Abstract
Cells of the central nervous system (CNS) are prone to the devastating consequences of trinucleotide repeat (TNR) expansion. Some CNS cells, including astrocytes, show substantial TNR instability in affected individuals. Since astrocyte enrichment occurs in brain regions sensitive to neurodegeneration and somatic TNR instability, immortalized SVG-A astrocytes were used as an ex vivo model to mimic TNR mutagenesis. Cultured astrocytes produced frequent (up to 2%) CAG·CTG contractions in a sequence-specific fashion, and an apparent threshold for instability was observed between 25 and 33 repeats. These results suggest that cultured astrocytes recapitulate key features of TNR mutagenesis. Furthermore, contractions were influenced by DNA replication through the repeat, suggesting that instability can arise by replication-based mechanisms in these cells. This is a crucial mechanistic point, since astrocytes in the CNS retain proliferative capacity throughout life and could be vulnerable to replication-mediated TNR instability. The presence of interruptions led to smaller but more frequent contractions, compared to a pure repeat, and the interruptions were sometimes deleted to form a perfect tract. In summary, we suggest that CAG·CTG repeat instability in cultured astrocytes is dynamic and replication-driven, suggesting that TNR mutagenesis may be influenced by the proliferative capacity of key CNS cells.
INTRODUCTION
Trinucleotide repeat (TNR) expansion causes at least 15 debilitating and often fatal neurodegenerative and neuromuscular diseases such as Huntington's disease (HD), myotonic dystrophy (DM1), dentatorubropallidoluysian atrophy (DRPLA) and many of the spinocerebellar ataxias (SCAs) [reviewed in Refs (1–3)]. Each disease has a unique clinical manifestation, but the underlying genetic mutation is the same—an expansion in the length of a TNR alters the function or expression of a nearby gene. The link between this unique form of mutagenesis and human disease makes understanding the mechanisms of TNR instability both medically relevant and scientifically important.
It is clear that new DNA must be synthesized to expand a TNR, but there are differing models as to the nature of the synthesis. One school of thought is that non-replicational mechanisms, such as error-prone DNA repair, are the major sources of instability in key affected tissues. Evidence from several model systems supports this contention. For example, intergenerational instability in male HD transgenic mice was traced to post-mitotic, haploid germ cells (4). Instability in cell lines derived from DM1 mice did not correlate with cell division (5). Moreover, a DM1 mouse line that showed remarkable somatic instability showed the greatest instability in organs that were not highly proliferative, namely kidney and brain (6). In addition, a transcription-based assay for instability in cultured human cells found that large contractions were independent of proliferation, occurred equally in confluent and non-confluent cultures, and were reduced by knockdowns of repair proteins Msh2, Msh3 and XPA (7).
In addition to post-mitotic mechanisms, a second idea is that TNR mutations can arise as a result of aberrant DNA replication in proliferating cells. Evidence to support replication-mediated instability again stems from several sources. Analysis of CAG repeat mutations in sperm tissue from HD men showed instability in proliferating pre-meiotic cells, and instability continued through meiosis into post-meiotic cells (8). Other studies have shown that TNR mutability can be modulated up to 10-fold by altering the direction of DNA replication through a repeat allele in human cells (9), human cell extracts (10) and nonhuman primate cells (9,11). Also, a repeat tract's proximity to the origin of replication appears to influence the bias for expansions or contractions in nonhuman primate cells (11) and replication inhibitors can modulate instability in primary human DM1 fibroblasts (12). When this evidence is placed in context with many microbial studies that invoke DNA replication as a major source of TNR expansions and contractions (13–16), these findings indicate that TNR instability likely has more than one mechanistic source and that repair, transcription and replication need to be evaluated for their contribution to TNR mutagenesis.
Since the brain is a target organ in many of the TNR diseases, an important consideration is that terminally differentiated neurons are outnumbered by other critical cell populations such as astrocytes, oligodendrocytes, and microglia that retain proliferative capacity throughout life. This consideration is important when interpreting studies in humans and transgenic mice showing age-dependent increases in TNR length in the central nervous system (CNS) [reviewed in Ref. (3)]. A main conclusion in some of these studies (6,17–19) is that non-replicational repair must be the key mechanism for triggering TNR expansion because neurons are non-dividing and thus do not replicate their DNA. A concern with this interpretation is that the affected brain regions include neurons and non-neuronal cells. Since these PCR analyses included DNA from cells such as glia that retain proliferative capacity, one cannot rule out replication as a source of the expansions. In fact, several labs have proposed that replication-derived TNR mutations in glial cells account for a large portion of the somatic mutations seen in autopsy tissue from TNR diseased patients (20–22). Moreover, a laser-capture microdissection study showed that cortical and cerebellar glial cells from DRPLA patients exhibit greater somatic TNR instability than surrounding neurons (23). This is especially significant because the cerebellum very rarely exhibits somatic TNR instability (18), implying that the cerebellum's rare TNR mutations are enriched in cells capable of proliferation. Taken together with recent evidence that glia help protect neurons from the degenerative effects of expansions in animal models and that glia themselves are sensitive to the toxic effects of TNR expansion (24,25), a clearer picture of the factors that promote or prevent somatic TNR instability in astrocytes and other glial cells is needed.
To better understand TNR instability in key CNS cells such as astrocytes, we took advantage of an immortalized human astrocytic cell line, SVG-A, and a previously described shuttle vector assay for CAG·CTG repeat contractions (9). The results from this model system indicate several striking similarities with human TNR genetics and provide new mechanistic insights into the fascinating but complex problem of triplet repeat mutagenesis. We investigated TNR contractions for several reasons. First, contractions usually outnumber expansions at allele lengths below the threshold for the CAG·CTG repeats at the HD locus (26), the androgen receptor gene and the DM1 locus (27), and the MJD1 gene (28). This observation also holds for subthreshold CGG·CCG repeats at the fragile X locus (29,30), suggesting that contractions help avoid attainment of the threshold. Second, a recent study found that HD transgenic animals produce somatic TNR deletions during development, dependent on cell proliferation, leading the authors to suggest that contractions may precede expansions in some circumstances (31). Third, several lines of evidence also suggest that at least some cellular factors are different in expansions versus contractions (31,32). If so, the contraction process might be exploited as a means to selectively shorten TNR alleles.
MATERIALS AND METHODS
Cell lines and cell culture
SVG-A cells, an astrocytic subclone of the astroglial SVG cell line (33,34), were obtained as a generous gift from Dr Ashok Chauhaun, Johns Hopkins University. These cells are immortalized with SV40 and express replication-competent SV40 Large T Antigen. Cells were grown adherently in DMEM (Mediatech, Inc.) supplemented with 10% fetal bovine serum, 50 U/ml Penicillin/Streptomycin and 2.5 µg/ml Amphoterecin B (all from Invitrogen Corporation).
Plasmids and cloning
All shuttle vectors (Figure 1A and B) were produced as described previously (9). Briefly, these plasmids are derivatives of pRS313 (35). The presence of the SV40 viral origin of replication (ori) enables plasmid replication in SVG-A cells. Yeast markers include a selectable promoter-TNR-URA3 reporter construct for identification of TNR contractions (9). The ARS/CEN sequence enables low-copy plasmid replication and segregation in yeast while the HIS3 gene allows selection for plasmid molecules independent of TNR length.
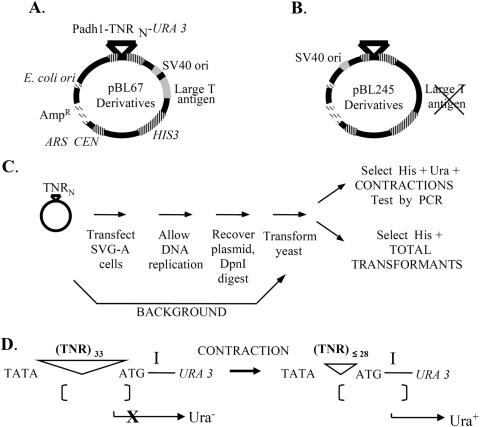
Figure 1.
Shuttle vector plasmids and assay outline. Details are provided in Materials and Methods. (A) The shuttle vector pBL67 contains several important genetic elements enabling analysis of TNR instability in cultured cells. Genetic elements tinted gray, such as the SV40 origin of replication, drive plasmid replication in SVG-A cells. A striped-pattern indicates yeast genetic elements for determination of contraction frequency by selection in Saccharomyces cerevisiae. Dashed bars indicate Escherichia coli elements that enable replication and large-scale preparation of plasmid by plasmid maxi-kit (Qiagen Inc). (B) The vector pBL245 follows the same pattern scheme, the only difference between the two vectors being the absence of the large T antigen and the relocation of the SV40 ori to the opposite side of the repeat tract. (C) Each TNR-containing vector was transfected into SVG-A cells, the cells were cultured for 2–3 days, then the plasmid DNA was isolated and transformed into yeast for analysis. Contraction frequency was calculated by dividing the number of yeast colonies with a contraction by the total number of transformants. Background contraction frequencies were measured as described in Materials and Methods. (D) The selection for contractions in yeast is based on spacing sensitivity of the Schizosaccharomyces pombe adh1 promoter to the distance between the TATA box and the preferred transcription initiation site, labeled ‘I’. Starting TNR lengths of 33 inhibit expression of the URA3 reporter gene and yield a Ura− phenotype (36). TNR contractions that remove at least five repeats restore promoter activity and result in a Ura+ phenotype.
Two different shuttle vector backbones were used in this study. The first backbone, pBL67 (Figure 1A), contains the SV40 origin of replication and large T antigen gene cloned into the BamHI site of pRS313. The promoter-TNR-URA3 cassettes were transferred into the vector backbone in two steps. First, complementary oligonucleotides containing TNRs and SphI sticky ends were annealed and cloned into the SphI site of pBL94 (16,36). This produced promoter-TNR-URA3 constructs which were then digested with EcoRI and XhoI (New England Biolabs, Inc.) and transferred en bloc to the EcoRI/XhoI sites of pBL67 (9). The second vector used here is pBL245 (Figure 1B). This backbone contains only the SV40 origin of replication (and not the large T antigen gene), cloned in the ApaI site of pRS313 to create pBL185 (9). SVG-A cells constitutively express SV40 large T antigen, allowing the elimination of large T sequences from the pBL67 vector. Another advantage of this approach was that it eliminated an SphI site from the vector backbone that was present in the large T antigen gene, making unique the SphI site in the promoter-URA3 cassette. The empty promoter-SphI-URA3 cassette, containing no TNRs, was transferred into pBL185 after digestion with the EcoRI and BamHI, producing pBL245. SphI digestion (New England Biolabs, Inc.) of pBL245 enabled annealed TNR oligonucleotides containing SphI sticky ends to be cloned into the shuttle vector backbone directly. Since cloning the SV40 origin of replication into the ApaI site of pRS313 resulted in its placement on the opposite side of the repeat tract versus pBL67, a standard system of nomenclature was adopted to ensure consistency between experiments. In all experiments, the named repeat, such as (CAG)33, refers to a plasmid molecule where CAG repeats comprise the template for lagging strand synthesis. As a further precaution, comparisons between repeats of different sequence, length or orientation were only made between plasmids derived from the same vector backbone.
Shuttle vector assay
The shuttle vector assay (Figure 1C) was slightly modified from the previous study (9). Briefly, the day before transfection ∼2.5 × 105 SVG-A cells were seeded in triplicate in 60 mm tissue culture dishes. The next day, cells were transfected using Lipofectamine reagent (Invitrogen Corporation), according to the manufacturer's protocol using 5 µg of shuttle vector DNA and 20 µl of Lipofectamine per dish. After transfection, cells were incubated for 72 h to enable plasmid replication. The cells were lysed and plasmid DNA was extracted using Hirt's technique (37). Lysates were concentrated by centrifugation through a Centricon 50 kDa MWCO filter (Millipore Corporation). Plasmid DNA was precipitated with ethanol and digested with DpnI (New England Biolabs, Inc.) DpnI digestion eliminates any plasmid molecules that did not replicate in human cells. DpnI-resistant DNA was then transformed into yeast strain S1502B (MATa leu2-3, leu2-112 his3-Δ trp1-289 ura3-52) using the lithium acetate method (38). A fraction of each transformation mixture (typically 0.5%) was plated onto SC-His plates (synthetic complete, lacking histidine), and the remainder onto SC-His-Ura (synthetic complete, lacking histidine and uracil) to score for contractions. Colonies on each plate were counted after 3 days of growth at 30°C. The frequency of contraction was determined as the number of colonies obtained on SC-His-Ura divided by the total number of transformants on SC-His, with appropriate correction for dilution factors. Statistical analyses were performed by applying a two-tailed Student's t-test with equal variance to the relevant datasets. Comparisons between datasets observed in SVG-A cells were made with no correction for background. Full details of the assay results are given in Table 1.
Table 1.
TNR contraction data in SVG-A cells
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Repeat sequence | Plasmid name | Vector backbone | SVG-A contractions observed (His+ Ura+ colonies) | Plasmid population size (His+ colonies × dilution) | N | SVG-A contraction frequency% (±SEM%)a | Background contraction frequency% (±SEM%) | Corrected contraction frequency | P-value: SVG-A versus Background | |
| A | (C,A,G)33= 0 rpts | pBL200 | pBL67 | 0 | 6000 | 3 | 0 (±0) | 0 (±0) | 0 | N/A |
| B | (CAG)15+18 | pBL213 | pBL67 | 24 | 392 400 | 6 | 0.0079 (±0.0029) | 0.0077 (±0.0086) | 0.0002 | 0.91 |
| C | (CAG)25+8 | pBL176 | pBL67 | 2923 | 789 000 | 5 | 0.34 (±0.09) | 0.16 (±0.02) | 0.19 | 0.0004 |
| D | (CTG)25+8 | pBL207 | pBL179 | 10 | 29 800 | 3 | 0.04 (±0.02) | 0.02 (0.04) | 0.02 | 0.56 |
| E | (CAG)30+3 | pBL227 | pBL67 | 204 | 70 800 | 6 | 0.82 (±0.28) | 0.43 (±0.12) | 0.39 | 0.18 |
| F | (CAG)33 | pBL217 | pBL67 | 622 | 37 600 | 4 | 2.3 (±0.4) | 0.31 (±0.11) | 1.9 | 0.01 |
| G | (CTG)25+8 | pBL199 | pBL67 | 8 | 22 200 | 2 | 0.034 (±0.006)a | 0.015 (±0.009) | 0.019 | 0.10 |
| H | (CTG)50 | pBL175 | pBL67 | 37 | 12 800 | 3 | 0.30 (±0.06) | 0.055 (±0.014) | 0.24 | 0.002 |
| I | (CAG)25+8 | pBL250 | pBL245 | 49 | 34 600 | 4 | 0.17 (±0.03) | 0.003 (±0.002) | 0.17 | 0.006 |
| J | (TAG)25+8 | pBL246 | pBL245 | 4 | 6000 | 2 | 0.067 (±0.009)a | 0.013 (±0.013) | 0.054 | 0.05 |
| K | (CTA)50 | pBL216 | pBL67 | 173 | 274 000 | 5 | 0.09 (±0.05) | 0.06 (±0.01) | 0.03 | 0.89 |
| L | (CTG)6(ATG)2(CTG)25 | pBL248 | pBL245 | 576 | 60 000 | 6 | 0.87 (±0.07) | 0.051 (±0.004) | 0.82 | 0.0001 |
| M | (CTG)33 | pBL249 | pBL245 | 50 | 12 800 | 3 | 0.39 (±0.01) | 0.12 (±0.02) | 0.27 | 0.05 |
| N | (CAG)33 | pBL247 | pBL245 | 2421 | 118 400 | 3 | 2.0 (±0.5) | 0.14 (±0.01) | 1.9 | 0.04 |
Column 1 lists the trinucleotide repeat sequence comprising the template for lagging strand synthesis (as defined in Materials and Methods). Column 2 gives each plasmid's name for reference. Column 3 indicates the vector backbone in which each TNR sequence was cloned. For (CAG)25+8 and (CAG)33, the repeats were cloned in both vector backbones—the pBL67 derivatives (pBL176 and pBL217) were used in the threshold study while the pBL245 vectors (pBL250 and pBL247) were used in the hairpin and orientation dependence studies. The pBL67 derivative, pBL217, was also used as the (CAG) 33-new ori vector in the orientation dependence study. Column 4 indicates the total number of contractions observed in triplicate transfections in SVG-A cells (the number of colonies counted on SC-His-Ura plates). Column 5 lists the total number of plasmids in the population (the number of colonies counted on SC-His plates multiplied by the appropriate dilution factor). Column 6 lists the number of samples recovered after transfection. Column 7 lists the cntraction frequency observed for plasmids replicated in SVG-A cells (%) ±1 SEM (%). The error for samples with two replicates (marked ‘a’) is listed as the absolute difference between the two observed values. Column 8 lists the background contraction frequency (%) ±1 SEM (%), as described in Materials and Methods. Column 9 lists the corrected contraction frequency, obtained by subtracting the background (column 8) from the contraction frequency observed in SVG-A cells (column 7). The corrected contraction frequency is used to produce the graphs shown in the figures. Column 10 lists the P-value for each sequence versus the background as determined by two-tailed student's t-test.
aError shown is the range between the two values.
A critical control for this assay involves determination of the background contraction frequency, defined as the percentage of contracted TNR tracts in the plasmid population that did not occur in human cells. The background was determined by transforming stock plasmid directly into yeast, bypassing replication in SVG-A cells (Figure 1C). The background provides quantitation of any TNR contractions that occurred in Escherichia coli during preparation stock plasmid, or in yeast before selection became effective. Background frequencies were calculated from at least three independent measurements.
Identification of TNR contractions
Yeast was used as a biosensor (9) for contractions arising in SVG-A cells (Figure 1D). Briefly, gene expression driven by the Schizosaccharomyces pombe adh1 promoter is dependent on the proper spacing between the TATA box and the downstream transcription initiation site (labeled ‘I’ in the figure). When a TNR-containing 33 triplet repeats is inserted between the TATA box and ‘I’, yeast transcription initiates upstream of ‘I’ and this causes an out-of-frame ATG codon to be encoded and a nonsense URA3 gene product to be produced. However, if a contraction to a TNR length ≤28 repeats occurs in human cells, upon transfer to yeast transcription will initiate at ‘I’, translation begins at the URA3 ATG codon, and the cells become phenotypically Ura+. Thus Ura+ yeast colonies reflect contraction events that occurred in SVG-A cells. Note that in some plasmids, the initial TNR tract was <33 repeats. In these instances, genetically inert, randomized sequences were appended to make the total DNA length 99 nt. For example, 25 CAG repeats +24 nt of randomized sequences are referred to as (CAG)25+8.
PCR analysis
To authenticate contractions and to determine their size, individual Ura+ yeast colonies were resuspended in water and heated to 95°C for 10 min. Subsequent PCR amplification used α-32P-dCTP or Cy5-labeled primers flanking the repeat tract for 30 cycles (1 min at 94°C, 1 min at 60°C and 1 min at 72°C), plus a final extension at 72°C for 5 min. The products were separated on a 6% denaturing polyacrylamide gel. PCR product sizes (±2 repeat units) were determined by comparison with a standard containing repeats of known size.
Restriction analysis of contractions of interrupted TNR alleles
Interrupted TNR alleles were created by introducing tandem ATG interruptions in a CTG repeat tract, resulting in a TNR tract with the sequence (CTG)6(ATG)2(CTG)25. The presence of either one or two ATG interruptions in a CTG repeat sequence introduces a recognition site for the restriction enzyme SfaNI (New England Biolabs). After passage through SVG-A cells and transformation into yeast, PCR products from contracted interrupted and perfect TNR alleles were isolated by the QIAQuick PCR Purification Kit (QIAGEN Inc.) and digested with SfaNI. The presence or absence of the interruptions (as determined by SfaNI sensitivity) was visualized after separation on a 6% sequencing gel.
Immunofluorescent detection of GFAP
The day before staining, 105 SVG-A cells were seeded in each well of a 24-well tissue culture plate. Cells were washed with PBS and fixed in ice-cold acetone/methanol. After fixation, cells were permeabilized and blocked with Triton-X 100/BSA for 20 min. Half of the wells were treated overnight with a 1:500 dilution of mouse anti-human GFAP (Dako Denmark A/S) in PBS and the other half treated with PBS alone. After staining, cells were washed with PBS and treated with a 1:100 dilution of fluorescein-tagged goat-anti-mouse IgG (Invitrogen Corporation) in PBS in the dark for 30 min. Cells were washed again with PBS and stained for 2 min with a 1:1000 dilution of DAPI (Sigma-Aldrich Company) in sterile deionized H2O. Cells were visualized under an inverted fluorescence microscope through either DAPI or fluorescein specific filters and the images merged. The primary and secondary antibodies as well as the DAPI reagent were provided as a gift from Dr Anuja Ghorpade, University Nebraska Medical Center.
RESULTS
In previous work (9), we developed a shuttle vector assay that quantitatively measures CAG·CTG repeat contractions (Figure 1). The assay allows facile analysis of contraction frequency as a function of key cis-elements known from human TNR genetics, such as the repeat length, the direction of DNA replication, the structure-forming capacity and the purity of the repeat. This assay utilizes shuttle vector plasmids (Figure 1A and B) that are passaged through SVG-A astrocytes and transferred into yeast, where genetic selection reveals the frequency of TNR contractions occurring during replication in cell culture. An outline of the assay is shown in Figure 1C and the mechanism of the yeast selection process is shown in Figure 1D. Yeast colonies, harboring a contracted TNR-containing shuttle vector plasmid, are lysed and the repeat tract is amplified with primers flanking the repeat to measure contraction size (Materials and Methods). The shuttle vector assay enables quantitative measurement of contraction frequencies (9), and is, to our knowledge, the only cell culture assay with suitable sensitivity to readily monitor TNR instability for alleles near the crucial threshold length of 30–40 repeats.
Replication and CAG·CTG repeat contractions occur readily in SVG-A cells
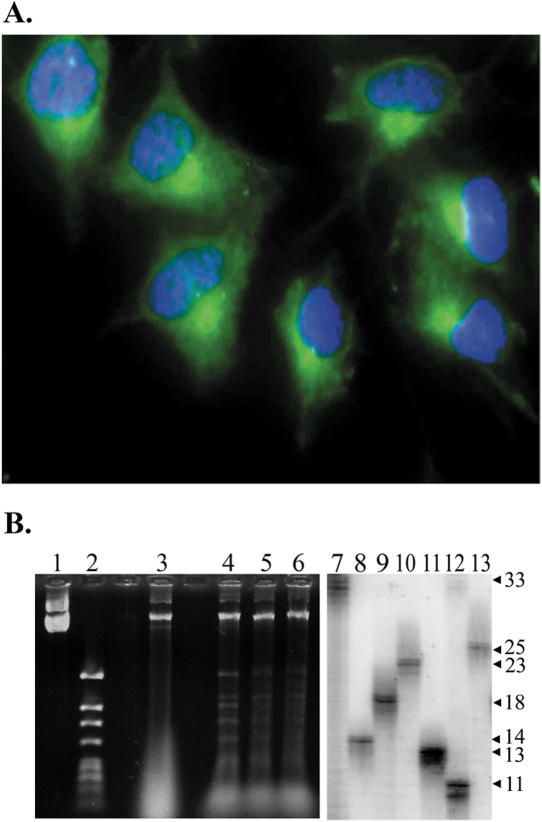
To confirm the CNS nature of the SVG-A cells under study, cells were stained for expression of glial fibrillary acidic protein (GFAP), an intermediate filament restricted to glial cells like astrocytes. Figure 2A shows that SVG-A cells express GFAP. More than 1000 cells were examined by fluorescence microscopy and 100% stained positively for GFAP. Control experiments (data not shown) proved that positive staining required the anti-GFAP primary antibody. Additionally, the GFAP stain in Figure 2A demonstrates a morphology consistent with that observed in another study of cultured astrocytes maintained in serum-supplemented media (39). Together, these findings support the astrocytic nature of SVG-A cells used here.
Figure 2.
Analysis of SVG-A cells and transfected TNR plasmid DNA. (A) SVG-A cells stained with anti-GFAP antibodies (green false color) and counterstained with DAPI (blue false color) at 400× magnification. (B) DpnI resistance assay after recovery of plasmid pBL248 from SVG-A cells and subsequent PCR analysis of contracted alleles after yeast selection. The left panel shows the result of DpnI digestion of stock plasmid used for background determinations (lane 1 = uncut, lane 2 = cut) and plasmid DNA that has been passaged through SVG-A cells (lane 3 = uncut, lanes 4–6 = cut). Sample wells between lanes 2 and 3 and lanes 3 and 4 were intentionally left empty. The right panel shows the analysis of individual contraction events from SVG-A cells. PCR amplification of starting plasmid DNA indicates the size of the 33-repeat parental allele (lane 7). Individual contractions are shown in lanes 8–13. Fragment sizes, stated as repeat units, were deduced by comparison with molecular weight standards (data not shown).
Since the role of replication in TNR mutagenesis is controversial, it is of particular importance to evaluate replication in cultured astrocytes. Shuttle vectors (Figure 1A and B) are predicted to replicate in SVG-A cells due to expression of large T antigen plus the presence of the SV40 ori. The SV40 system is a reasonable model because, once the origin is unwound, viral replication occurs with high fidelity using the host-cell replication machinery (40), suggesting our plasmid system will likely mimic replication events on chromosomal DNA. Furthermore, incubation with nuclear extracts causes SV40-derived plasmids to fold into chromatin structures in vitro, and this nucleosome folding is enhanced after plasmid replication (41). Following transfection and recovery from SVG-A cells, replication was assessed by challenging DNA with the dam methylation-sensitive restriction enzyme DpnI. DNA that has replicated at least once in SVG-A cells will be resistant to digestion. A representative DpnI analysis is shown in the left panel Figure 2B. Lanes 1–2 show that DpnI digestion proceeds to completion for stock plasmid prepared in E.coli, as expected. Lane 3 contains uncut DNA recovered from transfected SVG-A cells, while plasmid preparations from three independent transfections (lanes 4–6) show a bright band of DpnI-resistant plasmid DNA, consistent with substantial levels of DNA replication. DpnI-digested plasmid preparations were transformed into yeast and putative CAG repeat contractions were identified as Ura+ yeast colonies (Figure 1C and D). Contractions were confirmed by PCR amplification of the CAG repeat locus, as shown in Figure 2B, right panel. The starting plasmid gave a PCR product consistent with 33 repeats, whereas plasmid DNA from the Ura+ colonies yielded shorter PCR products corresponding to ultimate repeat tracts of 11–25 repeats (contractions of −8 to −22). Together, these results indicate that the glial SVG-A cells support shuttle vector replication and show instability of CAG·CTG repeat tracts.
Length and orientation dependence of CAG·CTG contractions in SVG-A cells
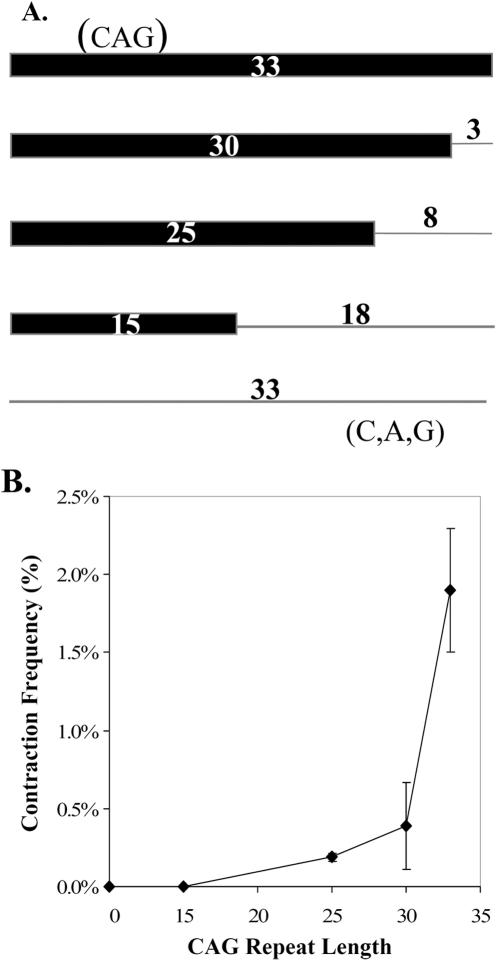
A signature characteristic of TNR instability in humans is a sharp increase in mutagenesis as the tract reaches or exceeds a threshold of ∼35 repeats. Consistent with the literature, we define a threshold as a demarcation between genetically stable and unstable alleles over a narrow range of repeat length. We sought to evaluate a potential threshold in astrocytic SVG-A cells. To accomplish this, CAG alleles of 0–33 repeats were created as shown in Figure 3A. Non-repeating sequence was used to normalize the length of all CAG tracts to 99 nt. Shuttle vectors harboring these repeat constructs were passaged through SVG-A cells and subsequent analysis in yeast produced the plot shown in Figure 3B. The frequency of CAG contractions was indistinguishable from background at 0 or 15 repeats (Table 1, entry 9B), but then rose sharply between 25 and 33 repeats. For the longest reporter, ∼2% of all the CAG alleles underwent contraction during a transient three-day passage in SVG-A cells. Furthermore, lengthening the CAG tract from 25 to 33 repeats (+32%) resulted in a 1000% increase in contraction frequency, consistent with a threshold at or near 35 repeats. The observed contraction frequency at 30 repeats did not achieve statistical significance over background, but the 25 and 33 repeat results were clearly distinguishable from background (Table 1, entries 10C, E and F). Thus CAG contraction frequencies suggest a threshold-like response in SVG-A cells.
Figure 3.
Repeat-length dependence for TNR contractions in SVG-A cells. (A) The top line represents a perfect (CAG)33 allele while the bottom indicates a scrambled (C,A,G)33 allele with no repeating nature (zero repeats). The thick bar indicates the length of the perfect CAG repeat while the thin line shows the number of scrambled C,A, and G triplet equivalents appended to each repeat in order to normalize all constructs to 99 nt. (B) Response of CAG contractions in SVG-A cells to changes in initial tract length. The contraction frequency (Table 1) on the ordinate was derived by subtracting the background contraction frequency from the contraction frequency observed in SVG-A cells. Error bars indicate ±1 SEM.
Given that the distance of the SV40 origin from the CAG·CTG tract is known to modulate repeat instability in primate cells (11), the position of the appended random sequence [e.g. in (CAG)25+8] is a potential complicating factor. However, the data in Table 1 show that (CAG)25, whether tested in pBL67 or pBL245, had indistinguishable mutation frequencies of 0.19 and 0.17%. The results for (CAG)33 were also consistently ∼10-fold higher in the two plasmids (1.9% for both). These data indicate that the appended random sequence does not affect the results, at least for these two locations relative to the SV40 origin.
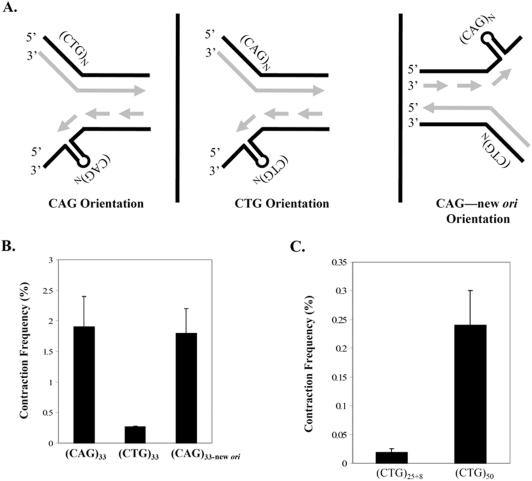
Replication-dependent TNR contractions in microbes often exhibit an orientation effect, where contractions are more frequent when CTG occupies the lagging template strand rather than the complementary sequence CAG. In a prior study using the shuttle vector assay, the opposite was true in human 293T and 293 kidney cells; contractions were ∼10-fold more frequent when CAG comprised the lagging strand template (9). This prompted us to examine orientation effects in SVG-A cells, as depicted schematically in Figure 4A. The only difference in the two plasmids (left and middle panels in Figure 4A) is the cloning orientation of the CAG·CTG tract. To add to the findings in our previous study (9), we examined the orientation dependence of (CAG·CTG)33 alleles, in addition to alleles containing 25 repeats. The result in Figure 4B indicates that (CAG)33 sequences contract ∼7-fold more often than (CTG)33 and the orientation effect was statistically significant (P = 0.02). Moreover, we took advantage of another series of shuttle vectors—with the SV40 ori relocated to the opposite side of the repeat tract—to test whether switching the direction of replication through otherwise identical repeats can influence TNR mutability. The plasmid, called (CAG)33–new ori harbored repeats in the same sequence context as the (CTG)33 vector, but movement of the replication origin resulted in fork movement through the repeat in the opposite direction, such that CAG repeats occupied the lagging template strand. This effect is depicted pictorially in the middle and right panels of Figure 4A. Reassignment of CAG repeats to the lagging template resulted in full restoration of instability in the (CAG)33–new ori vector (Figure 4B and Table 1 entry 9F). This restoration suggests that the orientation of the repeat with respect to replication is the most important factor, not the distance from the origin nor flanking DNA sequences. Furthermore, restoration also held true for (CAG)25+8 versus (CTG)25+8 repeat tracts (Table 1, entries 9C, 9G and 9I). To address explicitly the role of DNA replication in producing CAG·CTG contractions in SVG-A cells, the SV40 origin of replication was deleted from the shuttle vector backbone. When a 25-repeat CAG·CTG allele is cloned into this backbone and transfected into SVG-A cells, the contraction frequency was indistinguishable from background (Table 1, pBL207, entries 9D and 10D), confirming that astrocytes experience replication-mediated TNR instability in this model system, with similar orientation effects as kidney-derived cells (9).
Figure 4.
The repeat orientation relative to the direction of replication influences CAG·CTG contractions in SVG-A cells. (A) Complementary CAG and CTG repeats can differentially affect TNR instability based on the orientation of the repeat relative to the direction of replication. In the left panel, CAG repeats reside on the lagging strand template. In the middle panel, the reverse orientation places the CTG repeats on the lagging template strand. On the right, switching the position of the SV40 ori reverses the direction of replication and swaps the assignment of leading and lagging strand. The direction of replication was deduced from the position of the S40 ori relative to the TNR. (B) Contraction frequency (%) is plotted as given in Table 1 (column 9) for the indicated starting alleles. Error bars indicate ±1 SEM. (C) Contraction frequency (%) is plotted versus CTG repeat length. All values in (B and C) represent the corrected contraction frequency in SVG-A cells.
The relatively low frequency of (CTG)25+8 and (CTG)33 contractions raised the issue of whether CTG repeats require longer lengths to be mutable, or whether—for some unknown reason—they are largely immutable in our system. This question was answered by increasing the tract length to (CTG)50. Figure 4C shows that the contraction frequency for (CTG)50 increased significantly compared to (CTG)25+8 (Table 1, entries 9H and 9G). The absolute frequency for the (CTG)50 reporter approaches the level seen for (CAG)25+8, and is significantly above background (P = 0.002). However, the observed (CTG)50 contraction frequency likely underestimates the true contraction frequency in SVG-A cells because the 50-repeat allele must delete at least 22 repeats to register as Ura+ in the yeast selection step of our assay (Figure 1D). Therefore our assay does not register contractions of <22 repeats from the (CTG)50 reporter. By comparison, a 25 + 8 allele must lose only five repeats to score as a contraction in our assay. Nonetheless, we can conclude that CTG runs are mutable in SVG-A cells in a length-dependent manner (P = 0.04), but they require a longer tract than CAG repeats to achieve similar contraction frequencies.
Structure-forming capacity promotes TNR contractions in SVG-A cells
One of the seminal observations in the field of TNR mutagenesis was that disease-causing repeat sequences have the ability to form aberrant DNA secondary structures (42–44). This observation led to models in which TNR instability, either expansions or contractions, results from error-prone replication, recombination or repair of DNA structures that form at TNR loci. To determine whether structure-forming capacity influences the propensity for TNR contractions in SVG-A cells, we created shuttle vectors harboring either a (CAG)25+8 allele, which is predicted to form a stable hairpin under physiological conditions, or a (TAG)25+8 allele, which is not predicted to form stable secondary structure. Even with allele lengths below the putative threshold, hairpin-forming capacity stimulated contractions by ∼3-fold (P = 0.04; Table 1, entries 9I and 9J). The (CAG)25+8 allele contracted at a frequency that is statistically significantly above background (P = 0.006), while the (TAG)25+8 contraction frequency compared to background is only borderline significant (P = 0.05; entries 10I and J in Table 1). Hairpin-forming capacity also appears to be important for CAG·CTG contractions arising from longer 50-repeat alleles. The (CTG)50 run contracted more frequently than hairpin-incompatible (CTA)50 alleles (Table 1, entries 9H and 9K, P = 0.002). From these studies, it appears that TNR sequence compatible with hairpin formation is critical for promoting maximal TNR instability in CNS cells.
CTG repeat contractions arise from an interrupted allele
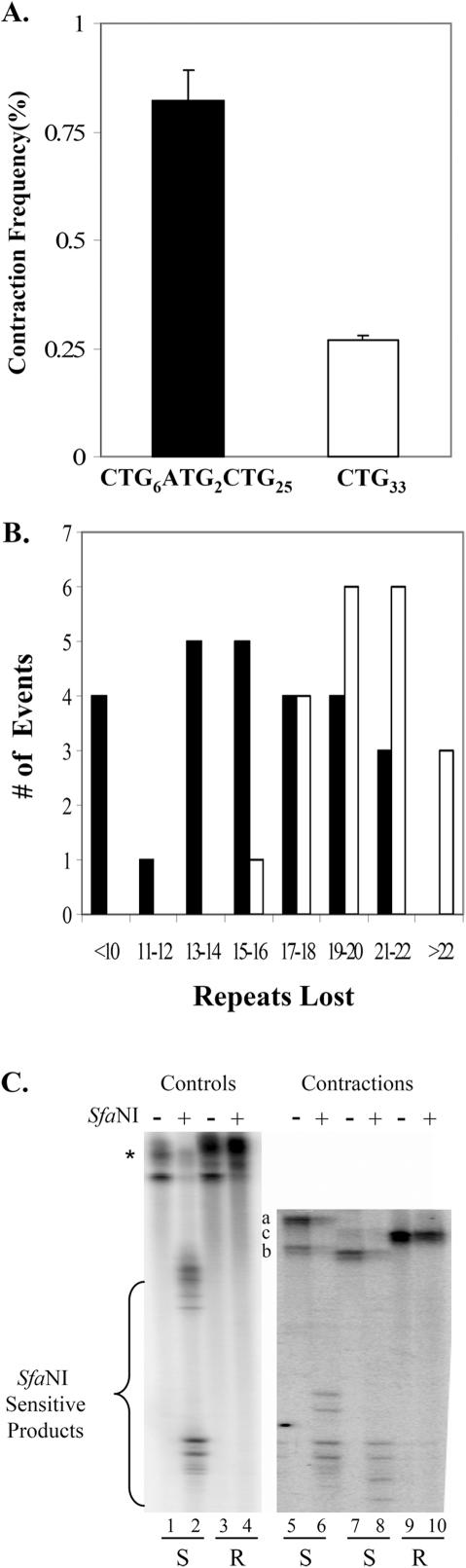
In humans, interrupted alleles harboring 1–3 bp substitutions do not expand, whereas perfect alleles do. The inference is that interruptions inhibit expansions, and that the interrupting nucleotide(s) must be mutated or deleted to yield a perfect, expandable repeat. However there is little or no contraction data available in human cells for interrupted alleles. We used the shuttle vector assay to investigate the contractibility of an interrupted allele in cultured human astrocytes. Shuttle vectors were created with an interrupted repeat, (CTG)6(ATG)2(CTG)25, and a perfect (CTG)33 repeat as a control. After passage of interrupted and perfect repeat harboring shuttle vectors through SVG-A cells, there was a 3-fold increase in contraction frequency for interrupted repeat alleles compared to perfect repeat controls (Figure 5A). The contraction frequency for the interrupted allele was statistically significant (P = 0.0001; entry 10L in Table 1) while the perfect (CTG)33 contraction frequency was borderline significant (P = 0.05; entry 10M in Table 1). The difference in contraction frequency between the interrupted allele and perfect allele, depicted in Figure 5A, was statistically significant (P = 0.005). Thus the interruption in this particular allele resulted in more contractions, not fewer.
Figure 5.
Influence of interruptions on CTG repeat contractions in SVG-A cells. (A) Corrected contraction frequencies (Table 1, entries 9L and 9M) are plotted for starting tracts of (CTG)6(ATG)2(CTG)25 (filled bar) and for (CTG)33 (unfilled bar). Error bars indicate ±1 SEM. (B) Mutation spectra for (CTG)6(ATG)2(CTG)25 (filled bars) versus (CTG)33 (unfilled bars). The number of observed events is plotted on the ordinate versus the size of the contraction (as determined by PCR) on the abscissa. (C) Representative gel showing results of the SfaNI-resistance assay. PCR products were analyzed on 6% denaturing polyacrylamide gels prior to (odd number lanes) or following (even number lanes) treatment with SfaNI, as described in Materials and Methods. Lanes 1–4 show results with PCR products from control 33-repeat plasmids that are interrupted (lanes 1 and 2) or perfect (lanes 3 and 4). The asterisk indicates the starting position of the 33-repeat uncut and SfaNI-resistant perfect repeat tracts. Lanes 5–10 show results with PCR products from contractions of the (CTG)6(ATG)2(CTG)25 allele in SVG-A cells. The letters a–c indicate the position of the uncut samples in lanes 5, 7 and 9, respectively. The letters below the gel image indicate the assignment of SfaNI sensitivity (S) or resistance (R). To become resistant, both ATG interruptions must be lost.
Though contractions occurred more frequently with the interrupted allele, PCR analysis (Figure 5B) revealed that the median contraction size was smaller for an interrupted allele (median contraction size −16 repeats) compared to the perfect repeat control (median contraction size −20 repeats). Consistent with a smaller median contraction size, the spectrum of contraction mutations for the interrupted allele is skewed towards smaller contractions, and the two spectra are statistically different by two-tailed Student's t-test (P < 0.0001). Retention or loss of the interruptions was deduced by treating PCR-amplified repeat tracts from contracted alleles with the restriction enzyme SfaNI. Contracted alleles that retain at least one ATG interruption will be sensitive to SfaNI, but deletions that remove both interruptions become enzyme-resistant. A representative gel obtained from this assay is shown in Figure 5C. SfaNI sensitivity was retained in 14 out of 20 contractions tested, suggesting a 70% bias towards retaining the interruption during contractions of interrupted CTG alleles. To confirm that the SfaNI cleavage assay was appropriately detecting contractions, DNA sequences were obtained for six samples, three each from the SfaNI sensitive and resistant categories. All three contractions which retained SfaNI sensitivity maintained both ATG interruptions, the deletions occurred within the 25 repeat block, and none of the contractions extended into flanking sequence. The three SfaNI resistant samples deleted the interruptions and did not involve flanking DNA. In conclusion, contractions from (CTG)25(ATG)2(CTG)6 were smaller and more frequent than a perfect repeat control, and the interruptions were usually retained.
DISCUSSION
The cells of the CNS are among the most prone to the devastating consequences of TNR expansion, and some CNS cells, including astrocytes, show substantial TNR instability in affected individuals. Furthermore, astrocytes are enriched in brain regions that are sensitive to neurodegeneration and somatic instability in TNR diseases (45,46). Based on this biological relevance, we used immortalized SVG-A astrocytes as an ex vivo model to mimic TNR mutagenesis. The culture model takes into account the proliferative capacity of astrocytes, and therefore enables us to address questions regarding replication-dependent sources of TNR instability in an important CNS cell type. Our results show that astrocytes in culture have substantial levels of CAG·CTG repeat contractions, and the genetic elements controlling this instability mimic signature features from human genetics. Together these findings suggest that replication-based models of TNR contractions are relevant to CNS cells with proliferative capacity, such as astrocytes, and that the molecular mechanisms of instability are likely retained in these cultured cells. To our knowledge, this study comprises the first genetic assay for TNR instability performed in cells derived from the human CNS. The data also suggest that the mechanistic features that govern TNR instability are consistent between CNS cells and other tissue types, such as the kidney (9), that show somatic instability in DM1.
A genetic hallmark of TNR disease is sequence specificity, such that sequences capable of aberrant secondary structure in vitro correlates strongly with instability and disease in human patients (42). In cultured astrocytes, hairpin-forming capacity was critical for producing statistically distinguishable TNR contractions. Furthermore, the unique sensitivity and flexibility of the shuttle vector assay enabled us to determine that this holds true in CNS cells for both short and disease-causing CAG·CTG alleles. Another signature characteristic of TNR instability in humans is the threshold, where mutagenesis increases strongly over a narrow range of repeat lengths, typically 30–40 repeats (1). SVG-A cells exhibit a striking 1000% increase in CAG tract contraction frequency between 25 and 33 repeats, a length change of only 32%. CTG sequences also showed a strong increase in contraction frequencies between 25 and 50 repeats. This is the first genetic evidence that TNR contractions display a threshold-like behavior in cells derived from the human nervous system. A similar result was reported earlier for COS-1 cells (9). Moreover, our observations are consistent with a contraction threshold in cultured astrocytes that is similar to the clinically relevant germline expansion threshold of ∼35 repeats (1). Perhaps as TNR length approaches the threshold in CNS cells, contractions can occur to reduce the allele size back to a more stable length and thereby reduce the risk of disease-causing expansions. We note that our astrocyte system models somatic instability moreso than germline events, and that much less is known about genetic thresholds in somatic tissues. Nonetheless the contraction frequency curve (Figure 3B) strongly suggests a threshold-like response.
The requirement for the SV40 origin of replication for contractions and the orientation effect observed during replication of the shuttle vector in SVG-A cells suggests that CAG·CTG contractions in this system are replication-dependent. This is important in light of the fact that astrocytes and other glia retain proliferative capacity throughout life, unlike terminally differentiated neurons. Therefore, replication could be a key source of somatic instability in glial cells. One indication of the important role of somatic instability in the disease process is a correlation between the amount of somatic instability in specific brain regions and susceptibility to neurodegeneration in HD (46). Since expression of expanded huntingtin in cultured astrocytes decreased their protection of co-cultured neurons against excitotoxicity (25), then replication-mediated somatic instability in glial cells could hasten neurodegeneration in susceptible brain regions.
In our astrocyte model, we observed a ∼10-fold orientation effect such that TNR contractions were strongly favored when CAG repeats form the template for lagging strand synthesis. The magnitude of this effect was constant for allele lengths of both 25 and 33 repeats. Similar findings were observed in 293T and 293 cells, using the same shuttle vector assay (9). However these results are somewhat surprising because contractions in most model systems are favored when CTG runs populate the template strand (13,15,16,47). Perhaps differential binding by trans-acting factors might impart greater contractibility to repeats that are replicated in the CAG orientation. For example, an unknown protein might bind preferentially to the CAG hairpin and stabilize it. Alternatively, RPA or another protein might bind preferentially to single-stranded CTG repeats and reduce their ability to form a hairpin. Whatever the mechanism, our findings highlight this unique feature of TNR instability in human cells that is not observed in yeast or E.coli.
We found that an interrupted CTG repeat allele is approximately three times more contraction prone than a perfect repeat control. Although our observations run contrary to the current belief that interruptions act in a wholesale fashion to prevent TNR instability, it is important to note that ours is the first study of the effect of interruptions on TNR contractions in a human system. In yeast, genetic studies indicated that interruptions prevent contractions primarily by destabilizing putative hairpin intermediates (48), yet our data implies a different mechanism. Since the majority (70%) of contractions from interrupted TNRs retained at least one of the interruptions, it suggests that the interruptions are excluded from any mutagenic hairpins, but somehow promote contraction in the nearby DNA. The 30% of contractions that delete interruptions also suggest that creation of a shortened but perfect TNR allele via a contraction also represents a potentially significant event. Consistent with this idea, when disease-free alleles at the SCA1 locus were analyzed, 98% contained interruptions, but the normal alleles that lacked interruptions were among the shortest observed (49), perhaps the byproduct of a TNR contraction. If interruptions indeed promote contraction of TNR alleles, this would further highlight their critical importance in maintaining short, expansion-averse TNRs in the human genome.
Acknowledgments
The authors gratefully acknowledge Dr Raisa Persidsky and Dr Anuja Ghorpade for assistance in immunostaining of SVG-A cells, and Dr Richard Pelletier for instruction (to B.T.F.) in the procedures for the shuttle vector assay and for cloning of some of the shuttle vectors used here. This work was supported by National Institutes of Health grant GM61961 (to R.S.L.), by a graduate fellowship from the University of Nebraska Medical Center and National Institutes of Health fellowship F30 NS046135 (to B.T.F.), and by National Cancer Institute Cancer Center Support Grant P30 CA36727 (to the Eppley Institute). Funding to pay the Open Access publication charges for this article was provided by NIH GM61961.
Conflict of interest statement. None declared.
REFERENCES
- 1.Paulson H.L., Fischbeck K.H. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin S.M. Molecular models for repeat expansions. Chemtracts-Biochem. Mol. Biol. 2004;17:639–662. [Google Scholar]
- 3.Pearson C.E., Edamura K.N., Cleary J.D. Repeat instability: mechanisms of dynamic mutations. Nature Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 4.Kovtun I.V., McMurray C.T. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 5.Gomes-Pereira M., Fortune M.T., Monckton D.G. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001;10:845–854. doi: 10.1093/hmg/10.8.845. [DOI] [PubMed] [Google Scholar]
- 6.Fortune M.T., Vassilopoulos C., Coolbaugh M.I., Siciliano M.J., Monckton D.G. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y., Dion V., Wilson J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nature Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 8.Yoon S.-R., Dubeau L., de Young M., Wexler N.S., Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl Acad. Sci. USA. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelletier R., Farrell B.T., Miret J.J., Lahue R.S. Mechanistic features of CAG·CTG repeat contractions in cultured cells revealed by a novel genetic assay. Nucleic Acids Res. 2005;33:5667–5676. doi: 10.1093/nar/gki880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panigrahi G.B., Cleary J.D., Pearson C.E. In vitro (CTG)·(CAG) expansions and deletions by human cell extracts. J. Biol. Chem. 2002;277:13926–13934. doi: 10.1074/jbc.M109761200. [DOI] [PubMed] [Google Scholar]
- 11.Cleary J.D., Nichol K., Wang Y.-H., Pearson C.E. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nature Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z., Lau R., Marcadier J.L., Chitayat D., Pearson C.E. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type I disease locus in human cells. Am. J. Hum. Genet. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang S., Jaworski A., Ohshima K., Wells R.D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in Escherichia coli. Nature Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 14.Samadashwily G.M., Raca G., Mirkin S.M. Trinucleotide repeats affect DNA replication in vivo. Nature Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 15.Maurer D.J., O'Callaghan B.L., Livingston D.M. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:6617–6622. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miret J.J., Pessoa-Brandao L., Lahue R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato T., Oyake M., Nakamura K., Nakao K., Fukusima Y., Onodera O., Igarashi S., Takano H., Kikugawa K., Ishida Y., et al. Transgenic mice harboring a full-length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients. Hum. Mol. Genet. 1999;8:99–106. doi: 10.1093/hmg/8.1.99. [DOI] [PubMed] [Google Scholar]
- 18.Watase K., Venken K.J.G., Sun Y., Orr H.T., Zoghbi H.Y. Regional differences of somatic CAG repeat instability do not account for selective neuronal vulnerability in a knock-in mouse model of SCA1. Hum. Mol. Genet. 2003;12:2789–2795. doi: 10.1093/hmg/ddg300. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy L., Evans E., Chen C.M., Craven L., Detloff P.J., Ennis M., Shelbourne P.F. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 20.Chong S.S., McCall A.E., Cota J., Subramony S.H., Orr H.T., Hughes M.R., Zoghbi H.Y. Gametic and somatic tissue-specific heterogeneity of the expanded SCA 1 CAG repeat in spinocerebellar ataxia type 1. Nature Genet. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- 21.Takano H., Onodera O., Takahashi H., Igarashi S., Yamada M., Oyake M., Ikeuchi T., Koide R., Tanaka H., Iwabuchi K., et al. Somatic mosaicism of expanded CAG repeats in brains of patients with dentatorubral-pallidoluysian atrophy: cellular population-dependent dynamics of mitotic instability. Am. J. Hum. Genet. 1996;58:1212–1222. [PMC free article] [PubMed] [Google Scholar]
- 22.Maciel P., Lopes-Cendes I., Kish S., Sequeiros J., Rouleau G.A. Mosaicism of the CAG repeat in CNS tissue in relation to age at death in spinocerebellar ataxia type 1 and Machado-Joseph disease patients. Am. J. Hum. Genet. 1997;60:993–996. [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe H., Tanaka F., Doyu M., Riku S., Yoshida M., Hashizume Y., Sobue G. Differential somatic CAG repeat instability in variable brain cell lineage in dentatorubral pallidoluysian atrophy (DRPLA): a laser-captured microdissection (LCM)-based analysis. Hum. Genet. 2000;107:452–457. doi: 10.1007/s004390000400. [DOI] [PubMed] [Google Scholar]
- 24.Kretzschmar D., Tschape J., Bettencourt Da Cruz A., Asan E., Poeck B., Strauss R., Pflugfelder G.O. Glial and neuronal expression of polyglutamine proteins induce behavioral changes and aggregate formation in Drosophila. Glia. 2005;49:59–72. doi: 10.1002/glia.20098. [DOI] [PubMed] [Google Scholar]
- 25.Shin J.Y., Fang Z.H., Yu Z.X., Wang C.E., Li S.H., Li X.J. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J. Cell. Biol. 2005;171:1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leeflang E.P., Zhang L., Tavare S., Hubert R., Srinidhi J., MacDonald M.E., Myers R.H., de Young M., Wexler N.S., Gusella J.F., et al. Single sperm analysis of the trinucleotide repeats in the Huntington's disease gene: quantification of the mutation frequency spectrum. Hum. Mol. Genet. 1995;4:1519–1526. doi: 10.1093/hmg/4.9.1519. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L., Leeflang E.P., Yu J., Arnheim N. Studying human mutations by sperm typing: instability of CAG trinucleotide repeats in the human androgen receptor gene. Nature Genet. 1994;7:531–535. doi: 10.1038/ng0894-531. [DOI] [PubMed] [Google Scholar]
- 28.Takiyama Y., Sakoe K., Soutome M., Namekawa M., Ogawa T., Nakano I., Igarashi S., Oyake M., Tanaka H., Tsuji S., et al. Single sperm analysis of the CAG repeats in the gene for Machado-Joseph disease (MJD1): evidence for non-Mendelian transmission of the MJD1 gene and for the effect of the intragenic CGG/GGG polymorphism on the intergenerational instability. Hum. Mol. Genet. 1997;6:1063–1068. doi: 10.1093/hmg/6.7.1063. [DOI] [PubMed] [Google Scholar]
- 29.Mornet E., Chateau C., Hirst M.C., Thepot F., Taillandier A., Cibois O., Serre J.-L. Analysis of germline variation at the FMR1 CGG repeat shows variation in the normal-premutated borderline range. Hum. Mol. Genet. 1996;5:821–825. doi: 10.1093/hmg/5.6.821. [DOI] [PubMed] [Google Scholar]
- 30.Kunst C.B., Leeflang E.P., Iber J.C., Arnheim N., Warren S.T. The effect of FMR1 CGG repeat interruptions on mutation frequency as measured by sperm typing. J. Med. Genet. 1997;34:627–631. doi: 10.1136/jmg.34.8.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovtun I.V., Thornhill A.R., McMurray C.T. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum. Mol. Genet. 2005;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya S., Lahue R.S. Yeast Srs2 DNA helicase selectively blocks expansions of trinucleotide repeats. Mol. Cell. Biol. 2004;24:7324–7330. doi: 10.1128/MCB.24.17.7324-7330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Major E.O., Miller A.E., Mourrain P., Traub R.G., de Widt E., Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc. Natl Acad. Sci. USA. 1985;82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gee G.V., Manley K., Atwood W.J. Derivation of a JC virus-resistant human glial cell line: implications for the identification of host cell factors that determine viral tropism. Virology. 2003;314:101–109. doi: 10.1016/s0042-6822(03)00389-1. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolfsmeier M.L., Dixon M.J., Pessoa-Brandao L., Pelletier R., Miret J.J., Lahue R.S. Cis-elements governing trinucleotide repeat instability in Saccharomyces cerevisiae. Genetics. 2001;157:1569–1579. doi: 10.1093/genetics/157.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 38.Gietz D., St Jean A., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heffron D.S., Mandell J.W. Opposing roles of ERK and p38 MAP kinases in FGF2-induced astroglial process extension. Mol. Cell. Neurosci. 2005;28:779–790. doi: 10.1016/j.mcn.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Roberts J.D., Nguyen D., Kunkel T.A. Frameshift fidelity during replication of double-stranded DNA in HeLa cell extracts. Biochemistry. 1993;32:4083–4089. doi: 10.1021/bi00066a033. [DOI] [PubMed] [Google Scholar]
- 41.Gruss C., Gutierrez C., Burhans W.C., DePamphilis M.L., Koller T., Sogo J.M. Nucleosome assembly in mammalian cell extracts before and after DNA replication. EMBO J. 1990;9:2911–2922. doi: 10.1002/j.1460-2075.1990.tb07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gacy A.M., Goellner G., Juranic N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structure in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 43.Pearson C.E., Sinden R.R. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 44.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2253. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roos R.A., Bots G.T., Hermans J. Neuronal nuclear membrane indentation and astrocyte/neuron ratio in Huntington's disease. A quantitative electron microscopic study. J. Hirnforsch. 1985;26:689–693. [PubMed] [Google Scholar]
- 46.Kono Y., Agawa Y., Watanabe Y., Ohama E., Nanba E., Nakashima K. Analysis of the CAG repeat number in a patient with Huntington's disease. Intern. Med. 1999;38:407–411. doi: 10.2169/internalmedicine.38.407. [DOI] [PubMed] [Google Scholar]
- 47.Freudenreich C.H., Stavenhagen J.B., Zakian V.A. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dixon M.J., Lahue R.S. DNA elements important for CAG·CTG repeat thresholds in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:1289–1297. doi: 10.1093/nar/gkh292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung M.-Y., Ranum L.P.W., Duvick L.A., Servadio A., Zoghbi H.Y., Orr H.T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nature Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]