Abstract
Hepatitis B virus (HBV) replication is initiated by HBV RT binding to the highly conserved encapsidation signal, epsilon, at the 5′ end of the RNA pregenome. Epsilon contains an apical stem–loop, whose residues are either totally conserved or show rare non-disruptive mutations. Here we present the structure of the apical stem–loop based on NOE, RDC and 1H chemical shift NMR data. The 1H chemical shifts proved to be crucial to define the loop conformation. The loop sequence 5′-CUGUGC-3′ folds into a UGU triloop with a CG closing base pair and a bulged out C and hence forms a pseudo-triloop, a proposed protein recognition motif. In the UGU loop conformations most consistent with experimental data, the guanine nucleobase is located on the minor groove face and the two uracil bases on the major groove face. The underlying helix is disrupted by a conserved non-paired U bulge. This U bulge adopts multiple conformations, with the nucleobase being located either in the major groove or partially intercalated in the helix from the minor groove side, and bends the helical stem. The pseudo-triloop motif, together with the U bulge, may represent important anchor points for the initial recognition of epsilon by the viral RT.
INTRODUCTION
More than 300 million people worldwide are estimated to be chronically infected by hepatitis B virus (HBV) (1) and chronic HBV infection carriers have a great risk to develop severe liver diseases, including cirrhosis and liver cancer, resulting in a million deaths annually (2). No treatment for the efficient elimination of HBV in infected patients exists as yet. Therefore more knowledge about HBV replication is needed to enable the design of more efficient antiviral drugs.
HBV is a member of the Hepadnaviridae family, consisting of hepatotropic DNA viruses which also includes related animal viruses such as duck HBV (DHBV) and heron hepatitis virus. HBV has a small (3.2 kb), relaxed circular, partially double-stranded DNA genome and replicates this DNA genome through an RNA intermediate, the pregenomic RNA (pgRNA), by reverse transcription [for reviews see (3–5)]. The RNA pregenome also serves as the mRNA for the capsid (or core) protein and the P protein. The P protein contains the evolutionarily conserved RT domain, a middle spacer region, a C-terminal RNase H (RH) domain and a unique terminal protein (TP) domain at its N-terminus, which acts as a protein primer for reverse transcription. Replication is initiated by the binding of P to epsilon (ɛ) (Figure 1), a 60 nt bulged stem–loop at the 5′ end of the pgRNA (6–8). This binding event triggers encapsidation of the P–ɛ complex by capsid proteins, resulting in a priming competent, encapsidated complex. The product of the priming reaction is a 4 nt DNA, synthesized off a template in the primer bulge in ɛ, whose 5′ end is covalently attached to a tyrosine residue in the TP domain. This complex subsequently translocates to a 3′-proximal RNA element in the pregenome where full-length (−)-DNA synthesis is primed by the 4 nt DNA oligonucleotide (9–12).
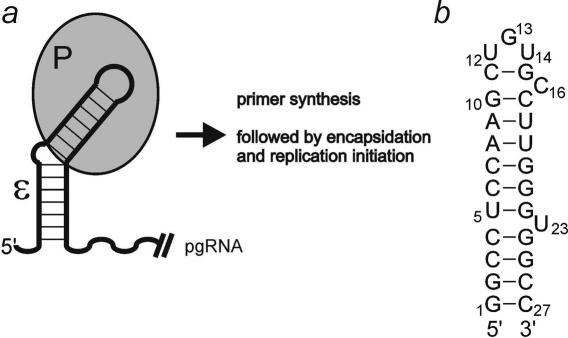
Figure 1.
(a) The ε stem–loop element is located in the 5′-UTR of the pgRNA of HBV. The viral reverse transcriptase (indicated by a P) recognizes and binds to the apical stem–loop of ε, thus triggering encapsidation and initiation of replication. (b) The apical stem–loop sequence used for NMR structure determination. The numbering scheme employed is indicated.
Detailed biochemical studies of the P–ɛ interaction have been made possible in recent years by the development of DHBV cell-free reconstitution systems consisting of P, ɛ and cellular chaperones (13–17). The system shows both P–ɛ binding and priming. Using truncated P protein constructs in these in vitro systems it was demonstrated that P–ɛ interaction requires sequences from both RT and TP protein domains (18). On the RNA side, the loop at the apical stem–loop of DHBV-ɛ is found to be essential for binding and primer synthesis (12). Recent SELEX experiments in such a system further defined the structure and sequence elements in the apical stem–loop of DHBV crucial for binding and/or priming (16). For instance, the middle of the stem underlying the loop should be weakly or not base paired at all. Most recently, a cell-free and chaperone dependent in vitro reconstitution system was developed also for human HBV (19). It shows P–ɛ binding but, in contrast to the DHBV system, not priming. Similar to DHBV, in human HBV sequences from both the RT and TP domains are required for binding of P to ɛ. Surprisingly, and in contrast to DHBV P–ɛ where the ɛ-apical loop is essential, in human HBV it is not needed for binding. The ɛ-apical loop is, however, required for encapsidation. Moreover, the structural features, requirement for base pairing in the stem part of the apical stem–loop, differ from those in DHBV. In human HBV, the upper part of the stem of the apical stem–loop needs to be base paired and the bulged out U is essential for binding. Although the structural basis and sequence requirements for P–ɛ binding and priming are emerging, a full understanding of the molecular basis for the specific interactions between P and ɛ awaits high-resolution structural studies.
Some high-resolution data have already been obtained on the human HBV ɛ apical stem–loop from NMR studies (20). We note in passing that the residues in the ɛ apical stem–loop are either totally conserved or show rare non-disruptive mutations (20). The tip of ɛ contains a CUGUGC sequence, for which secondary structure predictions have predicted a hexaloop structure (21–23). However, enzymatic probing studies have suggested a base pair between the first and fifth residue of this hexaloop (24). Our previous NMR studies confirmed the presence of this base pair indicating that the loop forms a pseudo-triloop motif (20). The pseudo-triloop is a recently proposed structural motif that consists of a hexaloop with transloop base pairing between residues 1 and 5 and a bulged out residue 6 (25). Hairpin loops with the potential to form pseudo-triloops are found in many RNA sequences, e.g. the brome mosaic virus (25,26), the iron responsive element (IRE) (27,28), domain IIId of the internal ribosomal entry site (IRES) of the hepatitis C virus (29,30), the 5′ terminal hairpin of R-U5 of simian foamy virus (31) and HIV-1 TAR (32). The common appearance of the pseudo-triloop motif in different RNA sequences suggests that it might be an important protein binding motif.
Here we present the high-resolution 3D structure of the human HBV ɛ apical stem–loop, i.e. of the 27 nt fragment which includes the pseudo-triloop and the conserved U bulge in the underlying stem (Figure 1b). Thanks to selective 2H/13C/15N-uridine-labelling, NMR spectral resolution could be increased and spectral overlap reduced (33–38), so that a set of highly reliable structural restraints for the structure derivation based on NOE, RDC and 1H chemical shift NMR data could be derived.
MATERIALS AND METHODS
Sample preparation
Preparation of unlabelled and labelled 27 nt RNA oligonucleotides, representing the apical stem–loop of epsilon, was done as described previously (20,36).
NMR spectroscopy
NMR spectroscopy was carried out as described previously (20,36). RDC measurements were done at 298 K, using a 0.5 mM unlabelled sample of the apical stem–loop in D2O. HSQC experiments without decoupling in the 13C dimension were acquired on a Bruker DRX600 spectrometer equipped with a HCN cryo-cooled probe. Reference spectra and spectra in Pf1 phages (15 mg/ml; ASLA Biotech) were recorded in in-phase and anti-phase mode, respectively, and were subsequently added or subtracted to obtain the chemical shifts of the two peaks of the doublet (39).
Structure calculations
Structure calculations were performed with the sander module of AMBER7 (40), whilst final refinement (step B3, see below) was carried out using X-PLOR (version 3.851) (41). All calculations were performed with the force field of Cornell et al. (42) with electrostatic interactions and a Lennard–Jones potential describing van der Waals interactions. Back calculations of chemical shift values were done using nuchemics (43). Here we will summarize the pertinent features of the calculations; a detailed description is included in Supplementary Data.
Two independent structure calculations were performed with different goals; calculation (A) to search conformational space efficiently for the loop region (nt 10–17, 8 nt), and calculation (B) to determine the global structure of the 27 nt molecule using RDCs to define the relative orientation of the helical parts of the apical stem–loop.
Structure calculation A: (A1) Exploration of loop conformations (residues G10–C17). An extended starting structure was randomized with dynamics at 1000 K, followed by high-temperature simulated annealing with NOE and torsion angle restraints. An ensemble of 200 structures was calculated. (A2) Ranking and selection of structures from the calculated ensemble. The 200 structures were evaluated by the sum of NOE violations, restraint energies and force field energies. The final selection step was composed of back-calculation of aromatic and H1′ chemical shifts for the loop nucleotides and selection of the structures that agreed best with chemical shift values while maintaining adequate restraint and force field features. With this approach, 12 structures were selected and their coordinates have been deposited in the Protein DataBank (PDB) (id code 2ixz).
Structure calculation B: (B1) Calculation of NOE structures of the whole apical loop using classical (NOE and torsion angle) restraints. In the starting structure, the two helical regions were A-type as indicated by analysis of 1H chemical shift values and the pseudo-triloop geometry was as determined in calculation A. An ensemble of 100 structures was generated using high-temperature simulated annealing by randomly varying initial atomic velocities. (B2) Determination of global structure. To define the global structure of the molecule using RDC restraints, semi rigid-body molecular dynamics was performed on each of the structures generated in step B1 with the local geometry of the stem regions and pseudo-triloop fixed by synthetic distance constraints. Synthetic distance constraints were generated for residues G1–C4 and G24–C27 (lower stem) and U5–G22 (upper stem). Constraints were included for all atom pairs consisting of one proton and one heavy atom within distances of 3–11Å. Classical restraints were included for the U23 bulge region and artificial restraints, to maintain tetrahedral geometry of C1′ and planar geometry of bases, were included to reinforce the force field where the RDC restraints were applied. Stem RDCs (28 in total) were included in the refinement with a single floating alignment tensor. The RDCs from the pseudo-triloop and U23 were excluded due to potential dynamics for these parts of the molecule. The alignment tensor was described by its five unique elements which all were free to fluctuate in the calculations. (B3) Reoptimization of the local structure. To optimize the local structure, low-temperature simulated annealing refinement, including all experimental data (NOEs, torsion angles and RDCs except those mentioned above), was performed. Except for planarity restraints for nucleobases (not base pairs), no synthetic distance restraints were included. The axial and rhombic components of the alignment tensor were fixed in this step, while the orientation of the alignment tensor was allowed to rotate as implemented in X-PLOR. For each structure from step B2, the rhombicity of the alignment tensor was calculated using the method proposed by Wijmenga and co-workers (44) and subsequently, the axial component was determined using the distribution of the RDCs. (B4) Selection of structures from the calculated ensemble based on comparison of predicted and experimental alignment tensors. For each structure from step B2, the predicted rhombicity (RPRED) was calculated using the gyration tensor method proposed by Wijmenga and co-workers (44). Independently, singular value decomposition (SVD) using PALES (45) was used to calculate the rhombicity of the alignment tensor (RSVD) from the set of experimental RDCs. Structures were selected which fulfilled the criteria |RSVD − RPRED| < 0.1 and fulfilment of NOE and torsion angle restraints. All molecules selected also displayed low force field energies. A total of 23 molecules were selected in this manner and their structures have been deposited in PDB (id code2ixy).
RESULTS
Structure determination
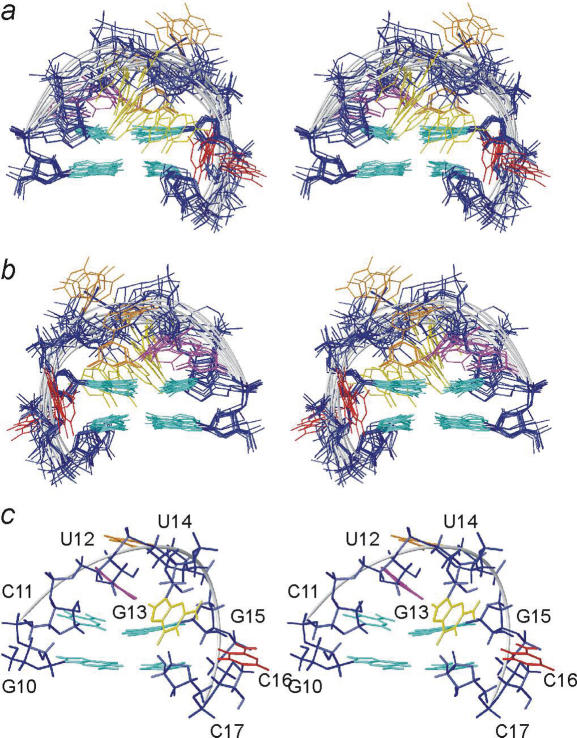
The structure calculation of the epsilon apical stem–loop structure was divided into two separate calculations. One with the aim of exploring conformational space for the loop at the tip of the apical stem–loop (calculation A) and the other with the aim of global structure determination for the whole apical stem–loop RNA (calculation B). The 112 NOE and 20 torsion angle restraints included in the structure determination of the loop region of the apical stem–loop (Table 1) did not define a single loop conformation and for U12, G13 and U14 no convergence was observed. Thus, we resorted to comparison with chemical shift values to define the class of structures in best agreement with all experimental data. In this manner an ensemble of 12 structures was generated with a root mean square deviation (r.m.s.d.) of 2.71 Å (Figure 2 and Table 2).
Table 1.
Number and distribution of restraints in calculationsa
| Structural restraints | 27 nt | 8 nt loop |
|---|---|---|
| Distance restraints | ||
| Intraresidue NOE | 109 | 57 |
| Interresidue NOE | 157 | 49 |
| Hydrogen bonding | 30 | 6 |
| Subtotal | 296 | 112 |
| Torsion angle restraints | ||
| Glycosidic | 27 | 8 |
| Sugar pucker | 42 | 5 |
| Backbone torsion angles | 90 | 7 |
| Subtotal | 159 | 20 |
| RDC restraints | 28b | —b |
aRestraints are deposited with the structures in the PDB.
bRDCs from the pseudo-triloop and U23 were excluded in calculations due to potential dynamics of these residues (these RDCs are also deposited in the PDB).
Figure 2.
Stereo views of an overlay of the 12 structures of the pseudo-triloop selected in step A2. (a) Viewed into the minor groove and (b) into the major groove. The sugar–phosphate backbone is coloured dark blue and the fold of the backbone is indicated as light grey tubes; colouring scheme of nucleobases is G10, C11, G15 and C17, light blue; U12, magenta; G13, yellow; U14, orange and C16, red. (c) The best structure as defined by the selection criteria.
Table 2.
Structural statisticsa
| 27 nt | 8 nt loop | |
|---|---|---|
| Violations of experimental restraints | ||
| Mean number of NOE violations >0.1 Å | 11.8 ± 1.3 | 5.0 ± 1.4 |
| Maximum NOE violation (Å) | 0.38 | 0.37 |
| Mean number of torsion angle violations >2° | 0.5 ± 0.5 | 0 |
| Maximum torsion angle violation (°) | 3.7 | 1.9 |
| The r.m.s.d. of RDC violation (Hz) | 1.61 ± 0.16 | |
| Alignment tensor statistics | ||
| Axial component, Da (Hz)b | −26.9 ± 1.8 | |
| Rhombicityb | 0.17 ± 0.06 | |
| Axial component, Da (Hz)c | −24.7 ± 1.1 | |
| Rhombicityc | 0.19 ± 0.04 | |
| The r.m.s.d. values from ideal covalent geometry | ||
| Bond lengths (Å) | 0.012 ± 0.000 | 0.011 ± 0.001 |
| Bond angles (°) | 2.81 ± 0.07 | 2.74 ± 0.25 |
| Atomic r.m.s.d. from average structure | ||
| Stem I (27 nt) (Å)d | 0.64 | |
| Stem II (27 nt) (Å)d | 0.79 | |
| Residues 10, 11, 15 and 17 (Å) | 1.31 | |
| Overall (Å) | 1.92 | 2.71 |
aFor the complete 27 nt molecule, 20 structures selected in step B3 are included in the analysis; for the 8 nt loop, 12 structures selected in step A2 are included.
bCalculated by SVD using PALES.
cCalculated using the gyration tensor method.
dFor the 27 nt molecule: stem I = residues 1–4 and 24–27; stem II = residues 5–10 and 17–22.
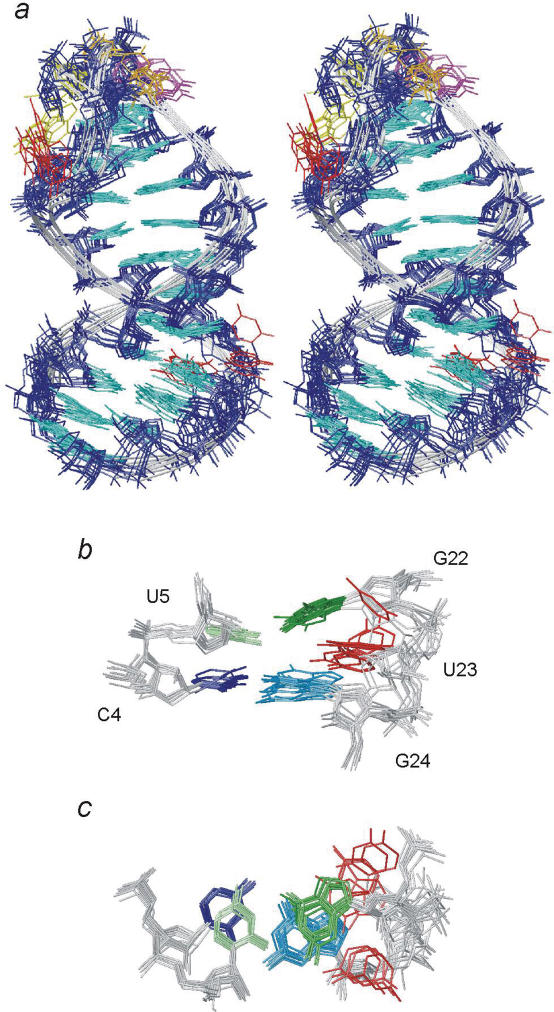
The global structure of the whole 27 nt apical stem–loop was determined in four steps to ascertain the correct global conformation, using 296 NOE, 159 torsion angle and 28 RDC restraints (Table 1). Initially, a classical NOE structure was calculated without RDC restraints (step B1). In this step, the local structure of each helical stem converged. The RDCs of both helical stems agreed with a single, common alignment tensor and in the next step, B2, RDCs were utilized to define the global structure of the 27 nt apical stem–loop sequence using rigid-body dynamics (46,47). During the final refinement step, B3, the local and to some extent global structure was reoptimized. For selection of molecules, we tested the consistency between the alignment tensor defined by the RDCs and the gyration tensor determined by the shape of the molecule. By doing so and requiring fulfilment of restraints and low force field energy, 23 structures were selected which we consider as the structural ensemble for the global structure of the apical stem–loop (Figure 3). The global structure is well defined and the all-atom r.m.s.d. is 1.92 Å (1.35 Å for helical residues). The structural statistics are presented in Table 2.
Figure 3.
The global structure of the apical loop. Colouring scheme as in Figure 2 and U23 is coloured red. (a) Stereo view of an overlay of 10 of the 23 selected structures. (b) Detailed side view of the U23 bulge. (c) The U23 bulge viewed along the helix axis.
Pseudo-triloop structure
The loop at the tip of the apical stem folds into a pseudo-triloop in which C11 and G15 form a Watson–Crick base pair (20). This base pair stacks onto the G10:C17 base pair at the top of the upper stem. Between U14 and G15 the phosphate backbone makes a turn and the γ angle of G15 adopts a trans conformation. This turn in the backbone probably facilitates the formation of the C11:G15 Watson–Crick base pair and contributes to restricting U14 to the major groove, albeit it can adopt multiple conformations within this groove. The bulged out residue, C16, is located in the major groove where it is unrestricted in its location and solvent accessible (Figure 2). The NMR data at hand do not specify a single conformation of the UGU triloop and the calculations with NOE and torsion angle data (step A1) yield several conformations of U12 and G13 that all fulfil these restraints. These conformations fall in four different groups as defined by the location of U12 and G13 in either the minor or major groove.
To validate the accuracy of the different conformations, we used back-calculated 1H chemical shifts from the structures generated in the conformational search (step A1). 1H chemical shifts depend on the local environment and are therefore an excellent tool for validation of local structure (43). In the validation procedure, we used aromatic and H1′ shifts. The aromatic nucleobase chemical shifts are mainly influenced by the position of the nucleobase itself and H1′ by the conformation of the glycosidic linkage (43). A wide variety of conformations was generated in step A1 of the calculations thus showing that conformational space indeed was searched thoroughly. Most of the loop conformations generated comply poorly with the experimental NMR data (1H chemical shifts and NOEs) or have excessively large force field energies (Supplementary Figure S2). A total of 12 loop structures with low chemical shift r.m.s.d. (<0.3 p.p.m.) and low constraint and force field energies were selected and they all display an arrangement where G13 is located in the minor groove (burying its Hoogsteen edge into the loop) and U12 and U14 in the major groove (Figure 2). In some structures, U12 stacks on top of C11 and U14 can stack on top of U12. Even though all UGU triloop conformations selected have a UGU: major–minor–major geometry, the calculations do not yield one single well-defined structure of the triloop. Furthermore, we observe a high degree of local structural heterogeneity in the sugar–phosphate backbone of the loop region, with several backbone angles populating multiple rotamers (Supplementary Figure S1).
With regard to the back-calculated chemical shifts, we note that we cannot make a population weighted average of the selected loop conformations that fit the experimental chemical shifts. For the majority of the aromatic protons in the loop region, the back-calculated shifts are higher than the observed ones. Thus successful conformation averaging is impossible which suggests that conformations other than those in the selected set are present, at least transiently. Such conformations could be high in force field energy or violate NOE restraints (which are r−6 averaged).
Global apical stem–loop structure
The structure of the whole apical stem–loop bends at the U23 bulge and the upper and lower stems converge to an average angle of 21 ± 9° (Figure 3). This standard deviation represents the uncertainty observed in calculations and is not a measure of the real amplitude of motion which could be larger. The bend at U23 is towards the major groove which consequently appears very deep and fairly narrow. The overall characteristics for the two stem parts are mostly A-helical as supported by chemical shifts, although the U23 bulge induces some buckling in the U5:G22 base pair. G22 stacks with G21 and the upper helix while U5 stacks with C4 and the lower helix (Figure 3). This buckling creates a wedge-like cavity for the U23 nucleobase. In the structural ensemble, U23 is found both destacked while turned into the major groove and partly stacked on G22 from the minor groove side (Figure 3). The exclusion of U23 from the helix is accomplished by adjustment of the sugar–phosphate backbone between G22 and U23. This is consistent with the G22pU23 phosphorus chemical shift being on the edge of the regular helical range and the mixed sugar pucker of G22 (20). Back-calculation of the H1′, H5 and H6 chemical shifts for U23 shows that no single conformation fulfils all experimental shifts. For the minor groove conformations, the H1′ shift is well predicted but not H5 (with a near random coil shift) and H6, whilst for the major groove conformations the situation is reversed. These observations are consistent with the analysis of NOE contacts which shows that the nucleobase of U23 cannot be fully intercalated (20). Even though the exact position of U23 cannot be defined by the experimental data, the global average structure of the molecule seems well defined.
DISCUSSION
Here we present the high-resolution 3D solution structure of the apical stem–loop of epsilon, the binding site of the viral reverse transcriptase in HBV. Chemical shift analysis showed that both helical regions are mainly A-type, and thus the structure determination has two main objectives, to determine the structure of the pseudo-triloop (PTL) motif at the tip of the apical stem–loop and the global structure of the whole molecule induced by the U23 interruption. As a consequence, we divided our structure calculation into two separate parts. For determination of the PTL conformation, only the top 8 nt were included in the calculations (calculation A). This enabled us to make a thorough sampling of conformational space. The dissection of the molecule for computational purposes also facilitated the analysis as we could evaluate the PTL conformations without considering whether the global geometry was optimal or not. Thus, fewer trial structures had to be calculated.
For the global structure, a protocol was designed to determine the geometry relying mainly on the RDC data. Here, we exploited the modular build of the apical stem–loop RNA and used semi rigid-body dynamics to reorient the two helical regions relative to each other (step B2) (47). During this step, the five independent parameters of the alignment tensor were optimized simultaneously with the stem orientation. This approach takes away the need for time consuming grid searches to determine the alignment tensor and hence increases computational efficiency. However, we noticed that the two stem regions had to be kept rigid as otherwise the alignment tensor had a tendency to ‘blow up’. In the initial structure, determined by NOE restraints (Step B1), the local geometry is only optimized with respect to the NOE restraints and not for the RDC restraints. Hence, initially there is a tendency to underestimate the axial component of the alignment tensor. If both the geometry of the molecule and the alignment tensor are optimized simultaneously, computations incorrectly satisfy the experimental RDCs by increasing the components of the alignment tensor excessively (resulting in the alignment tensor ‘blowing up’). The rigid-body dynamics step ensures that this does not happen. After realigning the two stem regions of the molecule, the local geometry is reoptimized in the final step (B3). Using this protocol, the RDCs are mainly used for determining the global structure of the molecule while the NOEs determine the local structure. The outline of our strategy resembles the local-to-global structure determination approach presented by McCallum and Pardi (47). Wijmenga and co-workers (44) have shown recently that for nucleic acids aligned with Pf1 phages, the rhombicity of the molecular alignment tensor can be predicted accurately from the shape of the molecule. We utilized this in the final selection of molecules by demanding that the rhombicities of the alignment tensor calculated by PALES and by the gyration tensor method should agree. In this way, structures which have a rhombicity, and hence alignment tensor, inconsistent with their overall structure are removed from the set.
Our methodology streamlines the structure determination by the subdivision of the problem into two separate and simpler problems. This strategy is only applicable if the molecule under study has a suitably modular build as the apical stem–loop of epsilon. For the global structure determination, we circumvent the problem of determining the components of the alignment tensor by grid searching. This strategy is less time consuming and if only fairly few RDCs are measured should be less prone to errors than a grid search which would overestimate the rhombicity and underestimate the axial component of the alignment tensor if the RDC space is sampled anisotropically (a common problem for elongated helical molecules) (48). Importantly, both strategies are applicable to a wide range of RNA molecules appropriate for structural studies using NMR spectroscopy.
The PTL at the tip of the apical stem–loop does not have a rigid and well-defined structure. The refinement with classical NOE and torsion angle restraints resulted in conformations of the triloop, where the two first residues, U12 and G13, alternate between the major and minor groove sides, showing all four possible permutations of minor–major groove conformations of U12 and G13. However, only conformations with U12 and G13 in the major and minor groove, respectively, were found to fit also 1H chemical shifts.
Besides the lack of convergence based on NOE and torsion angle data, there are some additional indications of flexibility within the HBV PTL. First, inclusion of RDCs in the loop refinement (using the magnitude of the alignment tensor determined from the full 27 nt molecule) did not improve the convergence of the loop structures. Second, an inspection of the RDCs shows that compared to the stem RDCs, the loop RDCs appear small (Supplementary Figure S3), which could be the result of averaging of RDCs due to motion of the loop residues. The same phenomenon is observed for the bulge residues in Loop B RNA from the IRES in Enterovirus (49). In addition, comparison of experimental and back-calculated 1H chemical shifts suggest the possible presence of flexibility. The loop conformations showing the best correspondence with the experimental 1H chemical shifts have an r.m.s.d. in the range of ≈0.25–0.30 p.p.m. (Supplementary Figure S2). However, full agreement of the 1H chemical shifts with the structure (i.e. a rigid structure) would yield an r.m.s.d. of less than ≈0.16 p.p.m. (43). The larger r.m.s.d. observed here could be an effect of chemical shift averaging due to internal motion. We finally note that ensemble averaging did not improve the 1H chemical shift correspondence for the loop protons and therefore speculate that other, transiently populated, conformations than those displayed in Figure 2 might exist for the triloop with the conformations shown being those with the highest probability of occurrence. Relaxation studies are in progress to further investigate the flexibility of the PTL.
The two helical stems of the apical loop are disrupted by a conserved, unpaired residue, U23. As determined from the stem RDCs, U23 induces a bend of ≈20° between the lower and upper helices which deepens and narrows the major groove. This angle is well defined as judged from its convergence in the RDC refined structures. Similar to the PTL, the bulged U-nucleotide is dynamic and switches between both the minor and major groove. The exclusion of U23 from the helical stack causes perturbations of the sugar–phosphate backbone in the 5′-direction of the strand which is also observed in molecular dynamics simulations of single uridine bulges (50). It is noteworthy that when U23 is located in the major groove, the PTL, C16 and U23 are all located on the same side of the structure (Figure 3a). In this manner, the elements important for recognition of the viral polymerase are accessible from one face of the apical stem–loop.
The sequence of the upper (apical) stem–loop of epsilon is conserved among all human HBV strains (20). Thus, this sequence is maintained in viable HBV, strongly suggesting that the sequence and PTL structure of this molecule are important for polymerase recognition. Hairpin loops with the potential to form PTLs are found in many RNA sequences, including viral genomes, and are therefore considered an important structural motif for protein recognition (25,26). Albeit constituting a general motif, the nucleotide sequences of the PTLs can be quite different. The most common closing base pair is C–G, but other base pairs can also occur, such as a trans-wobble U–G pair in domain IIId of HCV IRES (29,30).
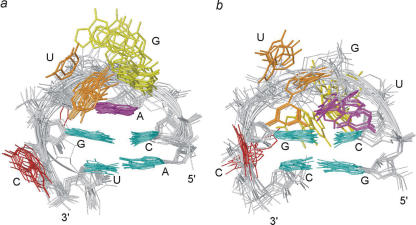
It is interesting to compare our PTL structure with that of the IRE PTL of sequence 5′-CAGUGC-3′, which differs from the HBV apical loop sequence only by the A highlighted in boldface (47). In the IRE structure, this A is structurally well defined, cross-strand stacking onto the guanine nucleobase of the C:G closing base pair, whilst the second and third residues, G and U, appear quite unrestricted in their motion (Figure 4). In the HBV apical loop, U12 and G13 are not structurally well defined, while U14 is. Perhaps the difference in structure between the HBV and IRE PTLs is dictated by the improved stacking capacity of the adenine nucleobase in the IRE PTL as compared to the uracil in the HBV PTL. However, fluorescence and stochastic dynamics simulation of the IRE PTL show that even though its A residue is rigidly stacked in the NMR structure, it possesses some potential for mobility as well (51).
Figure 4.
Comparison of the pseudo-triloops of IRE (a) and HBV (b). The colouring scheme is the same as in Figure 2. The nucleotide differing between the IRE and HBV sequences is coloured magenta.
Most knowledge of the P–ɛ interaction has been obtained from studies carried out on the DHBV and heron HBV cell-free in vitro reconstitution systems (12,16). The complex between epsilon and the DHBV polymerase was investigated by chemical probing in an arrested state obtained after a few primer nucleotides had been synthesized (12). In this state, the stem of the apical stem–loop of epsilon is melted and interacts with the polymerase. In addition, recent SELEX studies have further defined and distinguished the structure and sequence requirements for binding and priming for the DHBV in vitro system (16). Based on these biochemical studies, Nassal and Beck (12) proposed that the replication initiation is a two-step process in which the initial physical RNA binding (and recognition) is followed by a structural rearrangement for its use as template for the 4 nt DNA primer. Interestingly, in this in vitro system, the P protein binds both duck ɛ, with a well-defined upper stem–loop structure, and heron ɛ, where several of the base pairs in the upper stem are non-canonical and base pairing may be absent, but this P protein does not bind human ɛ. Thus, in the avian in vitro system the exact structure of the stem of the upper stem–loop of ɛ does not appear critical for binding. Instead, essential for binding are the loop at the tip of the stem as well as the bulged non-paired U residue further down the stem opposite to the primer loop. It is noteworthy that this P binding loop at the tip of the DHBV ɛ does not contain a PTL motif as in human HBV but a tetraloop motif.
Recently, an in vitro system has also been developed for human HBV (19). As for DHBV it comprises the P protein as well as chaperones. In contrast to the DHBV in vitro system, the human HBV in vitro system shows P binding to ɛ but is not priming competent. There are many similarities in the systems, but also several differences. The U23 bulge is essential for binding of ɛ to the P protein of human HBV while the corresponding bulged U in DHBV is dispensable. Furthermore, in contrast to DHBV, in the human HBV system, P binding requires base pairing in the upper part of the stem of the apical stem–loop. Surprisingly, binding of ɛ to P does not require the PTL at the tip of the apical stem–loop, while in DHBV this loop is essential for binding. In human HBV, the PTL is essential only for encapsidation. This suggests that the conserved PTL interacts with the capsid proteins rather than the RT. Similar to DHBV, in the human HBV system the apical stem–loop structure is expected to change conformation after initial binding to become priming competent. The scheme that emerges for human HBV is that after initial binding of ɛ to P, which must involve the U23 bulge and stem of the apical stem–loop of ɛ, the PTL of ɛ can still interact with the capsid protein.
Interestingly, there is a rare, viable U→A mutation in the apical loop of epsilon (U12 in the numbering scheme used in this paper) (20). This mutation makes the HBV PTL sequence identical to that of the IRE. P interacts with the stem and U23 which is unchanged in the U→A HBV mutant; however, viability also requires encapsidation capability of the mutant. As noted, it is likely that the capsid protein interacts with the PTL. The viability of the U→A mutant shows that the capsid protein is somewhat promiscuous in its recognition of the PTL at the tip of ɛ. Possibly, the interaction between the capsid protein and the PTL is required to induce the melting of the base paired ɛ apical stem, required for priming. Alternatively, the stem might be melted immediately upon recognition between P and ɛ in an induced-fit step, changing the structure of the triloop at the tip into a geometry appropriate for interaction with the capsid protein. If this suggestion is right, it would infer that the capsid protein binds primarily to the G13 and U14 residues of the triloop while U12 (or A in the mutant) is less important.
The conserved non-paired U23 located in the stem of the apical stem–loop could serve a dual role, to lower the energetic barrier for unfolding of the apical stem and to act as a recognition element. From our data U23 appears flexible, thus it may be that the exact position of this residue is not crucial for initial P–ɛ binding. On the other hand, the function of U23 could also be to guide the global structure of the apical stem–loop into a geometry favourable for initial P–ɛ interaction.
CONCLUSION
In conclusion, the 3D of the structure of the wild-type apical stem–loop of epsilon of human HBV has been derived based on NOE, RDC and 1H chemical shift NMR data. The apical stem–loop is capped by a PTL motif, while a U bulge is located in the underlying stem. Although the global structure of the apical stem–loop shows a well defined 20° angle between both helices, some local conformations, namely, the PTL and U bulge, are not well defined by the restraints used. In spite of this, the sequence of the upper stem–loop of epsilon is conserved among all known HBV strains, suggesting that the structure of the PTL and the U bulge are critical for viral viability. More studies are needed to define the exact nature of the steps in the P–ɛ binding and subsequent primer synthesis. Structure elucidation of the complete ɛ encapsidation motif is in progress. Irrespective of the exact nature of the binding process, the conservation of the structure of the upper stem–loop of free epsilon in human HBV makes it an outstanding target for potential antiviral drugs.
COORDINATES
Coordinates and restraints employed in calculations have been deposited in the PDB (accession codes: 2ixy and 2ixz).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Michael Nassal (Freiburg, Germany) for helpful discussion on different aspects of HBV P–ɛ recognition. This work was supported by grants from the Dutch Science Foundation (S.W.); the Swedish Research Council and the Medical Faculty of Umeå University (J.S.); the Faculty of Medicine, Lund University (K.K.L.); and the Danish National Research Council (M.P.). Funding to pay the Open Access publication charges for this article was provided by Dutch Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee W. Hepatitis B virus infection. N. Engl. J. Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Buendia M.A. Hepatitis B viruses and hepatocellular carcinoma. Adv. Cancer. Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 3.Nassal M. Hepatitis B virus replication: novel roles for virus–host interactions. Intervirology. 1999;42:100–116. doi: 10.1159/000024970. [DOI] [PubMed] [Google Scholar]
- 4.Nassal M. Macromolecular interactions in hepatitis B virus replication and particle assembly. In: Cann A.J., editor. DNA Virus Replication. Vol. 26. Oxford, UK: Oxford University Press; 2000. pp. 1–40. [Google Scholar]
- 5.Ganem D., Schneider R. Hepadnaviridae: the viruses and their replication. In: Fields B.N., et al., editors. Fields Virology. 4th edn. Philadelphia, PA: Lippincott, Williams & Wilkins; 2001. [Google Scholar]
- 6.Ganem D., Varmus H. Molecular biology of the Hepatitis B virus. Annu. Rev. Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 7.Tavis J.E., Perri S., Ganem D. Hepadnavirus reverse transcriptase initiates within the stem–loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassal M., Rieger A. A bulged region of the Hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G., Seeger C. Novel mechanism for reverse transcription in Hepatitis B viruses. J. Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem D., Pollack J., Tavis J. Hepatitis B virus reverse transcriptase and its many roles in hepadnaviral genomic replication. Infect. Agents Disc. 1994;3:85–93. [PubMed] [Google Scholar]
- 11.Rieger A., Nassal M. Specific Hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′ proximal direct repeat DR1. J. Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck J., Nassal M. Formation of a functional Hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 1998;18:6265–6272. doi: 10.1128/mcb.18.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J.M., Anselmo D. In vitro reconstitution of a functional duck Hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 2000;74:11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck J., Nassal M. Reconstitution of a functional duck Hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 2001;75:7410–7419. doi: 10.1128/JVI.75.16.7410-7419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck J., Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck Hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 2003;278:36128–36138. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- 16.Hu K., Beck J., Nassal M. SELEX-derived aptamers of the duck Hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 2004;32:4377–4389. doi: 10.1093/nar/gkh772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J., Flores D., Toft D., Wang X., Nguyen D. Requirement of heat shock protein 90 for human Hepatitis B virus reverse transcriptase function. J. Virol. 2004;78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeger C., Leber E.H., Wiens L.K., Hu J.M. Mutagenesis of a Hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology. 1996;222:430–439. doi: 10.1006/viro.1996.0440. [DOI] [PubMed] [Google Scholar]
- 19.Hu J., Boyer M. Hepatitis B virus reverse transcriptase and ɛ RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flodell S., Schleucher J., Cromsigt J., Ippel H., Kidd-Ljunggren K., Wijmenga S. The apical stem–loop of the Hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 2002;30:4803–4811. doi: 10.1093/nar/gkf603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack J., Ganem D. An RNA stem–loop structure directs Hepatitis-B virus genomic RNA encapsidation. J. Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskus R., Rakela J., Persing D. The stem–loop structure of the cis-encapsidation signal is highly conserved in naturally occurring Hepatitis B virus variants. Virology. 1994;200:809–812. doi: 10.1006/viro.1994.1247. [DOI] [PubMed] [Google Scholar]
- 23.Kidd A.H., Kidd-Ljunggren K. A revised secondary structure model for the 3′-end of Hepatitis B virus pregenomic RNA. Nucleic Acids Res. 1996;24:3295–3301. doi: 10.1093/nar/24.17.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knaus T., Nassal M. The encapsidation signal on the Hepatitis B virus RNA pregenome forms a stem–loop structure that is critical for function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haasnoot P.C.J., Brederode F.T., Olsthoorn R.C.L., Bol F. A conserved hairpin structure in alfamovirus and bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA. 2000;6:708–716. doi: 10.1017/s1355838200992471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haasnoot P.C.J., Olsthoorn R.C.L., Bol F. The brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem–loop. RNA. 2002;8:110–122. doi: 10.1017/s1355838202012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sierzputowska-Gracz H., McKenzie R.A., Theil E.C. The importance of a single G in the hairpin loop of the iron responsive element (IRE) in ferritin mRNA for structure: an NMR spectroscopic study. Nucleic Acids Res. 1995;23:146–153. doi: 10.1093/nar/23.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laing L.G., Hall K.B. A model of the iron responsive element RNA hairpin loop structure determined from NMR and thermodynamic data. Biochemistry. 1996;35:13596. doi: 10.1021/bi961310q. [DOI] [PubMed] [Google Scholar]
- 29.Klinck R., Westhof E., Walker S., Afshar M., Collier A., Aboul-ela F. A potential RNA drug target in the Hepatitis C virus internal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukavsky P.J., Otto G.A., Lancaster A.M., Sarnow P., Puglisi J.D. Structures of two RNA domains essential for Hepatitis C virus internal ribosome entry site function. Nature Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 31.Park J., Mergia A. Mutational analysis of the 5′ leader region of the Simian Foamy virus type 1. Virology. 2000;274:203–212. doi: 10.1006/viro.2000.0423. [DOI] [PubMed] [Google Scholar]
- 32.Critchley A.D., Haneef I., Cousens D.J., Stockley P.G. Modeling and solution structure probing of the HIV-1 TAR RNA stem–loop. J. Mol. Graphics. 1993;11:92–97. doi: 10.1016/0263-7855(93)87002-m. [DOI] [PubMed] [Google Scholar]
- 33.Tolbert T.J., Williamson J.R. Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatic synthesis. J. Am. Chem. Soc. 1996;118:7929–7940. [Google Scholar]
- 34.Tolbert T.J., Williamson J.R. Preparation of specifically deuterated and 13C-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc. 1997;119:12108. [Google Scholar]
- 35.Cromsigt J.A.M.T.C., Schleucher J., Kidd-Ljunggren K., Wijmenga S.S. Synthesis of specifically deuterated nucleotides for NMR studies on RNA. J. Biomol. Struct. Dyn. 2000;17:211–219. doi: 10.1080/07391102.2000.10506624. [DOI] [PubMed] [Google Scholar]
- 36.Flodell S., Cromsigt J., Schleucher J., Kidd-Ljunggren K., Wijmenga S. Structure elucidation of the HBV encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn. 2002;19:627–636. doi: 10.1080/07391102.2002.10506769. [DOI] [PubMed] [Google Scholar]
- 37.Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D. Structure of HCV IRES domain II determined by NMR. Nature Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 38.Lukavsky P.J., Puglisi J.D. Structure determination of large biological RNAs. Methods Enzymol. 2005;394:399–416. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 39.Kontaxis G., Bax A. Multiplet component separation for measurement of methyl 13C-1H dipolar couplings in weakly aligned proteins. J. Biomol. NMR. 2001;20:77–82. doi: 10.1023/a:1011280529850. [DOI] [PubMed] [Google Scholar]
- 40.Case D.A., Pearlman D.A., Caldwell J.W., Cheatham T.E., III, Wang J., Ross W.S., Simmerling C.L., Darden T.A., Merz K.M., Stanton R.V., et al. San Francisco, CA: University of California; 2002. AMBER 7. [Google Scholar]
- 41.Brunger A.T. New Haven, CT: Yale University Press; 1996. X-PLOR version 3.851: a system for X-ray crystallography and NMR. [Google Scholar]
- 42.Cornell W.D., Cieplak P., Bayly I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 43.Cromsigt J.A.M.T.C., Hilbers C.W., Wijmenga S.S. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- 44.Wu B., Petersen M., Girard F., Tessari M., Wijmenga S.S. Prediction of molecular alignment of nucleic acids in aligned media. J. Biomol. NMR. 2006;35:103–115. doi: 10.1007/s10858-006-9004-2. [DOI] [PubMed] [Google Scholar]
- 45.Zweckstetter M., Bax A. Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. [Google Scholar]
- 46.Sibille N., Pardi A., Simorre J.-P., Blackledge M. Refinement of local and long-range structural order in theophylline-binding RNA using 13C-1H residual dipolar couplings and restrained molecular dynamics. J. Am. Chem. Soc. 2001;123:12135–12146. doi: 10.1021/ja011646+. [DOI] [PubMed] [Google Scholar]
- 47.McCallum S.A., Pardi A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhou H., Vermeulen A., Jucker F.M., Pardi A. Incorporating residual dipolar couplings into the NMR solution structure determination of nucleic acids. Biopolymers. 1999;52:168–180. doi: 10.1002/1097-0282(1999)52:4<168::AID-BIP1002>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Du Z., Ulyanov N.B., Yu J., Andino R., James T.L. NMR structures of loop B RNAs from the stem–loop IV domain of the enterovirus internal ribosome entry site: a single C to U substitution drastically changes the shape and flexibility of RNA. Biochemistry. 2004;43:5757–5771. doi: 10.1021/bi0363228. [DOI] [PubMed] [Google Scholar]
- 50.Barthel A., Zacharias M. Conformational transition in RNA single uridine and adenosine bulge structures: a molecular dynamics free energy simulation study. Biophys. J. 2006;90:2450–2462. doi: 10.1529/biophysj.105.076158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall K.B., Williams D.J. Dynamics of the IRE RNA hairpin loop probed by 2-aminopurine fluorescence and stochastic dynamics simulations. RNA. 2004;10:34–47. doi: 10.1261/rna.5133404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.