Abstract
Lipopolysaccharide (LPS) is a potent stimulator of monocytes and macrophages, causing secretion of tumor necrosis factor α (TNF-α) and other inflammatory mediators. Given the deleterious effects to the host of TNF-α, it has been postulated that TNF-α gene expression must be tightly regulated. The nature of the nuclear factor(s) that control TNF-α gene transcription in humans remains obscure, although NF-κB has been suggested. Our previous studies pertaining to macrophage response to LPS identified a novel DNA-binding domain located from −550 to −487 in the human TNF-α promoter that contains transcriptional activity, but lacks any known NF-κB-binding sites. We have used this DNA fragment to isolate and purify a 60-kDa protein binding to this fragment and obtained its amino-terminal sequence, which was used to design degenerate probes to screen a cDNA library from THP-1 cells. A novel cDNA clone (1.8 kb) was isolated and fully sequenced. Characterization of this cDNA clone revealed that its induction was dependent on LPS activation of THP-1 cells; hence, the name LPS-induced TNF-alpha factor (LITAF). Inhibition of LITAF mRNA expression in THP-1 cells resulted in a reduction of TNF-α transcripts. In addition, high level of expression of LITAF mRNA was observed predominantly in the placenta, peripheral blood leukocytes, lymph nodes, and the spleen. Finally, chromosomal localization using fluorescence in situ hybridization revealed that LITAF mapped to chromosome 16p12–16p13.3. Together, these findings suggest that LITAF plays an important role in the activation of the human TNF-α gene and proposes a new mechanism to control TNF-α gene expression.
The innate host response to bacterial pathogens is characterized by an immediate release of biologically active compounds, including monokines and cytokines. These proinflammatory molecules, which are intended to enable the host to eliminate the pathogen, may adversely affect the host. Endotoxins, produced from the outer membrane of Gram-negative bacteria, and exotoxins, released from the cell wall of Gram-positive bacteria, are known to be potent inducers of the inflammatory response (1, 2). Lipopolysaccharide (LPS), extracted from the outer membrane of Gram-negative bacteria, has been identified as a principal endotoxic component. It is well known that LPS is one of the most potent stimulators of monocytes and macrophages, leading to the secretion of nitrogen intermediates, prostaglandins, and cytokines. Secretion of tumor necrosis factor α (TNF-α), IL-1, IL-6, and IL-12, demonstrated both in vivo and in vitro (3, 4), leads to rapid induction and amplification of the host response to infection (5–8). In turn, other cells are induced to produce large quantities of IL-1, TNF, IL-6, and IL-8, which mediate the induction of fever (6, 7, 9), synthesis of acute-phase proteins (10), release of collagenase (8), and synthesis of prostaglandin (11). In acute situations, the pathogen often is eliminated, with resolution of inflammation and minimal tissue damage. However, failure to control the pathogen often leads to a state of metabolic anarchy in which the inflammatory response is not controlled and significant tissue damage results.
Although the inflammatory response is mediated by a variety of secreted factors, the cytotoxic effects of LPS have been ascribed to TNF-α activity (4, 12, 13). TNF-α is a pleiotropic cytokine and may benefit the host or exert detrimental effects on the host (14–16). TNF-α helps prevent cancer (17), protects against infection (18–21), promotes tissue remodeling (22), and activates inflammatory responses (23). Conversely, TNF-α mediates septic shock in chronic infections (4, 24, 25), is responsible for cachexia in cancer patients (26), causes inflammation in rheumatoid arthritis patients (27, 28), and activates HIV (29–32). The pleiotropic effects of TNF-α are dose-dependent (33). Hence, the perceived need to control TNF-α production has raised interest into the understanding of the mechanisms that modulate TNF-α gene expression.
It is well known that gene transcription is controlled by DNA-binding proteins (34). Recently, several groups have examined the transcriptional regulation of TNF-α by various inducers, such as virus, LPS, and phorbol 12-myristate 13-acetate (PMA) (35–45). The human TNF-α promoter contains motifs that resemble NF-κB-binding sites; however, controversy exists as to the involvement of NF-κB in TNF-α gene regulation. These sequences do not seem to be necessary for virus or LPS induction, nor do they appear to be able to stimulate virus or LPS induction alone (35). However, it has been suggested that NF-κB is an important factor in TNF-α gene transcription in LPS-challenged monocytes and macrophages. NF-κB-binding motifs are found in the human TNF-α promoter region (46, 47) and were shown to translocate into the nuclei of LPS-stimulated monocytes (36, 48, 49). In mice, mutation(s) or deletion(s) of NF-κB-binding motifs on the TNF-α promoter failed to show reporter gene activation in transfected cells (37, 50, 51). However, in humans, TNF-α promoter activity in transfected cell lines was found to be independent of the NF-κB-binding motifs (52). Drouet et al. (37) offered an explanation for these conflicting data, suggesting that enough NF-κB is constitutively expressed to sustain high-level baseline expression of the human TNF-α gene (52) compared with the mouse. Nonetheless, the nature of the nuclear factor(s) involved in the regulation of LPS-induced TNF-α gene expression in humans remains unknown.
Our previous studies pertaining to macrophage response to LPS have identified a DNA sequence domain located from −550 to −487 in the human TNF-α promoter (53). Using electrophoretic mobility-shift assays and selective mutations, it was shown that a 64-bp fragment located within this region can bind unknown protein(s) and this binding is responsible for TNF-α transcriptional activity. Sequence analysis of this fragment revealed the absence of any potential NF-κB-binding sites (53). These intriguing results suggested the existence of a novel cis-acting regulatory protein, other than NF-κB, that is necessary for human TNF-α gene transcription.
In an effort to further our understanding of the molecular mechanisms of LPS-induced TNF-α gene regulation, we used this 64-bp fragment to isolate the purported native protein. Its amino-terminal sequence was obtained and used to design PCR primers to screen a THP-1 cDNA library. Using these probes, we isolated a novel full-length cDNA (1.8 kb) with a deduced amino acid sequence of the ORF encoding 229 aa. Characterization of this new gene revealed that its expression resulted from LPS stimulation of THP-1 cells; hence, the name LPS-induced TNF-alpha factor (LITAF). We investigated tissue distribution of the LITAF gene and found transcripts in a variety of human adult tissues. Northern blot hybridization revealed a high level of expression in the placenta, peripheral blood leukocytes, lymph nodes, and the spleen. Inhibition of LITAF mRNA translation using antisense DNA in naive THP-1 cells or those exposed to LPS resulted in a reduction of TNF-α transcripts. In addition, chromosomal localization using fluorescence in situ hybridization (FISH) revealed that LITAF mapped to chromosome 16p12–16p13.3. Together, these findings suggest that the LITAF gene may play an important role in human TNF-α gene activation.
MATERIALS AND METHODS
Cell Culture.
The human monocytic cell line, THP-1, was maintained in complete RPMI [C-RPMI; RPMI 1640 medium supplemented with 2 mM l-glutamine/25 mM Hepes/100 units/ml of penicillin/100 mg/ml of streptomycin/10% FBS (all from GIBCO)]. LPS-free tissue culture reagents were used for all experiments.
Preparation of Nuclear Extracts from Cultured THP-1 Cells.
THP-1 cells were induced to maturation by incubation in 200 nM PMA (Sigma) for 20 hr and then stimulated with 100 ng/ml of Porphyromonas gingivalis LPS (54) for 2 hr. Nuclear extracts were prepared as described in our previous work (53).
Preparation of DNA Affinity Beads.
The 64-bp initiator element located from –550 to −487 in the human TNF-α promoter (55) was amplified by PCR. The PCR mixture (50 ml) was prepared as described previously (55). The human TNF-α promoter (55) was used as the template for the PCR. We used the sequence 5′-TGAGGCCTCAAGCTGCCACCA-3′ for the forward primer sequence and 5′-XTGAGGCCTGTGTTTGGGTCTG-3′ for the reverse sequence (X = 5′ biotin). The cycling parameters (30 cycles) were as follows: an initial denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and elongation at 72°C for 1 min. The last cycle had an additional elongation time of 7 min. The PCR products were separated on agarose gel, and the 64-bp DNA bands were cut from the gel. The unincorporated biotinylated primers were removed by using the QIAEX II Gel Extraction Kit (Qiagen). The biotinylated DNA was desalted and dissolved in Tris⋅HCl/EDTA (2 mg/ml) and then used for the preparation of DNA affinity beads. Immobilization of labeled DNA to Dynabeads M-280 streptavidin (10 mg/ml; Dynal, Great Neck, NY) was performed as described by Gabrielsen and Huet (56). Briefly, a suspension of beads was coupled with the labeled DNA at room temperature for 30 min. The affinity beads were washed to remove unbound DNA and then used for purification of DNA-binding protein.
Purification of DNA-Binding Protein Using Affinity Beads.
Purification of the protein binding to the beads was performed as described previously (56). Briefly, the nuclear extracts were incubated with the beads in binding buffer containing poly (dI-dC) at room temperature. After washing (twice), the DNA-binding protein was eluted, desalted, and then used for further analysis.
Isolation of the DNA-Binding Protein and Sequence Analysis.
The eluted protein sample was separated on 12% SDS-polyacrylamide gel as described previously (53). The resolved proteins (approximately 2% of the protein of the total protein) were electroblotted onto a polyvinylidene difluoride membrane as described by Matsudaira (57) and stained with Coomassie blue. A predominant-stained protein band with an apparent molecular mass of 60 kDa was excised. After treatment with trypsin (58), the resultant peptides were separated by HPLC on a C18 column with a 0–100% acetonitrile gradient containing 0.01% trifluoroacetic acid (58). Fractions were collected, and a number of peptides were sequenced by established methods as described previously (57). All peptide sequences were found in the isolated cDNA clone. Degenerate oligonucleotides (16-mer) corresponding to either end of peptide 1, M-S-V-P-G-P-Y-Q-A-A-T-G, were used for PCR by using the LPS-induced THP-1 cDNA as a template to obtain a DNA fragment containing the exact nucleotide sequences encoding the middle part of this peptide. PCR products resolved in an 8% acrylamide gel were eluted by using the QIAEX II Gel Extraction Kit (Qiagen) and subcloned into a PCRII vector (Invitrogen). Sequencing verified the fidelity of the PCR products.
Screening the cDNA Library.
An LPS-stimulated, PMA-induced, custom-made human THP-1 Uni-ZAP XR cDNA library (Stratagene) was screened by using this PCR probe, and nine positive clones containing 0.8- to 1.8-kb inserts were obtained after screening 5 × 105 colonies. The longest cDNA insert (1.8 kb) was chosen for complete sequencing after the dideoxynucleotide chain-termination method by using a 373A sequencer (Applied Biosystems).
The blast program was used to search the database through the National Center for Biotechnology Information. The pileup and prettybox programs were used for sequence alignments and comparisons. The fasta program was used for calculating the protein identities. All these computer programs were from the Wisconsin Genetics Computer Group, Madison, WI. Finally, the sigscan program was used to search the Transcription Factor Database (TFD) through the bimas program of the Advanced Biosciences Computing Systems maintained at the University of Minnesota.
Detection of LITAF Gene Transcripts.
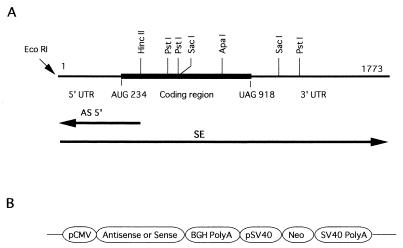
The transcripts for the new gene were detected by Northern blot analysis in cultured THP-1 cells and other human tissues. First, THP-1 cells (5 × 106) were induced to maturation by incubation in 200 nM PMA (Sigma) for 20 hr and then stimulated with 100 ng/ml of P. gingivalis LPS (54) for 2 hr. Noninduced THP-1 cells also were cultured. Total cellular RNA was collected by using RNA STAT-60 (Tel-Test, Friendswood, TX) by the method described in the instruction manual, and poly(A)+ RNA (mRNA) was recovered by using the Oligotex mRNA Kit (Qiagen). mRNA was size-fractionated on denaturing formaldehyde-agarose gel (1.1%) and transferred onto a Hybond-N+ membrane (Amersham) as described previously (59). Northern blot filters with mRNA from several different human tissues were obtained from CLONTECH. These filters were hybridized with an 35S-labeled cRNA probe at 60°C overnight in 50% formamide/5× SSC/5× Denhardt’s solution/0.1% SDS/50 mM sodium phosphate, pH 6.9/heat-denatured salmon sperm DNA. The filters were washed three times in 0.1× SSC/0.1% SDS at 68°C for 30 min and then autoradiographed with BIOMAX MR film (Kodak). The 511 bp of the HincII-ApaI cDNA fragment from the coding sequence in the new gene (Fig. 1) was subcloned into pGEM7Zf(+) (Promega). The antisense and sense riboprobes were prepared from this plasmid and labeled with [35S]UTP as described previously (59). These probes were used for Northern hybridizations.
Figure 1.
cDNA cloning of LITAF. (A) Physical map of the LITAF cDNA isolated. The dark, thicker line denotes the ORF (from AUG 234 to UAG 918) in LITAF. Restriction sites for EcoRI, HincII, PstI, SacI, and ApaI are shown. Note that the antisense RNA represented as AS5′ was designed to be complementary to the 5′ region of LITAF mRNA between the restriction sites (EcoRI and HincII). However, the full-length LITAF was used as sense RNA for overexpression of LITAF RNA and is shown as SE. (B) Schematic structure of antisense or sense constructs inserted downstream of the cytomegalovirus promoter.
Antisense and Sense Construct.
Eukaryotic expression vectors (pRc/CMV; Invitrogen) were constructed as described previously (60, 61) with slight modifications. Briefly, the LITAF cDNA fragments (indicated below and in Fig. 1) were placed in either an antisense or a sense orientation downstream of the human cytomegalovirus promoter element as the first ORF. The vector contained a neo-resistance gene for selection of stable transfectants. Plasmid constructions were as follows: AS5′, antisense cDNA corresponding to a 320-bp fragment that covered the 5′ untranslated region (UTR) and the AUG start codon; SE, sense full-length of cDNA; and CON, control vector without insert. These constructions are shown in Fig. 1. All plasmids were prepared by using Qiagen Plasmid Midiprep Kits.
Stable Transfection.
THP-1 cells (5 × 106/cuvette) were transfected by electroporation (Gene Pulser; Bio-Rad) with 20 mg of recombinant plasmid according to the manufacturer’s instructions. Mock cells were transfected similarly with a control vector (pRcCON). Two days later, cells were cultured in C-RPMI containing 0.8 mg/ml of G418 (Geneticin; GIBCO). This medium was changed every 3 days. The surviving cells were used for further experimentation.
Monitoring of Exogenous mRNA Expression.
Success of stable transformation was confirmed by Northern blot analysis with antisense and sense riboprobes. Total cellular RNA was recovered from transfected cells (2 × 106) and used for Northern blot analysis in a manner similar to that described previously. The 707 bp of the EcoRI-SacI cDNA fragment, which contained a 5′ UTR and a coding sequence for the new gene (Fig. 1), was subcloned into pBluescript SKII(−). The 35S-labeled riboprobes for hybridization were prepared from this plasmid as described previously (59).
Quantification of TNF-α mRNA Expression in Transfected Cells.
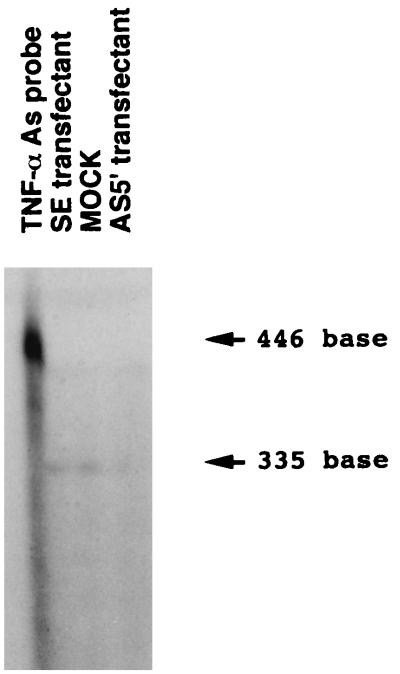
TNF-α mRNA expression was quantified by using an RNase protection assay. Transformed THP-1 cells (2 × 106) were incubated with 200 nM of PMA (Sigma) for 20 hr to induce differentiation and then stimulated with 100 ng/ml of P. gingivalis LPS (54) for 2 hr. mRNA was recovered from the cells as described previously. Successful recovery of mRNA from each sample was monitored by Northern blot hybridization with a β-actin probe. The 355-bp TNF-α cDNA was obtained from a PMA-induced human THP-1 Uni-ZAP XR cDNA library (Stratagene) by PCR with specific primers for TNF-α mRNA (Stratagene). After the cDNA was subcloned to a SmaI site of pGEM7Zf(+), the TNF-α cDNA was identified by sequencing. After the plasmid was linearized with XbaI and transcribed with SP6 polymerase, a 446-bp 35S-labeled antisense probe was constructed. The RNase protection assays were performed as described previously (59).
FISH.
The LITAF full-length cDNA probe was nick-translated with biotin by using kit no. S4099 (Oncor) according to the manufacturer’s instructions. One-half microgram of human Cot-1 DNA and 75 μg of salmon sperm DNA were resuspended in 50 μl of hybrisol VII (catalog no. S1390-10; Oncor) by sonication. The probe was denatured at 70°C for 5 min and incubated at 37°C for 30 min for biotin labeling before mixing with the D16Z1 probe. A digoxygenin-labeled D16Z1 Chromosome 16 α-satellite Probe (catalog no. P5035-DG-5; Oncor) was denatured at 70°C for 5 min and used to confirm the location of the LITAF gene on chromosome 16. Normal lymphocytes were treated with colcemid to arrest cell division, harvested, fixed, and placed onto slides according to standard cytogenetic protocols. The slides were denatured in 2× SSC (pH 7.0) plus 70% formamide at 72°C for 2 min, followed by dehydration in a series of ethanol rinses. Equal volumes of labeled, denatured probes were mixed, and 20 μl was added to each dehydrated slide. Hybridization proceeded for 15–17 hr at 37°C. The slides were washed for 15 min at 43°C in 2× SSC plus 65% formamide and then at 37°C for 8 min in 2× SSC. The biotin-labeled probe was detected by binding to avidin-Texas red, washed, amplified with antiavidin, washed, and amplified with avidin-Texas red by using the Biotin-Texas Red Detection Kit (catalog no. S1334-BTR; Oncor) according to the manufacturer’s instructions. The digoxygenin-labeled hybridized probe was detected with FITC-labeled antidigoxygenin antibody, washed, amplified with rabbit-anti-sheep antibody, washed, and amplified with FITC-anti-rabbit antibody (Digoxygenin-FITC Detection Kit, Cat. No. S1331-DF) according to the manufacturer’s instructions. The slides were counterstained with 4′,6-diamidino-2-phenylindole and photographed by using triple-excitation/emission bandpass filters (Cat. No 61002X; Olympus, New Hyde Park, NY).
RESULTS
Screening, Sequencing, and Structural Analysis of LITAF cDNA.
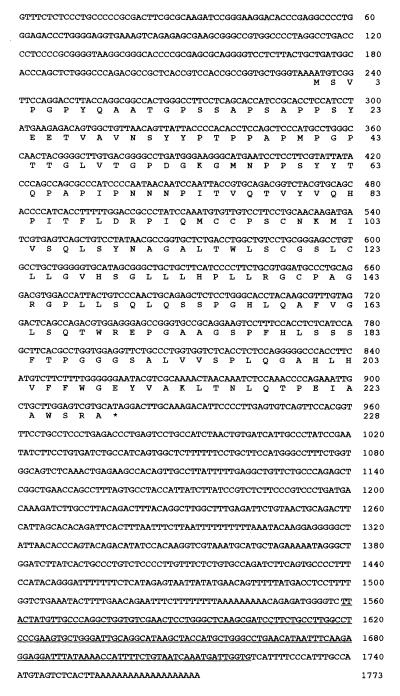
As shown in Fig. 2, the cDNA consists of a 234-nt 5′ noncoding region, a 687-nt ORF, and an 852-nt 3′ noncoding region that includes a poly(A) signal. The ORF encodes a 228-aa, 23.9-kDa protein (GenBank accession no. U77396). At the time that the DNA sequencing was almost completed, the clone was entered in GenBank as TNF-α-induced mRNA and did not have any homology with nucleotide and protein sequences available in all public databases. Polyak et al. (62) then examined transcripts markedly induced by the p53 gene. Among them, an expressed sequence tag named PIG7 was found to harbor 98% homology with TNF-α-induced mRNA (62). We have, in the present report, renamed this cDNA LITAF because it is induced by LPS and affects TNF-α gene expression.
Figure 2.
Nucleotide sequence of LITAF and its deduced amino acid sequence. The full-length LITAF cDNA was isolated from a cDNA library of PMA-differentiated THP-1 cells stimulated with LPS. This sequence has been submitted to GenBank and has been assigned accession no. U77396. The ORF encodes 228 aa with a predicted molecular mass of 23.9 kDa. The Alu sequence is underlined.
Induction of LITAF Gene mRNA.
Northern blot analysis clearly indicated that a single 1.8-kb mRNA encoding the LITAF gene was present in LPS-induced PMA-differentiated THP-1 cells. The size of the transcript was consistent with the sequence data. The expression of the LITAF gene occurred only in LPS-induced, PMA-differentiated THP-1 cells (Fig. 3). No signal was found in PMA-differentiated cells in the absence of LPS stimulation, in LPS-stimulated cells in the absence of PMA differentiation, or in unstimulated THP-1 cells (Fig. 3).
Figure 3.

Northern blot hybridization of mRNA from THP-1 cells. The cells (2 × 107) were cultured in various conditions: stimulation with P. gingivalis LPS (LPS); differentiation with PMA (PMA); differentiation with PMA followed by stimulation with LPS (PMA+LPS); and no stimulation (plain). The mRNA was recovered from cells, run on denaturing formaldehyde/1.2% agarose gel, and transferred to a Hybond-N+ filter. The filter was hybridized with the antisense RNA probe that corresponded to the coding region of the LITAF gene, as described in Materials and Methods. The hybridized filter was exposed to x-ray film for 24 hr. A 1.8-kb transcript was observed only in PMA+LPS-stimulated THP-1 cells. Similar amounts of β-actin mRNA were found in all mRNA tested (data not shown).
Expression Patterns of the Novel Gene in Human Tissues.
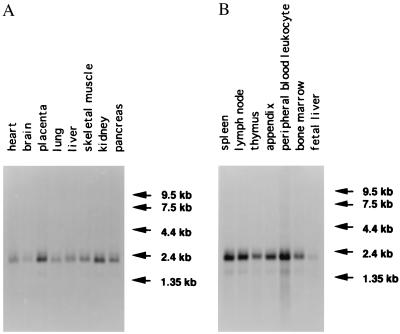
As shown in Fig. 4, a 1.8-kb transcript was significantly expressed in spleen, lymph node, and peripheral blood leukocytes. Moderate expression was observed in thymus, appendix, bone marrow, kidney, and placenta. Little expression was found in pancreas, skeletal muscle, liver, and lung whereas a very minor expression was observed in heart, brain, and fetal liver. The size of the transcripts again was consistent with the sequence results and confirms that the cDNA isolated was the full-length clone.
Figure 4.
Northern blot hybridization of mRNA (2 μg) from different adult tissues. (A and B) Expression of the LITAF gene. The preblotted filters (CLONTECH) were hybridized with the antisense RNA probe that corresponded to the coding region of the LITAF gene, as described in Materials and Methods.
Antisense and Sense LITAF mRNA Constructs and Expression.
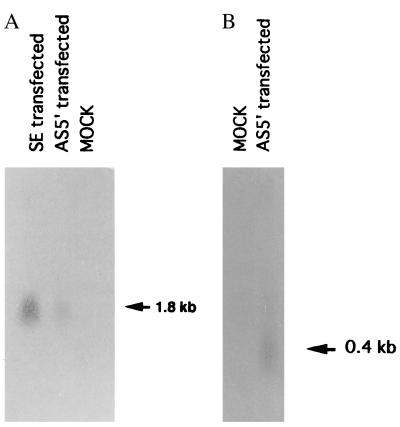
To determine the biological activity of the isolated LITAF gene, THP-1 cells were transfected with the following constructs: LITAF antisense RNA (AS5′), LITAF sense RNA construct(s), or vector alone. Using LITAF antisense RNA as a probe, a 1.8-kb transcript was detected in sense-transfected cells (Fig. 5A), and using LITAF sense RNA as a probe, we detected a band of approximately 400 bp in antisense-transfected cells (Fig. 5B). No signal was detected in mock-transfected cells (Fig. 5). These results are consistent with the antisense construction design.
Figure 5.
Northern blot hybridization of RNA from THP-1 cells transfected with AS5′, SE, or control vector (MOCK). Total RNA was recovered from 2 × 106 cells after differentiation with PMA and stimulation with P. gingivalis LPS. (A) RNAs were hybridized with the LITAF sense RNA probe. (B) RNAs were hybridized with the LITAF antisense RNA probe. Similar amounts of β-actin mRNA were found in LITAF antisense-expressing cells, LITAF sense-expressing cells, and mock-transfected cells (data not shown).
Subsequent examination of the effect of inhibiting LITAF RNA using LITAF antisense constructs on the transcription of the human TNF-α gene revealed that TNF-α mRNA signals were reduced more strongly in LITAF antisense-expressing cells than in mock-transfected cells (Fig. 6). No changes in TNF-α mRNA signals were observed with LITAF sense-expressing cells compared with mock-transfected cells (Fig. 6). Similar amounts of β-actin mRNA were found in LITAF antisense-expressing cells, LITAF sense-expressing cells, and mock-transfected cells (data not shown).
Figure 6.
RNase protection assay of RNA from THP-1 cells transfected with AS5′, SE, or control vector (MOCK). Total RNA was recovered from 2 × 106 cells after differentiation with PMA and stimulation with P. gingivalis LPS. The RNAs were hybridized with the TNF-α antisense RNA probe. After treatment with RNase A and T1, protected bands were electrophoresed through an 8% polyacrylamide gel, and the dried gel was exposed to x-ray film. No protected band is observed in AS5′-transfected cells. The band density observed in SE-transfected cells was found to be similar to the band in MOCK-transfected cells. Left lane, native TNF-α antisense probe.
Chromosome Localization.
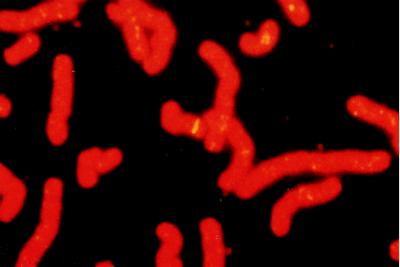
FISH of the LITAF cDNA probe to human metaphase spreads resulted in specific labeling on chromosome 16 (Fig. 7). The location on chromosome 16 α-satellite was confirmed by double-labeling with a commercial probe, D16Z2 (Oncor). The probe had specificity for this site, because symmetrical signals were not observed on other chromosomes. The FISH signals were localized relative to the chromosome bands and the LITAF gene locus assigned to chromosome 16 p12–16p13.3.
Figure 7.
FISH localization of human LITAF gene. Metaphase chromosome spread after hybridization with a biotinylated LITAF probe. The banding is observed on chromosome 16p12–16p13.3.
DISCUSSION
The present paper reports the cloning, characterization, and chromosomal assignment of an LPS-induced gene named LITAF that affects TNF-α gene expression in cells of monocytic lineage. The cDNA encoding LITAF was isolated by screening an LPS-induced THP-1 expression library with degenerate oligonucleotides corresponding to the native nuclear protein isolated. Functional studies aimed at interfering with LITAF mRNA expression by using antisense RNA in PMA-differentiated THP-1 cells exposed to LPS resulted in a reduction of TNF-α transcripts. In humans this gene has been localized to chromosome 16p12–16p13.3.
All the public nucleotide and protein sequence databases were searched. None of the sequences in the coding region were found to be similar to typical DNA-binding motifs. We did find sequences in two other regions of the gene (5′ and 3′ UTRs) that displayed some homology with sequences reported in the databases: several amino acid sequences were found to be similar to the Bicaudal-C gene of Drosophila melanogaster. Although the Bicaudal-C gene product contains a motif called the KH domain, which is found in many RNA and single-stranded DNA-binding proteins (63), we did not find a typical KH domain in the LITAF gene. In addition, sequencing of the LITAF gene revealed Alu elements in the 3′ noncoding region (Fig. 2). Several investigators have reported that Alu elements constitute approximately 5% of the human genome and often are found in introns or 3′ UTRs. However, the Alu region mistakenly may be incorporated into ORFs (84). To determine whether the presence of the Alu element was a result of alternative splicing from an adjacent intron, the other two cognate clones were sequenced. We found that the nucleotide sequences of the cognate clones significantly overlapped, indicating that the presence of the Alu region in the cDNA probably was not caused by a splicing error.
Recently, Neuenchwander et al. (64) reported that stable antisense RNA expression inhibits the translation of sense RNA. In a number of studies (65–68), stable antisense RNA expression has resulted in a decrease in endogenous mRNA levels. Although the mechanism responsible for this decrease remains unclear, it may be related to the inhibition of RNA synthesis, RNA splicing, mRNA export, binding of initiation factors, assembly of ribosome subunits, and sliding of the ribosome along the mRNA coding sequence, all of which result in translation arrest (69). Therefore, we attempted to interfere with LITAF gene activity at the mRNA level by transfecting antisense oligonucleotide-producing vectors into THP-1 cells. The most common mRNA target site reported in the literature is the AUG translation initiation codon (70). However, we have found in many cases that oligonucleotides that target other regions in the mRNA, such as 5′ and 3′ UTRs, were more effective (71–74). Therefore, to interfere with LITAF mRNA, we transfected vectors producing antisense oligonucleotides that target the 5′ region of the LITAF mRNA, and, for the overexpression of the LITAF mRNA, a vector producing the full-length LITAF mRNA was transfected. Using sense LITAF RNA as a probe, a band of approximately 400 bp was detected in AS5′-transfected cells, consistent with the design of the antisense vector. No significant signal was detected in mock-transfected cells. When the full-length sense LITAF mRNA was transfected in THP-1 cells, an intense signal for LITAF mRNA was detected at 1.8 kb by using an antisense LITAF mRNA probe, reflecting the overexpression of LITAF mRNA, which confirmed the stable expression of RNA from each construct. However, when the same probe was used, AS5′-transfected cells weakly expressed the LITAF gene (Fig. 5). It is possible that the interference of LITAF expression with the antisense was not complete and LITAF antisense-transfected cells continued to exhibit little expression of the LITAF gene partly as a compensatory mechanism to restore the function, although the mechanisms involved in this phenomena remain unknown.
Using the RNase protection assay, examination of the effect of antisense RNA on transcription of the human TNF-α gene revealed that the signal indicating the TNF-α mRNA level was weaker in antisense-expressing cells than in mock cells. However, similar amounts of β-actin mRNA were found in both cells. The reduction of TNF-α mRNA that occurs when LITAF gene mRNA activity is arrested strongly suggests that LITAF gene products play an important role in the regulation of human TNF-α gene transcription. In addition, the expression level of TNF-α mRNA was not enhanced in the sense-expressing cells (Fig. 6). The scope of our functional study is not broad enough for us to rule out the possibility that exogenous LITAF gene products may regulate TNF-α gene transcription in other ways than what occurs naturally. We posit three possibilities for how the LITAF gene product may regulate gene transcription: (i) the LITAF gene may have to be processed as a protein to activate the TNF-α gene; (ii) the endogenous LITAF gene product is enough to activate the TNF-α gene; and (iii) the exogenous LITAF gene product acts after protein processing because a large amount of LITAF mRNA is constitutively transcribed in the cells (Fig. 5). Nonetheless, the present data support recent data suggesting that LPS induction of TNF-α promoter may be mediated by the concerted participation of at least two separate, cis-acting regulatory elements (75).
Distribution of LITAF mRNA on various tissue blots revealed that the LITAF gene is expressed in most of the tissues tested; however, it is expressed more predominantly in hematolymphopoeitic tissues and placenta, kidney, and pancreas. This distribution of LITAF transcripts seems to parallel TNF-α tissue distribution during endotoxemia (76, 77).
The potential role of LITAF in human disease is implicated by the recent finding that p53 induces the expression of the LITAF gene (62) and by its chromosomal localization at 16p12–16p13.3. The p53 tumor-suppressor protein is thought to play a major role in the defense of the cell against agents that damage DNA (78, 79). Loss of function of the p53 tumor-suppressor gene is a frequent and important event in the genesis or progression of many human malignancies. Loss of p53-dependent apoptosis is believed to be critical to carcinogenesis in many of these cases, suggesting the possibility to therapeutically restore this pathway and directly eliminate malignant cells or increase or restore their sensitivity to chemotherapeutic agents (78, 79). The regulation of p53-dependent responses is complex and variable between cell types, and whether a cell undergoes apoptosis after activation of p53 is highly sensitive to signal context, including environmental and cell-intrinsic influences. Further insight has been provided into the activation of latent p53, the biochemical mechanisms involved in growth arrest and apoptosis, and the influence of various signals on these cellular effects. Additionally, roles for p53 have been described in cell senescence, suppressing teratogenesis, and processes that may contribute directly to the maintenance of genomic stability (78, 79). Recently, various monocytic cell lines have been shown to respond to LPS and interferon γ with endogenous nitric oxide (NO) formation (80), which, in turn, activates the expression of p53, leading to apoptosis (81). Furthermore, p53 has been shown recently to modulate the activity of various inflammatory cytokines such as IL-6 (82), IL-2, and IL-4 (83). In light of the present data, a potential, new p53-dependent pathway affecting TNF-α gene expression can be proposed: LPS stimulation of monocyte/macrophages can induce NO production. In turn, NO can up-regulate p53, which would stimulate the expression of the LITAF gene. Ultimately, the induction of the LITAF gene product would affect TNF-α gene expression. The validity of this pathway remains to be substantiated.
Altogether, the present data suggest that the novel LITAF gene product might be a crucial factor for the control of TNF-α gene expression.
Acknowledgments
This work was supported by National Institute of Craniofacial and Dental Research Grant DE10709 to S.A.
ABBREVIATIONS
- TNF
tumor necrosis factor
- PMA
phorbol 12-myristate 13-acetate
- LPS
lipopolysaccharide
- LITAF
LPS-induced TNF-alpha factor
- FISH
fluorescence in situ hybridization
- UTR
untranslated region
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U77396).
References
- 1.Morrison D C, Ryan J L. Annu Rev Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 3.Bazzoni F, Beutler B. J Inflamm. 1995;45:106–114. [PubMed] [Google Scholar]
- 4.Beutler B, Milsark I W, Cerami A C. Science. 1985;229:869–871. [PubMed] [Google Scholar]
- 5.Bone R C. Ann Intern Med. 1991;115:457–469. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- 6.Cannon J G, Tompkins R G, Gelfand J A, Michie H R, Stanford G G, van der Meer J W, Endres S, Lonnemann G, Corsetti J, Chernow B, et al. J Infect Dis. 1990;161:79–84. doi: 10.1093/infdis/161.1.79. [DOI] [PubMed] [Google Scholar]
- 7.Chai Z, Gatti S, Toniatti C, Poli V, Bartfai T. J Exp Med. 1996;183:311–316. doi: 10.1084/jem.183.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayer J M, Beutler B, Cerami A. J Exp Med. 1985;162:2163–2168. doi: 10.1084/jem.162.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng H, Fletcher D, Kozak W, Jiang M, Hofmann K J, Conn C A, Soszynski D, Grabiec C, Trumbauer M E, Shaw A. Immunity. 1995;3:9–19. doi: 10.1016/1074-7613(95)90154-x. [DOI] [PubMed] [Google Scholar]
- 10.Gordon A H, Koj A. In: The Acute-Phase Response to Injury and Infection. Gordon A H, Koj A, editors. Amsterdam: Elsevier; 1985. pp. 139–149. [Google Scholar]
- 11.Blatteis C M, Sehic E. Ann N Y Acad Sci. 1998;840:608–618. doi: 10.1111/j.1749-6632.1998.tb09600.x. [DOI] [PubMed] [Google Scholar]
- 12.Tracey K J, Beutler B, Lowry S F, Merryweather J, Wolpe S, Milsark I W, Hariri R J, Fahey T J, Zentella A, Albert J D, et al. Science. 1986;234:470–474. doi: 10.1126/science.3764421. [DOI] [PubMed] [Google Scholar]
- 13.Miethke T, Wahl C, Heeg K, Echtenacher B, Krammer P H, Wagner H. J Exp Med. 1992;175:91–98. doi: 10.1084/jem.175.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beutler B, Cerami A. Nature (London) 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 15.Beutler B, Krochin N, Milsark I W, Leudke C, Cerami A. Science. 1986;232:977–980. doi: 10.1126/science.3754653. [DOI] [PubMed] [Google Scholar]
- 16.Beutler B, Cerami A. N Engl J Med. 1987;316:379–385. doi: 10.1056/NEJM198702123160705. [DOI] [PubMed] [Google Scholar]
- 17.Sugarman B J, Aggarwal B B, Hass P E, Figari I S, Palladino M A, Jr, Shepard H M. Science. 1985;230:943–945. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 18.Alexander H R, Sheppard B C, Jensen J C, Langstein H N, Buresh C M, Venzon D, Walker C E, Fraker D L, Strovroff M C, Norton J A. J Clin Invest. 1991;88:34–39. doi: 10.1172/JCI115298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross A S, Sadoff J C, Kelly N, Bernton E, Gemski P. J Exp Med. 1989;169:2021–2027. doi: 10.1084/jem.169.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakane A, Minagawa T, Kato K. Infect Immun. 1988;56:2563–2569. doi: 10.1128/iai.56.10.2563-2569.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roll J T, Young K M, Kurtz R S, Czuprynski C J. Immunology. 1990;69:316–322. [PMC free article] [PubMed] [Google Scholar]
- 22.Beutler B, Cerami A. Annu Rev Immunol. 1989;7:625–656. doi: 10.1146/annurev.iy.07.040189.003205. [DOI] [PubMed] [Google Scholar]
- 23.Talmage J E, Philips H, Scheider M, Rowe T, Pennington R, Brwersax O, Lenz B. Cancer Res. 1988;48:544–553. [PubMed] [Google Scholar]
- 24.Mathison J C, Wolfson E, Ulevitch R J. J Inf Dis. 1988;81:1925–1937. doi: 10.1172/JCI113540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tracey K J, Fong Y, Hesse D G, Manogue K R, Lee A T, Kuo G C, Lowry S F, Cerami A. Nature (London) 1987;330:662–664. doi: 10.1038/330662a0. [DOI] [PubMed] [Google Scholar]
- 26.Beutler B, Cerami A. Biochemistry. 1988;27:7575–7582. doi: 10.1021/bi00420a001. [DOI] [PubMed] [Google Scholar]
- 27.Neale M L, Williams B D, Matthews N. Br J Rheumatol. 1989;28:104–108. doi: 10.1093/rheumatology/28.2.104. [DOI] [PubMed] [Google Scholar]
- 28.Manicourt D H, Triki R, Fukuda K, Devogelaer J P, Nagant de Deuxchaisnes C, Thonar E J. Arthritis Rheum. 1993;36:490–499. doi: 10.1002/art.1780360409. [DOI] [PubMed] [Google Scholar]
- 29.Duh E J, Maury W J, Folks T M, Fauci A S, Rabson A B. Proc Natl Acad Sci USA. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn L, Kunkel S, Nabel G J. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popik W, Pitha P M. J Virol. 1993;67:1094–1099. doi: 10.1128/jvi.67.2.1094-1099.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vesanen M, Wessman M, Salminen M, Vaheri A. J Gen Virol. 1992;73:1753–1760. doi: 10.1099/0022-1317-73-7-1753. [DOI] [PubMed] [Google Scholar]
- 33.Tracey K J, Wei H E, Manogue K R. J Exp Med. 1988;167:1221–1227. doi: 10.1084/jem.167.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleif R. Science. 1988;241:1182–1187. doi: 10.1126/science.2842864. [DOI] [PubMed] [Google Scholar]
- 35.Goldfeld A E, Doyle C, Maniatis T. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincenti M P, Burrel T A, Taffet S M. J Cell Physiol. 1992;150:204–213. doi: 10.1002/jcp.1041500127. [DOI] [PubMed] [Google Scholar]
- 37.Drouet C, Shakhov A N, Jongeneel C V. J Immunol. 1991;147:1694–1700. [PubMed] [Google Scholar]
- 38.Shakhov A N, Collart M A, Vassalli P, Nedospasov S A, Jongeneel C V. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taffet S M, Singhel K J, Overholtzer J F, Shurtleff S A. Cell Immunol. 1989;120:291–300. doi: 10.1016/0008-8749(89)90198-6. [DOI] [PubMed] [Google Scholar]
- 40.Sung S S J, Walters J A, Hudson J, Gimble J M. J Immunol. 1991;147:2047–2054. [PubMed] [Google Scholar]
- 41.Economou J S, Rhoades K, Essner R, McBride W H, Gasson J C, Morton D L. J Exp Med. 1989;170:321–326. doi: 10.1084/jem.170.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hensel G, Meiche A, Pfizenmaier K, Krönke M. Lymphokine Res. 1989;8:347–351. [PubMed] [Google Scholar]
- 43.Rhoades K L, Golub S H, Economou J S. J Biol Chem. 1992;267:22102–22107. [PubMed] [Google Scholar]
- 44.Goldfeld A E, Strominger J L, Doyle C. J Exp Med. 1991;174:73–81. doi: 10.1084/jem.174.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leitman D C, Mackow E R, Williams T, Baxter J D, West B L. Mol Cell Biol. 1992;12:1352–1356. doi: 10.1128/mcb.12.3.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nedospasov A, Shakhov A N, Turetskaya R L, Mett V A, Azizov M M, Georgiev G P, Korobko V G, Dobrynin V N, Filippov S A, Bystrov N S, et al. Cold Spring Harbor Symp Quant Biol. 1986;551:611–624. doi: 10.1101/sqb.1986.051.01.073. [DOI] [PubMed] [Google Scholar]
- 47.Nedwin G E, Nayler S L, Sakaguchi A Y, Smith D, Jarrett-Nedwin J, Pennica D, Goeddel D V, Gray P. Nucleic Acids Res. 1985;13:6361–6373. doi: 10.1093/nar/13.17.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cordle S R, Donald R, Read M A, Hawiger J. J Biol Chem. 1993;268:11803–11810. [PubMed] [Google Scholar]
- 49.Müller J M, Löms Ziegler-Hettbrock H W, Baeuerle P A. Immunobiology. 1993;187:233–256. doi: 10.1016/S0171-2985(11)80342-6. [DOI] [PubMed] [Google Scholar]
- 50.Collart M A, Berlin D, Vassalli J D, Dekossodo S, Vassalli P. J Exp Med. 1986;164:2113–2118. doi: 10.1084/jem.164.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shakhov A N, Collart M A, Vassalli P, Nedospasov S V, Jongeneel C V. J Exp Med. 1990;171:35–47. doi: 10.1084/jem.171.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldfeld A E, Doyle C, Maniatis T. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takashiba S, Van Dyke T E, Shapira L, Amar S. Infect Immun. 1995;63:1529–1534. doi: 10.1128/iai.63.4.1529-1534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapira L, Takashiba S, Amar S, Van Dyke T E. Oral Microbiol Immunol. 1994;9:112–117. doi: 10.1111/j.1399-302x.1994.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 55.Takashiba S, Shapira L, Amar S, Van Dyke T E. Gene. 1994;131:307–308. doi: 10.1016/0378-1119(93)90314-s. [DOI] [PubMed] [Google Scholar]
- 56.Gabrielsen O S, Huet J. Methods Enzymol. 1993;218:508–525. doi: 10.1016/0076-6879(93)18038-e. [DOI] [PubMed] [Google Scholar]
- 57.Matsudaira P. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 58.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 59.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. 3rd Ed. New York: Greene & Wiley; 1992. [Google Scholar]
- 60.Maret A, Galy B, Arnaud E, Bayard F, Prats H. Cancer Res. 1995;55:5075–5079. [PubMed] [Google Scholar]
- 61.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 62.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B A. Nature (London) 1997;18:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 63.Mahone M, Saffman E E, Lasko P F. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neuenchwander S, Roberts C T, LeRoith D. Endocrinology. 1995;136:4298–4303. doi: 10.1210/endo.136.10.7664648. [DOI] [PubMed] [Google Scholar]
- 65.Stein C A. Antisense Nucleic Acid Drug Dev. 1998;8:129–132. doi: 10.1089/oli.1.1998.8.129. [DOI] [PubMed] [Google Scholar]
- 66.Helene C, Giovannangeli C, Guieysse-Peugeot A L. CIBA Found Symp. 1997;209:94–102. doi: 10.1002/9780470515396.ch8. [DOI] [PubMed] [Google Scholar]
- 67.Morales A V, de Pablo F. Curr Topics Dev Biol. 1998;36:37–49. doi: 10.1016/s0070-2153(08)60494-9. [DOI] [PubMed] [Google Scholar]
- 68.Agrawal S, Iyer R P. Pharmacol Ther. 1997;76:151–60. doi: 10.1016/s0163-7258(97)00108-3. [DOI] [PubMed] [Google Scholar]
- 69.Kim S K, Wold B J. Cell. 1985;42:129–138. doi: 10.1016/s0092-8674(85)80108-2. [DOI] [PubMed] [Google Scholar]
- 70.Crooke S T, Lebleu B. Antisense Research and Therapeutic. Boca Raton, FL: CRC; 1993. [Google Scholar]
- 71.Bennett C F, Condon T P, Grimm S, Chan H, Chiang M Y. J Immunol. 1994;152:3530–3540. [PubMed] [Google Scholar]
- 72.Dean N M, McKay R, Condon T P. J Biol Chem. 1994;269:16416–16124. [PubMed] [Google Scholar]
- 73.Perlaky L, Saijo Y, Busch R K, Bennett C F, Mirabelli C K, Crooke S T. Anti-Cancer Drug Design. 1993;8:3–14. [PubMed] [Google Scholar]
- 74.Chiang M Y, Chan H, Zounes M A, Freier S M, Lima W F, Bennett C F. J Biol Chem. 1991;266:18162–18171. [PubMed] [Google Scholar]
- 75.Yao J, Mackman N, Edgington T S, Fan S T. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 76.Giroir B P, Johnson J H, Brown T, Allen G L, Beutler B. Br J Clin Invest. 1992;90:693–698. doi: 10.1172/JCI115939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruco L P, Stoppacciaro A, Pomponi D, Boraschi D, Santoni A, Tagliabue A, Uccini S, Baroni C D. Am J Pathol. 1989;135:889–897. [PMC free article] [PubMed] [Google Scholar]
- 78.Carson D A, Lois A. Lancet. 1995;346:1009–1011. doi: 10.1016/s0140-6736(95)91693-8. [DOI] [PubMed] [Google Scholar]
- 79.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 80.Brune B, Gotz C, Messmer U K, Sandau K, Hirvonen M R, Lapetina E G. J Biol Chem. 1997;272:7253–7258. doi: 10.1074/jbc.272.11.7253. [DOI] [PubMed] [Google Scholar]
- 81.Shapira L, Champagne C, Van Dyke T E, Amar S. Infect Immun. 1998;66:2736–2742. doi: 10.1128/iai.66.6.2736-2742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Margulies L, Sehgal P B. J Biol Chem. 1993;268:15096–15100. [PubMed] [Google Scholar]
- 83.Pesch J, Brehm U, Staib C, Grummt F. J Interferon Cytokine Res. 1996;16:595–600. doi: 10.1089/jir.1996.16.595. [DOI] [PubMed] [Google Scholar]
- 84.Claverie J-M, Makalowski W. Nature (London) 1994;371:752. doi: 10.1038/371752a0. [DOI] [PubMed] [Google Scholar]