berglund J.A., Chua K., Abovich N., Reed R., Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 13.Ruskin B., Zamore P.D., Green M.R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 14.Singh R., Valcárcel J., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 15.Merendino L., Guth S., Bilbao D., Martinez C., Valcárcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 16.Wu S., Romfo C.M., Nilsen T.W., Green M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 17.Zorio D.A., Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 18.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 19.Smith C.W., Chu T.T., Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988;2:1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- 21.Kol G., Lev-Maor G., Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- 22.Gooding C., Clark F., Wollerton M., Grellscheid S.-N., Groom H., Smith C.W. A class of human exons with predicted distant branch points revealed by analysis of AG dinucleotide exclusion zones. Genome Biol. 2006;7:R1. doi: 10.1186/gb-2006-7-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro M.B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senapathy P., Shapiro M.B., Harris N.L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 25.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 26.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 27.Rogan P.K., Faux B.M., Schneider T.D. Information analysis of human splice site mutations. Hum. Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 28.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 29.Thanaraj T.A. Positional characterisation of false positives from computational prediction of human splice sites. Nucleic Acids Res. 2000;28:744–754. doi: 10.1093/nar/28.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nature Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- 31.Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 2006;78:291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendig I., Mohr N., Krämer F., Weber B.H. Identification of novel TP53 mutations in familial and sporadic cancer cases of German and Swiss origin. Cancer Genet. Cytogenet. 2004;154:22–26. doi: 10.1016/j.cancergencyto.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Newman P.J., Seligsohn U., Lyman S., Coller B.S. The molecular genetic basis of Glanzmann thrombasthenia in the Iraqi-Jewish and Arab populations in Israel. Proc. Natl Acad. Sci. USA. 1991;88:3160–3164. doi: 10.1073/pnas.88.8.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eng L., Coutinho G., Nahas S., Yeo G., Tanouye R., Babaei M., Dork T., Burge C., Gatti R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 35.Chen L.L., Sabripour M., Wu E.F., Prieto V.G., Fuller G.N., Frazier M.L. A mutation-created novel intra-exonic pre-mRNA splice site causes constitutive activation of KIT in human gastrointestinal stromal tumors. Oncogene. 2005;24:4271–4280. doi: 10.1038/sj.onc.1208587. [DOI] [PubMed] [Google Scholar]
- 36.Hovnanian A., Rochat A., Bodemer C., Petit E., Rivers C.A., Prost C., Fraitag S., Christiano A.M., Uitto J., Lathrop M., et al. Characterization of 18 new mutations in COL7A1 in recessive dystrophic epidermolysis bullosa provides evidence for distinct molecular mechanisms underlying defective anchoring fibril formation. Am. J. Hum. Genet. 1997;61:599–610. doi: 10.1086/515495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abramowicz M.J., Targovnik H.M., Varela V., Cochaux P., Krawiec L., Pisarev M.A., Propato F.V., Juvenal G., Chester H.A., Vassart G. Identification of a mutation in the coding sequence of the human thyroid peroxidase gene causing congenital goiter. J. Clin. Invest. 1992;90:1200–1204. doi: 10.1172/JCI115981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ejima Y., Yang L., Sasaki M.S. Aberrant splicing of the ATM gene associated with shortening of the intronic mononucleotide tract in human colon tumor cell lines: a novel mutation target of microsatellite instability. Int. J. Cancer. 2000;86:262–268. doi: 10.1002/(sici)1097-0215(20000415)86:2<262::aid-ijc17>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 39.Boot R.G., Renkema G.H., Verhoek M., Strijland A., Bliek J., de Meulemeester T.M., Mannens M.M., Aerts J.M. The human chitotriosidase gene. Nature of inherited enzyme deficiency. J. Biol. Chem. 1998;273:25680–25685. doi: 10.1074/jbc.273.40.25680. [DOI] [PubMed] [Google Scholar]
- 40.Webb J.C., Patel D.D., Shoulders C.C., Knight B.L., Soutar A.K. Genetic variation at a splicing branch point in intron 9 of the low density lipoprotein (LDL)-receptor gene: a rare mutation that disrupts mRNA splicing in a patient with familial hypercholesterolaemia and a common polymorphism. Hum. Mol. Genet. 1996;5:1325–1331. doi: 10.1093/hmg/5.9.1325. [DOI] [PubMed] [Google Scholar]
- 41.Ohno K., Tsujino A., Shen X.M., Milone M., Engel A.G. Spectrum of splicing errors caused by CHRNE mutations affecting introns and intron/exon boundaries. J. Med. Genet. 2005;42:e53. doi: 10.1136/jmg.2004.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher C.W., Lau K.S., Fisher C.R., Wynn R.M., Cox R.P., Chuang D.T. A 17-bp insertion and a Phe215Cys missense mutation in the dihydrolipoyl transacylase (E2) mRNA from a thiamine-responsive maple syrup urine disease patient WG-34. Biochem. Biophys. Res. Commun. 1991;174:804–809. doi: 10.1016/0006-291x(91)91489-y. [DOI] [PubMed] [Google Scholar]
- 43.Li S.S., Tseng H.M., Yang T.P., Liu C.H., Teng S.J., Huang H.W., Chen L.M., Kao H.W., Chen J.H., Tseng J.N., et al. Molecular characterization of germline mutations in the BRCA1 and BRCA2 genes from breast cancer families in Taiwan. Hum. Genet. 1999;104:201–204. doi: 10.1007/s004390050936. [DOI] [PubMed] [Google Scholar]
- 44.Stasia M.J., Bordigoni P., Martel C., Morel F. A novel and unusual case of chronic granulomatous disease in a child with a homozygous 36-bp deletion in the CYBA gene (A220) leading to the activation of a cryptic splice site in intron 4. Hum. Genet. 2002;110:444–450. doi: 10.1007/s00439-002-0720-8. [DOI] [PubMed] [Google Scholar]
- 45.Podkrajšek K.T., Bratanič N., Kržišnik C., Battelino T. Autoimmune regulator-1 messenger ribonucleic acid analysis in a novel intronic mutation and two additional novel AIRE gene mutations in a cohort of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients. J. Clin. Endocrinol. Metab. 2005;90:4930–4935. doi: 10.1210/jc.2005-0418. [DOI] [PubMed] [Google Scholar]
- 46.Smyth I., Wicking C., Wainwright B., Chenevix-Trench G. The effects of splice site mutations in patients with naevoid basal cell carcinoma syndrome. Hum. Genet. 1998;102:598–601. doi: 10.1007/s004390050747. [DOI] [PubMed] [Google Scholar]
- 47.Cooper D.N., Krawczak M. Human Gene Mutation. Oxford: BIOS Scientific Publishers; 1993. [Google Scholar]
- 48.Krawczak M., Cooper D.N. Single base-pair substitutions in pathology and evolution: two sides to the same coin. Hum. Mutat. 1996;8:23–31. doi: 10.1002/(SICI)1098-1004(1996)8:1<23::AID-HUMU3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 49.Parker R., Siliciano P.G. Evidence for an essential non-Watson–Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993;361:660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- 50.Dietrich R.C., Fuller J.D., Padgett R.A. A mutational analysis of U12-dependent splice site dinucleotides. RNA. 2005;11:1430–1440. doi: 10.1261/rna.7206305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deirdre A., Scadden J., Smith C.W. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995;14:3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaur R.K., Beigelman L., Haeberli P., Maniatis T. Role of adenine functional groups in the recognition of the 3′-splice-site AG during the second step of pre-mRNA splicing. Proc. Natl Acad. Sci. USA. 2000;97:115–120. doi: 10.1073/pnas.97.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weaving L.S., Christodoulou J., Williamson S.L., Friend K.L., McKenzie O.L., Archer H., Evans J., Clarke A., Pelka G.J., Tam P.P., et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am. J. Hum. Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnevie-Nielsen V., Leigh Field L., Lu S., Zheng D.J., Li M., Martensen P.M., Nielsen T.B., Beck-Nielsen H., Lau Y.L., Pociot F. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 Gene. Am. J. Hum. Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chavanas S., Bodemer C., Rochat A., Hamel-Teillac D., Ali M., Irvine A.D., Bonafe J.L., Wilkinson J., Taieb A., Barrandon Y., et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nature Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 56.Yokoi T., Shinoda K., Ohno I., Kato K., Miyawaki T., Taniguchi N. A 3′ splice site consensus sequence mutation in the intron 3 of the alpha-galactosidase A gene in a patient with Fabry disease. Jinrui Idengaku Zasshi. 1991;36:245–250. doi: 10.1007/BF01910542. [DOI] [PubMed] [Google Scholar]
- 57.Matsumura T., Osaka H., Sugiyama N., Kawanishi C., Maruyama Y., Suzuki K., Onishi H., Yamada Y., Morita M., Aoki M., et al. Novel acceptor splice site mutation in the invariant AG of intron 6 of alpha-galactosidase A gene, causing Fabry disease. Mutations in brief no. 146. Hum. Mutat. 1998;11:483. doi: 10.1002/(SICI)1098-1004(1998)11:6<483::AID-HUMU17>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A.R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992;20:1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varley J.M., Attwooll C., White G., McGown G., Thorncroft M., Kelsey A.M., Greaves M., Boyle J., Birch J.M. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene. 2001;20:2647–2654. doi: 10.1038/sj.onc.1204369. [DOI] [PubMed] [Google Scholar]
- 60.Xia K., Zheng D., Pan Q., Liu Z., Xi X., Hu Z., Deng H., Liu X., Jiang D., Deng H., et al. A novel PRPF31 splice-site mutation in a Chinese family with autosomal dominant retinitis pigmentosa. Mol. Vis. 2004;10:361–365. [PubMed] [Google Scholar]
- 61.Pasmooij A.M., Pas H.H., Deviaene F.C., Nijenhuis M., Jonkman M.F. Multiple correcting COL17A1 mutations in patients with revertant mosaicism of epidermolysis bullosa. Am. J. Hum. Genet. 2005;77:727–740. doi: 10.1086/497344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schimpf S., Schaich S., Wissinger B. Hum. Genet. Vol. 118. 2005. Activation of cryptic splice sites is a frequent splicing defect mechanism caused by mutations in exon and intron sequences of the OPA1 gene; pp. 767–771. [DOI] [PubMed] [Google Scholar]
- 63.Rickard S.J., Wilson L.C. Analysis of GNAS1 and overlapping transcripts identifies the parental origin of mutations in patients with sporadic Albright hereditary osteodystrophy and reveals a model system in which to observe the effects of splicing mutations on translated and untranslated messenger RNA. Am. J. Hum. Genet. 2003;72:961–974. doi: 10.1086/374566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schloesser M., Hofferbert S., Bartz U., Lutze G., Lammle B., Engel W. The novel acceptor splice site mutation 11396(G→A) in the factor XII gene causes a truncated transcript in cross-reacting material negative patients. Hum. Mol. Genet. 1995;4:1235–1237. doi: 10.1093/hmg/4.7.1235. [DOI] [PubMed] [Google Scholar]
- 65.Weber Y., Steinberger D., Deuschl G., Benecke R., Muller U. Two previously unrecognized splicing mutations of GCH1 in Dopa-responsive dystonia: exon skipping and one base insertion. Neurogenetics. 1997;1:125–127. doi: 10.1007/s100480050018. [DOI] [PubMed] [Google Scholar]
- 66.Hartikainen J.M., Pirskanen M.M., Arffman A.H., Ristonmaa U.K., Mannermaa A.J. A Finnish BRCA1 exon 12 4216-2nt A to G splice acceptor site mutation causes aberrant splicing and frameshift, leading to protein truncation. Hum. Mutat. 2000;15:120. doi: 10.1002/(SICI)1098-1004(200001)15:1<120::AID-HUMU31>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 67.O'Neill J.P., Rogan P.K., Cariello N., Nicklas J.A. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat. Res. 1998;411:179–214. doi: 10.1016/s1383-5742(98)00013-1. [DOI] [PubMed] [Google Scholar]
- 68.Nichols K.E., Houseknecht M.D., Godmilow L., Bunin G., Shields C., Meadows A., Ganguly A. Sensitive multistep clinical molecular screening of 180 unrelated individuals with retinoblastoma detects 36 novel mutations in the RB1 gene. Hum. Mutat. 2005;25:566–574. doi: 10.1002/humu.20184. [DOI] [PubMed] [Google Scholar]
- 69.Satokata I., Tanaka K., Miura N., Miyamoto I., Satoh Y., Kondo S., Okada Y. Characterization of a splicing mutation in group A xeroderma pigmentosum. Proc. Natl Acad. Sci. USA. 1990;87:9908–9912. doi: 10.1073/pnas.87.24.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rizzo W.B., Carney G., Lin Z. The molecular basis of Sjögren-Larsson syndrome: mutation analysis of the fatty aldehyde dehydrogenase gene. Am. J. Hum. Genet. 1999;65:1547–1560. doi: 10.1086/302681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beghini A., Castorina P., Roversi G., Modiano P., Larizza L. RNA processing defects of the helicase gene RECQL4 in a compound heterozygous Rothmund-Thomson patient. Am J. Med. Genet. A. 2003;120:395–399. doi: 10.1002/ajmg.a.20154. [DOI] [PubMed] [Google Scholar]
- 72.Bromidge T., Lowe C., Prentice A., Johnson S. p53 intronic point mutation, aberrant splicing and telomeric associations in a case of B-chronic lymphocytic leukaemia. Br. J. Haematol. 2000;111:223–229. doi: 10.1046/j.1365-2141.2000.02335.x. [DOI] [PubMed] [Google Scholar]
- 73.Jin Y., Dietz H.C., Montgomery R.A., Bell W.R., McIntosh I., Coller B., Bray P.F. Glanzmann thrombasthenia. Cooperation between sequence variants in cis during splice site selection. J. Clin. Invest. 1996;98:1745–1754. doi: 10.1172/JCI118973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villa A., Notarangelo L.D., Di Santo J.P., Macchi P.P., Strina D., Frattini A., Lucchini F., Patrosso C.M., Giliani S., Mantuano E., et al. Organization of the human CD40L gene: implications for molecular defects in X chromosome-linked hyper-IgM syndrome and prenatal diagnosis. Proc. Natl Acad. Sci. USA. 1994;91:2110–2114. doi: 10.1073/pnas.91.6.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burge C.B. Modeling dependencies in pre-mRNA splicing signals. In: Salzberg S. L., Searls D. B., Kasif S., editors. Computational methods in molecular biology. Amsterdam: Elsevier Science; 1998. pp. 129–164. [Google Scholar]
- 76.Hollins C., Zorio D.A., MacMorris M., Blumenthal T. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA. 2005;11:248–253. doi: 10.1261/rna.7221605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valcárcel J., Gaur R.K., Singh R., Green M.R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Erratum in: Science. 1996;274:21. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 78.Shen H., Kan J.L., Green M.R. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 79.Shen H., Green M.R. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Z., Luyten I., Bottomley M.J., Messias A.C., Houngninou-Molango S., Sprangers R., Zanier K., Krämer A., Sattler M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 81.Královičová J., Houngninou-Molango S., Krämer A., Vořechovský I. Branch sites haplotypes that control alternative splicing. Hum. Mol. Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 82.Zhuang Y., Weiner A.M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 83.Wu J., Manley J.L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 84.Mount S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saeys Y., Degroeve S., Aeyels D., Rouze P., Van de Peer Y. Feature selection for splice site prediction: a new method using EDA-based feature ranking. BMC Bioinformatics. 2004;5:64. doi: 10.1186/1471-2105-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo G., Hoon S., Venkatesh B., Burge C.B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15000–15005. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banerjee H., Rahn A., Davis W., Singh R. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA. 2003;9:88–99. doi: 10.1261/rna.2131603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kent O.A., Reayi A., Foong L., Chilibeck K.A., MacMillan A.M. Structuring of the 3′ splice site by U2AF65. J. Biol. Chem. 2003;278:50572–50577. doi: 10.1074/jbc.M307976200. [DOI] [PubMed] [Google Scholar]
- 89.Roscigno R.F., Weiner M., Garcia-Blanco M.A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem. 1993;268:11222–11229. [PubMed] [Google Scholar]
- 90.Singh R., Banerjee H., Green M.R. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA. 2000;6:901–911. doi: 10.1017/s1355838200000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith C.W., Porro E.B., Patton J.G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 92.Sickmier E.A., Frato K.E., Shen H., Paranawithana S.R., Green M.R., Kielkopf C.L. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Urban Z., Michels V.V., Thibodeau S.N., Donis-Keller H., Csiszar K., Boyd C.D. Supravalvular aortic stenosis: a splice site mutation within the elastin gene results in reduced expression of two aberrantly spliced transcripts. Hum. Genet. 1999;104:135–142. doi: 10.1007/s004390050926. [DOI] [PubMed] [Google Scholar]
- 94.Janssen R.J., Wevers R.A., Haussler M., Luyten J.A., Steenbergen-Spanjers G.C., Hoffmann G.F., Nagatsu T., Van den Heuvel L.P. A branch site mutation leading to aberrant splicing of the human tyrosine hydroxylase gene in a child with a severe extrapyramidal movement disorder. Ann. Hum. Genet. 2000;64:375–382. doi: 10.1046/j.1469-1809.2000.6450375.x. [DOI] [PubMed] [Google Scholar]
- 95.Fujimaru M., Tanaka A., Choeh K., Wakamatsu N., Sakuraba H., Isshiki G. Two mutations remote from an exon/intron junction in the beta-hexosaminidase beta-subunit gene affect 3′-splice site selection and cause Sandhoff disease. Hum. Genet. 1998;103:462–469. doi: 10.1007/s004390050851. [DOI] [PubMed] [Google Scholar]
- 96.Lucarini L., Giusti B., Zhang R.Z., Pan T.C., Jimenez-Mallebrera C., Mercuri E., Muntoni F., Pepe G., Chu M.L. A homozygous COL6A2 intron mutation causes in-frame triple-helical deletion and nonsense-mediated mRNA decay in a patient with Ullrich congenital muscular dystrophy. Hum. Genet. 2005;117:460–466. doi: 10.1007/s00439-005-1318-8. [DOI] [PubMed] [Google Scholar]
- 97.Mayer K., Ballhausen W., Leistner W., Rott H. Three novel types of splicing aberrations in the tuberous sclerosis TSC2 gene caused by mutations apart from splice consensus sequences. Biochim. Biophys. Acta. 2000;1502:495–507. doi: 10.1016/s0925-4439(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 98.Thomas P.M., Cote G.J., Wohllk N., Haddad B., Mathew P.M., Rabl W., Aguilar-Bryan L., Gagel R.F., Bryan J. Mutations in the sulfonylurea receptor gene in familial persistent hyperinsulinemic hypoglycemia of infancy. Science. 1995;268:426–429. doi: 10.1126/science.7716548. [DOI] [PubMed] [Google Scholar]
- 99.Chua K., Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marchetti C., Patriarca P., Solero G.P., Baralle F.E., Romano M. Genetic studies on myeloperoxidase deficiency in Italy. Jpn. J. Infect. Dis. 2004;57:S10–12. [PubMed] [Google Scholar]
- 101.Cladaras C., Hadzopoulou-Cladaras M., Felber B.K., Pavlakis G., Zannis V.I. The molecular basis of a familial apoE deficiency. An acceptor splice site mutation in the third intron of the deficient apoE gene. J. Biol. Chem. 1987;262:2310–2315. [PubMed] [Google Scholar]
- 102.Antonarakis S.E., Irkin S.H., Cheng T.C., Scott A.F., Sexton J.P., Trusko S.P., Charache S., Kazazian H.H., Jr beta-Thalassemia in American Blacks: novel mutations in the ‘TATA’ box and an acceptor splice site. Proc. Natl Acad. Sci. USA. 1984;81:1154–1158. doi: 10.1073/pnas.81.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Atweh G.F., Anagnou N.P., Shearin J., Forget B.G., Kaufman R.E. Beta-thalassemia resulting from a single nucleotide substitution in an acceptor splice site. Nucleic Acids Res. 1985;13:777–790. doi: 10.1093/nar/13.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong C., Antonarakis S.E., Goff S.C., Orkin S.H., Forget B.G., Nathan D.G., Giardina P.J., Kazazian H.H., Jr Beta-thalassemia due to two novel nucleotide substitutions in consensus acceptor splice sequences of the beta-globin gene. Blood. 1989;73:914–918. [PubMed] [Google Scholar]
- 105.Otterness D.M., Szumlanski C.L., Wood T.C., Weinshilboum R.M. Human thiopurine methyltransferase pharmacogenetics. Kindred with a terminal exon splice junction mutation that results in loss of activity. J. Clin. Invest. 1998;101:1036–1044. doi: 10.1172/JCI1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wassif C.A., Maslen C., Kachilele-Linjewile S., Lin D., Linck L.M., Connor W.E., Steiner R.D., Porter F.D. Mutations in the human sterol delta7-reductase gene at 11q12-13 cause Smith-Lemli-Opitz syndrome. Am. J. Hum. Genet. 1998;63:55–62. doi: 10.1086/301936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bulman M.P., Harries L.W., Hansen T., Shepherd M., Kelly W.F., Hattersley A.T., Ellard S. Abnormal splicing of hepatocyte nuclear factor 1 alpha in maturity-onset diabetes of the young. Diabetologia. 2002;45:1463–1467. doi: 10.1007/s00125-002-0919-1. [DOI] [PubMed] [Google Scholar]
- 108.Darling T.N., Yee C., Koh B., McGrath J.A., Bauer J.W., Uitto J., Hintner H., Yancey K.B. Cycloheximide facilitates the identification of aberrant transcripts resulting from a novel splice-site mutation in COL17A1 in a patient with generalized atrophic benign epidermolysis bullosa. J. Invest. Dermatol. 1998;110:165–169. doi: 10.1046/j.1523-1747.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 109.Shah A.B., Chernov I., Zhang H.T., Ross B.M., Das K., Lutsenko S., Parano E., Pavone L., Evgrafov O., Ivanova-Smolenskaya I.A., et al. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype-phenotype correlation, and functional analyses. Am. J. Hum. Genet. 1997;61:317–328. doi: 10.1086/514864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bouma P., Cabral W.A., Cole W.G., Marini J.C. COL5A1 exon 14 splice acceptor mutation causes a functional null allele, haploinsufficiency of alpha 1(V) and abnormal heterotypic interstitial fibrils in Ehlers-Danlos syndrome II. J. Biol. Chem. 2001;276:13356–13364. doi: 10.1074/jbc.M011742200. [DOI] [PubMed] [Google Scholar]
- 111.Verselis S.J., Rheinwald J.G., Fraumeni J.F., Jr, Li F.P. Novel p53 splice site mutations in three families with Li-Fraumeni syndrome. Oncogene. 2000;19:4230–4235. doi: 10.1038/sj.onc.1203758. [DOI] [PubMed] [Google Scholar]
- 112.Vockley J., Rogan P.K., Anderson B.D., Willard J., Seelan R.S., Smith D.I., Liu W. Exon skipping in IVD RNA processing in isovaleric acidemia caused by point mutations in the coding region of the IVD gene. Am. J. Hum. Genet. 2000;66:356–367. doi: 10.1086/302751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mardy S., Miura Y., Endo F., Matsuda I., Sztriha L., Frossard P., Moosa A., Ismail E.A., Macaya A., Andria G., et al. Congenital insensitivity to pain with anhidrosis: novel mutations in the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor. Am. J. Hum. Genet. 1999;64:1570–1579. doi: 10.1086/302422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teng Y.N., Wang T.R., Hwu W.L., Lin S.P., Lee-Chen G.J. Identification and characterization of -3c-g acceptor splice site mutation in human alpha-L-iduronidase associated with mucopolysaccharidosis type IH/S. Clin. Genet. 2000;57:131–136. doi: 10.1034/j.1399-0004.2000.570207.x. [DOI] [PubMed] [Google Scholar]
- 115.Hamed S., Sutherland-Smith A., Gorospe J., Kendrick-Jones J., Hoffman E. DNA sequence analysis for structure/function and mutation studies in Becker muscular dystrophy. Clin. Genet. 2005;68:69–79. doi: 10.1111/j.1399-0004.2005.00455.x. [DOI] [PubMed] [Google Scholar]
- 116.Bunge S., Steglich C., Zuther C., Beck M., Morris C.P., Schwinger E., Schinzel A., Hopwood J.J., Gal A. Iduronate-2-sulfatase gene mutations in 16 patients with mucopolysaccharidosis type II (Hunter syndrome) Hum. Mol. Genet. 1993;2:1871–1875. doi: 10.1093/hmg/2.11.1871. [DOI] [PubMed] [Google Scholar]
- 117.Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol. Cell. Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morgan N.V., Tipping A.J., Joenje H., Mathew C.G. High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am. J. Hum. Genet. 1999;65:1330–1341. doi: 10.1086/302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Messiaen L.M., Callens T., Mortier G., Beysen D., Vandenbroucke I., Van Roy N., Speleman F., Paepe A.D. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 120.Maquat L.E. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 121.Resch A., Xing Y., Alekseyenko A., Modrek B., Lee C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation; Nucleic Acids Res.; 2004. pp. 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gudikote J.P., Imam J.S., Garcia R.F., Wilkinson M.F. RNA splicing promotes translation and RNA surveillance. Nature Struct. Mol. Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- 123.Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouze P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang M., Marin A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.