Abstract
With the aim of investigating how motor proteins negotiate DNA nanostructures, we produced test circuits based on recombination intermediates in which 1D translocation across a Holliday junction (HJ) could be assessed by subsequent triplex displacement signals on each DNA arm. Using the EcoR124I restriction-modification enzyme, a 3′–5′ double-strand DNA (dsDNA) translocase, we could show that the motor will tend to follow its translocated strand across a junction. Nonetheless, as the frequency of junction bypass events increases, the motor will occasionally jump tracks.
INTRODUCTION
DNA, by virtue of its self-assembly properties, has proven to be an excellent scaffolding material for the assembly of 2D and 3D structures on the nanometer scale [e.g. see Ref. (1) for recent, striking examples]. These structures may eventually provide the architecture for nanoscale devices or circuits. Another advantage of DNA is the availability of numerous nucleic acid processing enzymes which could be used to manipulate the building blocks both before and after construction. One important class of DNA enzymes are the energy-dependent translocases that catalyse directional 1D motion along the DNA lattice. These motors could be used to explore the nanostructure; e.g. delivering a cargo or enzyme activity from one location to another (2).
A classical view of the DNA translocases is that they move along a single isolated polynucleotide strand—they are single-strand DNA (ssDNA) motors (3). Such enzymes, by their very nature, must unwind duplex DNA to access a ssDNA track. This necessity for a double-strand DNA (dsDNA) to ssDNA transition may cause significant problems in utilizing these enzymes in the context of dsDNA circuits: protein motion will dissemble the nanostructure. However, it is becoming increasingly clear that a large number of translocases move on DNA without the necessity for unwinding—they are true dsDNA motors (4). These enzymes could be utilized to explore nanostructures without altering the underlying track. However, the molecular mechanism of dsDNA translocation has not been examined in as much detail as ssDNA translocation. For example, it is not clear how these proteins might deal with non-linear regions of DNA, such as kinks and branches, which are necessary for the construction of nanostructures (1). Here we have addressed this by examining what happens when a dsDNA translocase—the Type I restriction enzyme EcoR124I—comes up against a four branch junction.
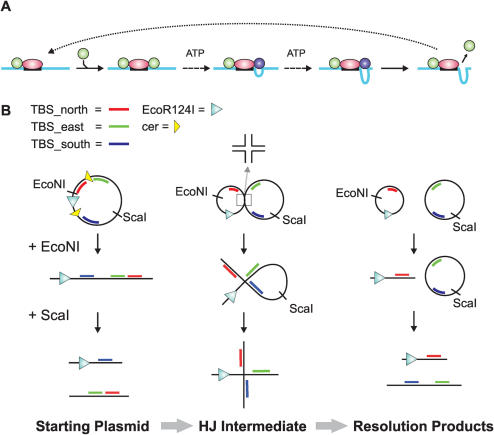
EcoR124I is a hetero-oligomeric DNA processing motor involved in protecting bacteria from infection by viruses. It comprises two main protein components: a core methyltransferase (MTase), which undertakes sequence-specific DNA recognition and modification; and two HsdR subunits which are loaded by the MTase onto the adjacent non-specific DNA and which carry out ATP hydrolysis, DNA translocation and endonuclease activities [Figure 1A, Refs. (4–6)]. ATP hydrolysis and DNA translocation are conferred by superfamily 2 (SF2) helicase motifs in HsdR (7). However, the helicase motifs are not involved in DNA unwinding. Instead, they are used to contact and translocate the 3′–5′ strand of intact dsDNA (4). Contacts to the 5′–3′ strand are not directly utilized for motion but are used to stabilize the motor on the DNA. Stretches of ssDNA can be traversed, albeit with a higher probability of dissociation (4). Each HsdR motor translocates independently on the DNA away from the core MTase. Because the HsdR subunits remain bound to the MTase, which in turn remains bound to the EcoR124I binding site, two DNA loops are extruded [Figure 1A, Ref. (8)]. Termination of a translocation event results in HsdR dissociation from both the DNA and the MTase, so that each new motor event is initiated by the MTase loading a new HsdR molecule from solution (6). Consequently, the frequency of motor events is simply a function of the relative HsdR concentration.
Figure 1.
DNA substrates for the analysis of junction bypass by a Type I restriction enzyme. (A) DNA translocation by a Type I restriction enzyme. DNA shown as a blue line with a restriction site as a black box; EcoR124I MTase shown as a pink oval; HsdR shown as a green (static) or blue (translocating) circle. For the sake of clarity, translocation on only one side of the site is shown. See main text for more details. (B) Generation of DNA junction substrates. E.coli RM40 cells were transformed with pLKS7 and recombination induced (see Materials and Methods for further details). Cells were harvested and the DNA purified as a mixture of starting plasmid (unrecombined pLKS7), HJ intermediate and resolution products (recombined circular products). Treatment with either EcoNI or EcoNI plus ScaI produced the substrates shown. The DNA was further purified using standard techniques, without separation of the different species. Approximate locations of the TFOs are given by the blue, red and green lines, respectively. More detailed information on spacings is given in Figures 3 and 5.
EcoR124I is a prototype dsDNA translocase (4), that has a number of advantages for nanotechnological studies: the protein components are readily prepared, easy to handle and can be analysed using both bulk solution and single molecule techniques; the motor events are long-lived so allowing long DNA distances to be explored [the average translocated distance is ∼5000 bp at 25°C and saturating ATP, Ref. (5)]; the core MTase which loads each new HsdR motor event is also long-lived, remaining at the recognition site for ∼30 min before dissociating (6); and, DNA cleavage and translocation can be readily uncoupled by using endonuclease mutants, which have the same motor properties as the wild-type enzyme (9).
In a previous study of Type I restriction enzymes (10), endonuclease activity was measured on substrates containing DNA junctions. It was suggested based on the data that junction bypass could occur, although translocation was not directly measured and the direction of bypass could not be assessed. However, we can now accurately measure translocation of Type I enzymes in bulk solution using a triplex displacement assay, in which a specifically bound DNA triplex is displaced by motion of the HsdR past the binding site [(4,6,9,11), and see text below]. To directly test what happens when EcoR124I reaches a junction, we utilized an inducible Xer recombinase system described in Ref. (12) to generate, in vivo, DNA substrates containing an EcoR124I binding site, a Holliday junction (HJ) and three independent triplex binding sites (Figure 1B and Supplementary Data). Our results show that bypass of an HJ can occur, but that, in some cases, the EcoR124I motor will jump tracks. This conclusion has important implications for the application of motor proteins in DNA circuits.
MATERIALS AND METHODS
Construction of pLKS7
Unless stated otherwise, all DNA manipulations were carried out using standard procedures. Oligonucleotides were supplied by Sigma Genosys. Manipulation of the pJB43a-based plasmids was carried out in an Escherichia coli xer strain, FC33 (pJB43a and FC33 were kindly provided by Dr Sean Colloms). A full description of the design and testing of the triplex binding sites (TBSs #14, #125 and #127) and the triplex forming oligonucleotides (TFOs #14, #125 and #127) used in this study is given in the Supplementary Data. For clarity, the triplex sequences are re-named according to their nominal location on the final HJ substrates as follows: TBS14 and TFO14 as TBS_south and TFO_south, respectively; TBS125 and TFO125 as TBS_north and TFO_north, respectively; and, TBS127 and TFO127 as TBS_east and TFO_east, respectively.
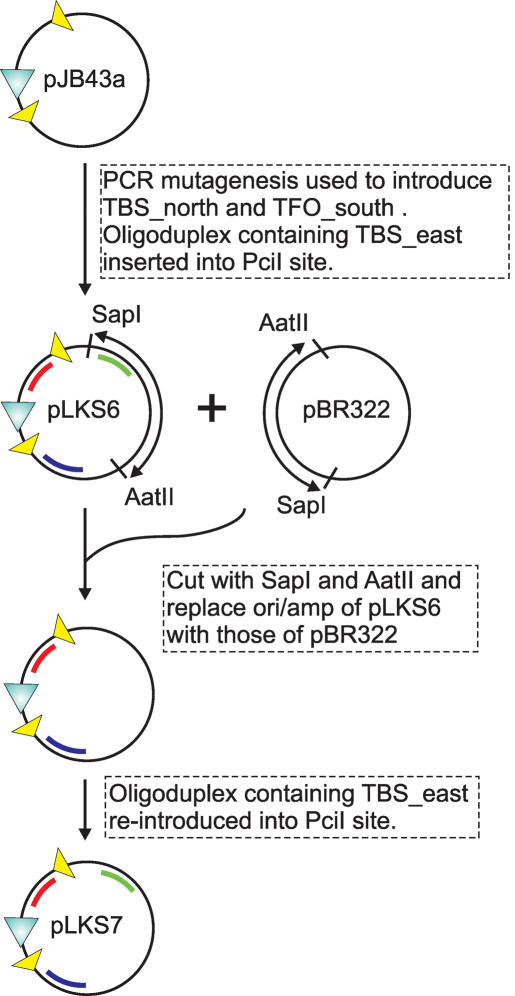
pLKS7 was created from pJB43a as follows (Figure 2): Primers ls41up (5′-GATATCAAGAAAAGAAAGAAGAAAGAAACCAGGCGCCATTCGCCATTCAG-3′) and ls41low (5′-GTTTCTTTCTTCTTTCTTTTCTTGATATCATGCGGTATTTTCTCCTTACGC-3′) were used to introduce TBS_south (in bold) at position 56 of pJB43a by ExSite PCR-based site-directed mutagenesis (Stratagene). TBS_north (in bold) was then introduced at position 1649 by a further round of ExSite mutagenesis with primers ls42up (5′-CCATGGCAAAGAAGAAGAAAAGAAAAGAACCTCGAGCAAGACGTTTCCGTTG-3′) and ls42low (5′-GTTCTTTTCTTTTCTTCTTCTTTGCCATGGGCCGCGATTAAATTCCAACATG-3′). TBS_east (in bold) was introduced into the PciI site of the resulting plasmid by insertion of the annealed oligoduplexes ls33F (5′-CATGCTCTAGACAAAGAAAGAAAAGAAGAAAGAAC-3′) and ls33R (5′-CATGGTTCTTTCTTCTTTTCTTTCTTTGTCTAGAG-3′) to generate pLKS6. However, pLKS6 did not undergo successful isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible recombination irrespective of induction conditions (data not shown). This was most likely a result of the high copy number of the plasmid (pJB43a is based on pUC19; Dr Sean Colloms, personal communication). To overcome this, the region between the AatII and SapI sites of pLKS6 were replaced with the equivalent region of pBR322. This reduces the plasmid copy number by introducing a regulated ColE1 origin. TBS_east was then re-introduced into the PciI site of the new pBR322 segment, as described above, to generate pLKS7, which showed the expected levels of IPTG-inducible recombination (see below).
Figure 2.
Construction of pLKS7. For full details, see Materials and Methods.
Recombination of pLKS7
E.coli RM40 cells [(12); kindly provided by Dr Sean Colloms] were transformed with pLKS7 and incubated overnight on 1.6% (w/v) Luria–Bertani (LB)-Agar supplemented with 50 μg/ml ampicillin and 1% (w/v) glucose. LB media [supplemented with 1% (w/v) glucose and 50 μg/ml ampicillin] was inoculated with the transformed cells at a density of 1 colony/ml and incubated at 37°C and 250 r.p.m. until the cells reached an OD600 of ∼0.5. The culture was then inoculated with 1 mM IPTG and incubation continued for a further 70 min [this time was determined empirically to give the maximum yield of HJ intermediate, Ref. (12)—data not shown]. The total plasmid DNA was then purified using standard procedures (Qiagen maxiprep). DNA substrates for the triplex assays were prepared by digestion with EcoNI and ScaI (as indicated in the text), followed by extraction with phenol/chloroform and chloroform, and precipitation with isopropanol.
EcoR124I-directed displacement of triplexes analysed by agarose gel electrophoresis
Triplexes were formed by incubating 50 nM DNA and 25 nM 32P-labelled-TFO in buffer MM [10 mM MES and 12.5 mM MgCl2 (pH 5.5)] overnight at 20°C. Triplex DNA 5 nM was incubated with 40 nM MTase and either 1 or 90 nM HsdR at 20°C in 1× buffer R [50 mM Tris–HCl, 10 mM MgCl2 and 1 mM DTT (pH 8.0)], and the reactions initiated by the addition of 4 mM ATP and 100 μM S-adenosyl methionine (AdoMet). Aliquots were removed at the times indicated and analysed as described previously (11). In brief, reactions were quenched using GSMB buffer [15% (w/v) glucose, 3% (w/v) SDS, 250 mM MOPS (pH 5.5), 0.4 mg/ml bromophenol blue] and analysed in 1% (w/v) agarose gels [40 mM Tris–acetate, 5.0 mM sodium acetate and 1.0 mM MgCl2 (pH 5.5)] at 10 V/cm for 2 h at 4°C. Gels were fixed in 5% (v/v) acetic acid, 50% (v/v) methanol for at least 1 h and air-dried overnight between sheets of cellophane. Dry gels were scanned in a Molecular Dynamics PhosphorImager, and the image analysed with ImageQuant software (MD) to determine the volume of each ‘band’. The amount of TFO bound to the HJ DNA in each sample was first calculated as a fraction of the total volume from all bound and free species. The relative amount of TFO bound to the HJ DNA was then calculated as described in the figure legends. The principals behind the triplex displacement assay are discussed further in the Results section.
RESULTS
Experimental design
For a 3′–5′ dsDNA translocase, such as EcoR124I [Ref. (4)], a number of events could occur upon reaching a junction (10): (i) the motor may stall; (ii) the motor may bypass the junction whilst remaining in tight contact with the translocated 3′–5′ strand, so following just one downstream DNA arm (the Southern arm as defined for the HJ in Figure 3) or (iii) the motor may bypass the junction but jump DNA tracks so following any downstream DNA arm. These alternative events can de differentiated using the triplex dissociation assay (9,11). This assay can be used in both discontinuous [(11), as here] or continuous modes (9), and has been used to successfully analyse motion on DNA by a number of different dsDNA translocases. The general principal of the assay is that a DNA ‘roadblock’ placed in the path of the translocating motor is displaced upon collision with the enzyme. The CT-rich TFOs used here will only bind stably under acidic conditions (pH < 6). Yet upon subsequent dilution into reaction buffer (pH 8), the triplexes are stabilized by the presence of Mg2+ ions and can remain bound indefinitely (11). Upon dissociation, the cytosines are deprotonated and the TFO is unable to re-bind. This irreversibility means that the triplex assay gives an unambiguous binary ‘on/off’ signal that an enzyme has passed a given point on the DNA. Therefore, by placing triplexes on each DNA arm downstream of a junction, we can assess whether the motor can bypass the junction and, if so, which tracks have been followed. Note that, Type I enzymes do not displace TFOs ‘at a distance’—there must be a collision event.
Figure 3.
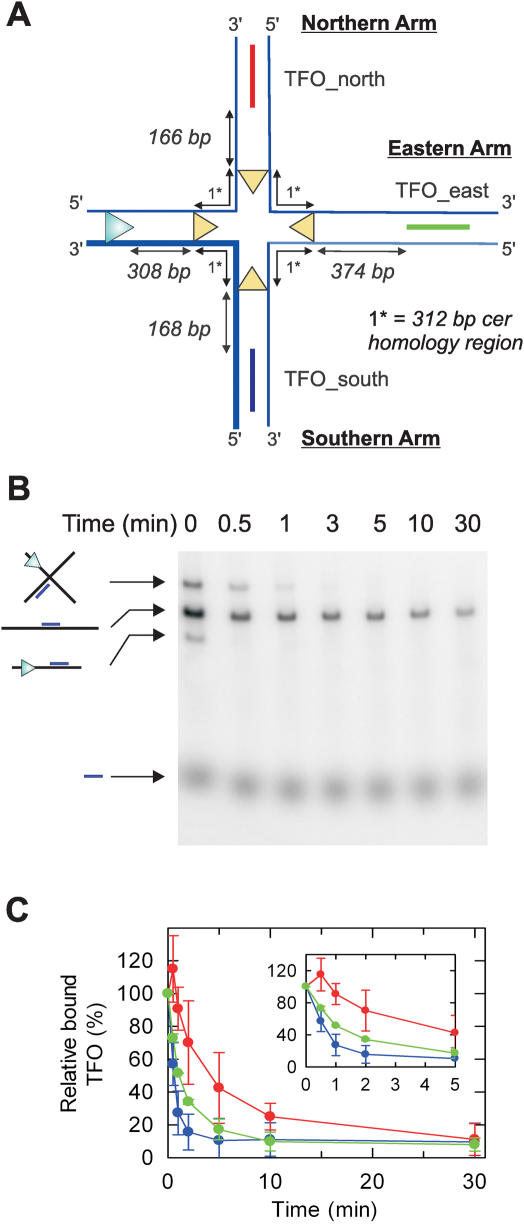
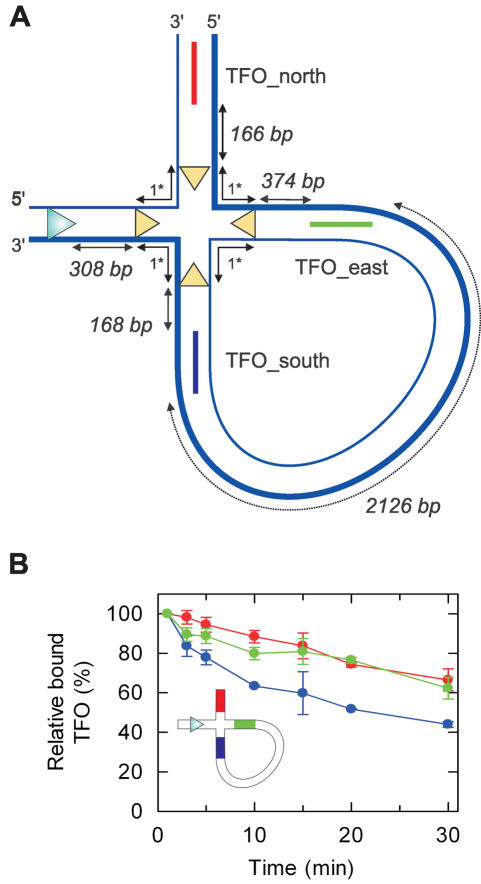
Translocation on χ-substrate DNA at high HsdR concentrations. (A) Structure of the HJ region of the χ-intermediate. The continuous 3′–5′ translocated strand is indicated as a thicker line. Note, the HJ is mobile within the homologous regions bounded by the cer sites (yellow triangles). Translocation by EcoR124I away from the HJ does not interfere with the reaction and is not considered. (B) Representative agarose gel showing separation of the DNA species produced by EcoNI/ScaI digestion and bound by TFO_south. DNA (5 nM) was pre-bound with 32P-labelled TFO_south as described (11), and then treated with 90 nM HsdR and 40 nM MTase in buffer R for the times shown. Images were captured using a Molecular Dynamics Typhoon 9200 PhosphorImager and quantified using ImageQuant software. Note the rapid displacement of TFO_south from the linear DNA. (C) TFO-binding to the χ-species was calculated relative to the zero time point sample, which was set up separately. Because the χ species represent only a fraction of the total sample DNA, experimental variation in sample composition leads to the percentage of bound TFO on the HJ substrate exceeding 100% in some cases. Each triplex was analysed in a separate reaction. Data represents the average of ≥2 independent experiments. The inset shows the first 5 min of the reactions.
Janscak et al. (10) previously analysed the DNA cleavage activity of the Type I enzyme EcoAI on HJ substrates created by Xer recombination in vivo (12). We chose to use the same approach to generate the DNA substrates for our study. To allow triplex binding to each arm of the HJ downstream of the EcoR124I binding site on a single DNA construct, we designed a family of TFOs that had mutually exclusive binding to their respective TBSs and which had similar displacement kinetics upon collision with EcoR124I (in this study we used three sequences; #14, #125 and #127 from the Supplementary Data). The resulting plasmid, pLKS7 (Figure 2, Materials and Methods), was isolated as a mixture of unrecombined substrate DNA, the HJ intermediate and recombined products (12). We then linearized the DNA mixture, as indicated, with commercially available restriction enzymes using standard procedures (Figure 1B). To avoid losses in DNA yields that would accompany further separation of the species, the mixture of linear DNA was purified and used directly. Accordingly, the triplexes formed were bound to a mixture of different DNA, some, which have Type I recognition sites, some that do not (Figure 1). This ruled out using the continuous assay (9) to monitor triplex binding as the signals from each DNA species would merge. Instead, the different DNA species were resolved using gel electrophoresis, allowing us to follow the occupancy of each individual DNA species relative to the free triplex. Since we are only concerned with events at DNA junctions in this study, and because events on the DNA species are independent of each other, we have only presented binding data for the TFOs attached to the HJ substrates. We did not fit the data to kinetic models to extract translocation rate constants as, due to model complexity, indistinguishable profiles would be returned using completely different schemes [similar limitations were encountered in Ref. (4)].
The locations of the TBSs relative to the EcoR124I binding site and branch point in the final χ- and α-structures are shown in Figures 3A and 5A, respectively. It should be noted that the branch point is mobile within the cer crossover region (∼312 bp), but cannot passively enter the heterologous region beyond the sites. The occupancy of each TBS was examined in individual reactions.
HJ bypass at high HsdR concentrations
We first examined the translocation activity of EcoR124I on the DNA mixture generated by cleaving the recombination intermediates with EcoNI and ScaI (Figure 1B). This mixture contained the χ-substrate shown in Figure 3A. Cleavage at a χ structure on linear DNA by a Type I enzyme has been observed, albeit with slow kinetics; on the order of <1 per hour (10). We also observed χ-dependent cleavage with very similar slow kinetics (data not shown). However, for the translocation studies here, we analysed our substrates over time courses of 30 min, during which very little cleavage occurred. Moreover, the same results could be obtained using nuclease-deficient versions of EcoR124I (data not shown).
32P-labelled triplexes were bound to the DNA mixtures as described [(11), Materials and Methods), and then mixed with 90 nM HsdR, 40 nM MTase, 4 mM ATP and 100 μM AdoMet. Reactions were initiated by the addition of ATP and AdoMet. With the concentrations of proteins used here, termination of a translocation event by dissociation of the HsdR will be followed almost immediately by re-initiation of a new event by the recruitment, from solution, of a new HsdR molecule (6). Agarose gel electrophoresis was used to separate the different DNA species, an example of which is shown in Figure 3B.
By analysing the relative proportions of TFO bound to each DNA species, we could calculate the absolute amount of triplex bound to the HJ species as a function of time. These data are presented for each TFO in Figure 3C. The triplex that was displaced most rapidly from the HJ DNA was TFO_south, which is located in the Southern arm of the branch point. This observation makes sense given that EcoR124I is a 3′–5′ translocase, as motor events that remain in close contact with the translocated strand will, by necessity, follow this route. However, the timescale of displacement was significantly slower than expected given the direct distance between the EcoR124I site and TBS (more than 80% triplex would be displaced within 10 s on an equivalent linear DNA, [Refs. (5,6,9,11)]. The simplest explanation for this observation is that the presence of the HJ introduces a slow translocation step. Similar results were obtained when other roadblocks, such as gaps in the DNA, were examined (4). We therefore suggest that EcoR124I can bypass a junction but that doing so requires a stall in translocation that increases the apparent translocation time.
We also observed displacement of TFO_east and TFO_north, albeit with slower kinetics than TFO_south. This indicates that in some stall events the motor did not translocate the Southern arm of the χ site but instead jumped tracks and translocated along either the Eastern or Northern arms. The kinetics of displacement suggests that there is an order of preference for junction bypass of PSouthern > PEastern> PNorthern.
HJ bypass at low HsdR concentrations
In the experiments above, the HsdR motor was at a high concentration relative to the DNA-MTase complex, such that association with the MTase is not rate-limiting and the motor event frequency is high (6). With the average lifetime of a motor event being ∼10 s [Ref. (5)], this means that displacement of TFO_east and TFO_north will have occurred only after multiple HsdR termination/dissociation/re-initiation events. Accordingly, many motor events will have bypassed the junction and visited the Southern arm before a rare track jumping event occurred. To investigate this further, we tested triplex displacement using a limiting concentration of HsdR (i.e. 1 nM [HsdR], 5 nM [MTase-DNA]). When a motor event terminates under these conditions, it is unlikely that another HsdR molecule will initiate immediately on the same DNA molecule. The result is that the kinetics of triplex displacement is slow and is limited by the HsdR concentration and by motor processivity (6). Because of their low probability, it is unlikely that track hopping events will be sampled under these conditions and the majority of events observed will be those that follow the translocated strand. Indeed, the results in Figure 4 clearly show that whilst displacement of TFO_south from the HJ DNA exceeded 50% in 30 min, virtually no displacement of TFO_east or TFO_north above background levels was observed in the same time. Therefore, in a single bypass event the motor is significantly more likely to follow the intact 3′–5′ strand. The displacement of TFO_east and TFO_north observed in Figure 3 must represent rare events that are only observed because the elevated HsdR concentrations result in a correspondingly high number of motor events.
Figure 4.
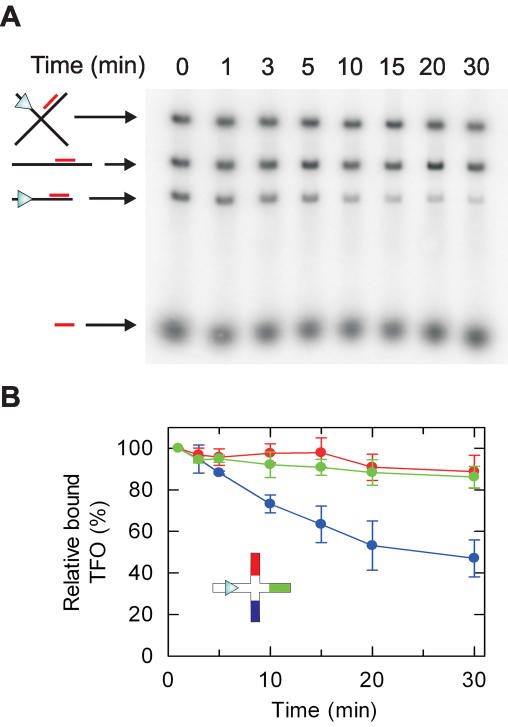
Translocation on χ-substrate DNA at low HsdR concentrations. (A) Representative agarose gel showing separation of the different DNA produced by EcoNI/ScaI digestion and species bound by TFO_north. DNA (5 nM) was pre-bound with 32P-labelled TFO_north as described (11), and then treated with 1 nM HsdR and 40 nM MTase in buffer R for the times shown. Images were captured using a Molecular Dynamics Typhoon 9200 PhosphorImager and quantified using ImageQuant software. (B) TFO-binding to the χ-species was calculated relative to the 1 min sample (with the bound TFO at t = 60 s set to 100%). Each triplex was analysed in a separate reaction. Data represents the average of ≥2 independent experiments.
EcoR124I translocation around an alpha structure
From the results above we can conclude that the HsdR motor can bypass an HJ, in the majority of cases following the 3′–5′ DNA strand along the Southern arm. If we were to covalently connect this Southern arm to one of the other arms, then we might predict that the motor will continue along the 3′–5′ strand and displace another triplex with enhanced efficiency. To test this we cleaved our HJ intermediate with only one Type II restriction enzyme, EcoNI, to generate an α-structure (Figure 1B). In this geometry (Figure 5A), translocation down the Southern arm could result in some long-lived motor events reaching TFO_east on the Eastern arm, possibly followed by TFO_north on the Northern arm. We tested this substrate with limiting HsdR concentrations where track jumping at the junction will be highly unlikely (viz. Figure 4). Using these conditions we now observed ∼40% displacement of both TFO_east and TFO_north in 30 min (Figure 5B); significantly more than observed on the χ-substrate (Figure 4B). TFO_south was displaced with the same rate and amplitude as before.
Figure 5.
Translocation on α-substrate DNA at low HsdR concentrations. (A) Structure of the HJ region of the α-intermediate. (B) Triplex displacement from the α-structures was analysed as in Figures 3 and 4. TFO-binding to the α-species was calculated as in Figure 4. Each triplex was analysed in a separate reaction. Data represents the average of ≥2 independent experiments.
The simplest explanation for the increased displacement of triplexes on the Eastern and Northern arms of a α-structure is that the connection of the Southern and Eastern arms produces a continuous 3′–5′ tracking strand starting at the EcoR124I binding site and passing, in order, TFO_south, TFO_east and TFO_north (Figure 5A). The slower displacement of TFO_east and TFO_north reflects the longer translocation distance from the EcoR124I site. However, the difference in distance between the EcoR124I binding site and TFO_east and TFO_north (852 bp, compared to 2126 bp between TFO_south and TFO_east) is probably not sufficient to clearly resolve the order of displacement of the Eastern and Northern TFOs from the kinetics obtained using the gel assay.
DISCUSSION
We demonstrate here that the EcoR124I restriction enzyme is a motor protein that is capable of bypassing an HJ, as suggested previously for the EcoAI Type I restriction enzyme (10). In the same study it was also suggested that EcoAI may cause branch migration of the HJ through regions of DNA homology (10). We can rule out migration of the HJ into the heterologous DNA as if, this were to occur, the Northern and Southern arms would be pumped through the motor at the same time with the result that TFO_north and TFO_south would be displaced simultaneously, whilst TFO_east would move away from the motor and would never be displaced; this outcome was never observed. Moreover, mapping of the HJ post-EcoR124I treatment did not indicate that the HJ had been translocated to any significant extent (see Supplementary Data).
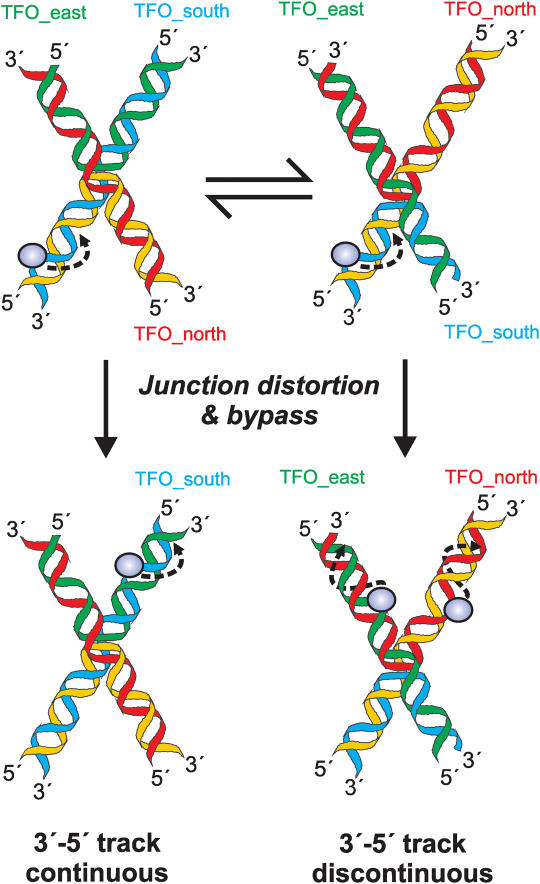
In the solution conditions used here (i.e. in the presence of millimolar concentrations of divalent cations), an HJ will adopt a stacked-X structure that exists in equilibrium between two conformers (13,14), as shown in Figure 6. However, it is more than likely that collision of the HsdR with the branch point will strain and deform the HJ into an alternative structure. Since EcoR124I can readily bypass bulky lesions in the non-tracked, complementary strand (4), we suggest that by remaining in close contact with the 3′–5′ strand the motor domain can negotiate around a distorted junction. The step size of EcoR124I has been estimated as 0.3–0.7 nm [Ref. (4)] and it may be that this relatively short distance constrains the motor domain into stepping along the covalently continuous 3′–5′ strand. The occasional track jumping event may reflect alternative HJ conformations in which DNA distortion causes the motor domain to step onto a distal, non-covalently attached strand.
Figure 6.
Before and after states of a dsDNA translocase during bypass of a branched junction. Schematics of the stacked-X conformers are based on the crystal structure of Ortiz–Lombardia et al. (13). Translocation of a dsDNA motor (blue circle) in a 3′–5′ direction towards the HJ occurs from the DNA arm containing the motor initiation site (blue/yellow strands). The black broken lines show the direction of motion. Upon reaching the junction, the motor will stall and, most likely, distort the DNA. Translocation will then continue on one of three tracks dependent upon the geometry of the branchpoint. The direction chosen by the motor reflects which strand is closest in space when the motor makes a forward step. As a simple example: In one stacked-X conformer (left-hand structures), the 3′–5′ translocated strand (blue) follows a continuous axial path approximating that found in B-form DNA. Here, forward steps by the motor favours translocation along the Southern arm; In the other conformer (right-hand structures), the 3′–5′ strand is kinked and folded-back and the translocated strand now follows a discontinuous path, coaxial with a strand from the Northern arm (red strand). Here, forward steps by the motor favours track jumping and translocation along either the Northern or Eastern arm (both possibilities shown on the same DNA).
From our analysis of EcoR124I translocation, we can show that a protein motor can remain in close contact with its DNA track and can be made to follow a DNA junction without disrupting significantly the nucleic acid structure. However, there were also instances where EcoR124I readily jumped tracks. Whilst we could favour one event over another by reducing the frequency of motor events, this, of course, has kinetic consequences as the frequency of bypass events also reduces (compare time bases in Figures 3 and 4). Nonetheless, it is clear that the helicase-related dsDNA translocases, of which EcoR124I is an archetype, could be adapted to faithfully explore an intact DNA nanostructure. Indeed, we were able to show that EcoR124I will follow an α-structure, despite the topological complexity of its loop translocation mechanism (15). The high processivity of the EcoR124I motor has clear advantages in this regard (5).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
The authors thank Dr Sean Colloms (University of Glasgow) for supplying the pJB43a plasmid and associated strains, and Prof. Malcolm White (University of St. Andrews) for supplying the Hje enzyme. This work was supported by grants from the Wellcome Trust (067439 and 071432). M.D.S. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences. L.K.S. is a Wellcome Trust Prize Ph.D. student. Funding to pay the Open Access publication charges for this article was provided by a Wellcome Trust ‘Value in People’ Grant to the University of Bristol, reference 078595.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rothemund P.W. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Pomerantz R.T., Ramjit R., Gueroui Z., Place C., Anikin M., Leuba S., Zlatanova J., McAllister W.T. A tightly regulated molecular motor based upon T7 RNA polymerase. Nano Lett. 2005;5:1698–1703. doi: 10.1021/nl0509125. [DOI] [PubMed] [Google Scholar]
- 3.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 4.Stanley L.K., Seidel R., van der Scheer C., Dekker N.H., Szczelkun M.D., Dekker C. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. EMBO J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seidel R., van Noort J., van der Scheer C., Bloom J.G., Dekker N.H., Dutta C.F., Blundell A., Robinson T., Firman K., Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nature Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 6.Seidel R., Bloom J.G., van Noort J., Dutta C.F., Dekker N.H., Firman K., Szczelkun M.D., Dekker C. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:4188–4197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClelland S.E., Szczelkun M.D. Molecular motors that process DNA. In: Pingound A., editor. Restriction Endonucleases, Nucleic Acids and Molecular Biology. Germany: Springer Verlag; 2004. pp. 111–135. Vol. 14. [Google Scholar]
- 8.van Noort J., van der Heijden T., Dutta C.F., Firman K., Dekker C. Initiation of translocation by Type I restriction-modification enzymes is associated with a short DNA extrusion. Nucleic Acids Res. 2004;32:6540–6547. doi: 10.1093/nar/gkh999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClelland S.E., Dryden D.T., Szczelkun M.D. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J. Mol. Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Janscak P., MacWilliams M.P., Sandmeier U., Nagaraja V., Bickle T.A. DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J. 1999;18:2638–2647. doi: 10.1093/emboj/18.9.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firman K., Szczelkun M.D. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCulloch R., Coggins L.W., Colloms S.D., Sherratt D.J. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortiz-Lombardia M., Gonzalez A., Eritja R., Aymami J., Azorin F., Coll M. Crystal structure of a DNA Holliday junction. Nature Struct. Biol. 1999;6:913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- 14.McKinney S.A., Declais A.C., Lilley D.M., Ha T. Structural dynamics of individual Holliday junctions. Nature Struct. Biol. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 15.Halford S.E., Welsh A.J., Szczelkun M.D. Enzyme-mediated DNA looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]