Abstract
The Escherichia coli DNA mismatch repair (MMR) protein MutS is essential for the correction of DNA replication errors. In vitro, MutS exists in a dimer/tetramer equilibrium that is converted into a monomer/dimer equilibrium upon deletion of the C-terminal 53 amino acids. In vivo and in vitro data have shown that this C-terminal domain (CTD, residues 801–853) is critical for tetramerization and the function of MutS in MMR and anti-recombination. We report the expression, purification and analysis of the E.coli MutS-CTD. Secondary structure prediction and circular dichroism suggest that the CTD is folded, with an α-helical content of 30%. Based on sedimentation equilibrium and velocity analyses, MutS-CTD forms a tetramer of asymmetric shape. A single point mutation (D835R) abolishes tetramerization but not dimerization of both MutS-CTD and full-length MutS. Interestingly, the in vivo and in vitro MMR activity of MutSCF/D835R is diminished to a similar extent as a truncated MutS variant (MutS800, residues 1–800), which lacks the CTD. Moreover, the dimer-forming MutSCF/D835R has comparable DNA binding affinity with the tetramer-forming MutS, but is impaired in mismatch-dependent activation of MutH. Our data support the hypothesis that tetramerization of MutS is important but not essential for MutS function in MMR.
INTRODUCTION
DNA mismatch repair (MMR) is critical for avoiding mutations, anti-recombination and cell checkpoint activation leading to apoptotic responses (1–3). In Escherichia coli, MutS recognizes mismatches in DNA and subsequently recruits MutL in an ATP-dependent manner. This ternary complex activates downstream effector proteins such as the strand-discrimination endonuclease MutH and UvrD helicase. MutH nicks a hemimethylated GATC site in the unmethylated, erroneous strand followed by unwinding of this strand by UvrD helicase and exonucleolytic digestion by one of several exonucleases. Repair is completed by DNA polymerase III holoenzyme and DNA Ligase I [reviewed in Ref. (2)].
The mismatch-recognizing MutS belongs to a family of proteins whose members are found in organisms ranging from bacteria to eukaryotes. In eukaryotes, several MutS homologs exist that form heterodimers (e.g. MSH2-MSH6 and MSH2-MSH3), with no indication of higher oligomers (2). Likewise, several heterodimeric MutL homologs have been studied from both human (MLH1-PMS2) and yeast (MLH1-PMS1) (3). In humans, mutations in the MMR genes hMSH2, hMSH6 or hMLH1 cause predisposition to a common form of cancer called hereditary non-polyposis colorectal cancer (HNPCC)/Lynch-syndrome (4).
The co-crystal structures of both E.coli and Thermus aquaticus MutS bound to heteroduplex DNA have been solved, using C-terminal truncated protein variants of MutS (MutS800, residues 1–800 E.coli numbering) (5,6). MutS800 crystallizes as a dimer, wherein only one monomer has specific contacts to the mismatch and bound ADP. Using analytical ultracentrifugation it was shown that in solution MutS800 exists in monomer/dimer equilibrium while the full-length MutS exist in a dimer/tetramer equilibrium (7,8). The physiological effect of truncating MutS is controversially reported in the literature, with data obtained using a multicopy plasmid resulting in either no effect or impaired MMR function (6,9,10). Recently, E.coli with a truncated chromosomal mutS gene (mutSΔ800) was shown to exhibit a MutS null phenotype for mutation avoidance, anti-recombination and sensitivity to cytotoxic agents in a dam mutant (10,11). However, the function of the tetrameric form still remains unclear (12).
In vitro, MutS800 is impaired in DNA binding and mismatch-provoked MutH activation (7). Deletion of a β sliding clamp binding motif (812QMSLL816) of MutS abolishes the interaction between MutS and β sliding clamp proteins in vitro, although tetramerization and in vivo MMR activity of MutS are unaffected (13). Since MutS800 lacks residues involved in dimerization, tetramerization and binding to the β sliding clamp, the primary cause of the observed defects of MutS800 in MMR and anti-recombination remain to be identified. Without structural data, the determinants of the dimerization and tetramerization within the MutS-CTD are unknown.
Here we describe the expression, purification and biophysical characterization of the MutS-CTD (residues 801–853). We demonstrate that a single point mutation (D835R) in MutS-CTD has a strong effect on the tetramer/dimer equilibrium, mimicking results observed for MutS800. In addition, we analyzed the effect of this point mutation in context of the full-length MutS and assayed MutSCF/D835R for MMR, DNA binding and mismatch-provoked activation of MutH. The implications of our results concerning the function of the MutS tetramer are discussed.
MATERIALS AND METHODS
Sequence analysis
To search for MutS homologs sequences five rounds of PSI-BLAST were performed on the NCBI non-redundant database (November 2005) using E.coli MutS as query sequence (gi: 16130640). Sequences were then clustered with CLANS (14), multiple alignments of the different clusters performed with MUSCLE (15) and visualized with BioEdit (16). Secondary structure prediction was performed with the genesilico metaserver (17). Mutations in the human MutS homologs were taken from the InSiGHT database (http://www.insight-group.org/) (18) and the Human Gene Mutation Database (19).
Strains, plasmids, enzymes and reagents
E.coli K12 strains CC106 (P90C [araΔ[lac-proXIII] [F'laciZ proB+]) (20), TX2929 (CC106 mutS201::Tn5; Kmr) and the pET-15b (Novagen) derived plasmids pTX412 and pTX418 containing the mutS and mutL genes, respectively, under control of the T7 promoter were kindly provided by Dr M. Winkler (21). Plasmid pMQ402 (His6-MutH), a pBAD18 derivative, was a kind gift of Dr M. Marinus (22). For MutS and MutL protein expression E.coli strain HMS174 (λDE3) (Novagen) and for MutH the E.coli strain XL1 blue MRF′ were used (Stratagene).
Site-directed mutagenesis
pTX412/Cys-free containing the gene for a cysteine-free MutS variant (referred to as MutSCF) was generated by replacing all six codons of the native cysteine residues. The mutations are, in order from the first cysteine in the sequence to the last, C93A, C235S, C239A, C297S, C569S and C711V with the numbering referring to the E.coli MutS sequence (L. Manelyte and P. Friedhoff, manuscript in preparation). Variants of MutS were generated with modifications of the Quikchange protocol (Stratagene) as described previously (23) using pTX412/Cys-free as a template and the respective mutagenesis oligodeoxynucleotides: CTD 5′-TTG CGT AGC GGC GGC GTT CGG ATG GCT GCC GCG CG GCA CCA G-3′; MutS800 5′-TAG CGG CCG CGT TCT ACG AAA TGC TTT-3′; MutSD835R 5′-GGT GAG TGA TCT CGG GTC CAG ATT TTC CA-3′.
E.coli XL1-blue MRF′ were transformed with the full-length PCR product. Marker positive clones were inoculated in LB medium containing ampicillin for overnight growth. Plasmid DNA was isolated using the QIAprep Spin Miniprep (Qiagen) and the whole mutS gene sequenced.
Complementation mutator assay
Cells lacking a functional chromosomal mutS gene show a mutator phenotype, which is analyzed by the frequency of rifampicin-resistant clones arising from unrepaired polymerase errors in the rpoB gene (22). Single colonies of mutS-deficient TX2929 cells transformed with vector control or plasmids carrying the indicated gene were grown overnight at 37°C in 3 ml LB cultures containing 100 μg/ml ampicillin. Aliquots of 50 μl from the undiluted or 10−6 diluted culture were plated on LB agar containing 25 μg/ml ampicillin with or without 100 μg/ml rifampicin. Colonies were counted after overnight incubation at 37°C.
Purification of proteins
Recombinant His6-tagged proteins were expressed and purified by Ni-NTA chromatography essentially as described elsewhere (21,23,24). MutH was stored in 10 mM HEPES–KOH (pH 7.9), 500 mM KCl, 1 mM EDTA, 1 mM DTT and 50% glycerol at −20°C while MutL, MutS and MutS-CTD proteins in 10 mM HEPES–KOH (pH 7.9), 200 mM KCl and 1 mM EDTA (for MutS 10% glycerol was added) were snap-frozen in liquid nitrogen and stored at −70°C. Protein concentrations were determined using the theoretical extinction coefficients calculated from amino acid composition (25). In order to remove N-terminal His6-tag 10 mU/μldiaminopeptidase DAPase (Qiagen) was incubated with MutS-CTD in phosphate-buffered saline for 16 h at room temperature. DAPase was removed following manufacture protocol using Ni-NTA (Qiagen).
MutH endonuclease assay
MutH endonuclease was assayed on heteroduplex DNA substrate (484 bp) containing a G/T mismatch at position 385 and a single unmethylated GATC site at position 210 (26). DNA (10 nM) was incubated with 200 nM MutH, 1 μM MutL and 0, 50, 100, 200, 400 and 1 μM (monomer equivalents) MutS in 10 mM Tris–HCl (pH 7.9), 5 mM MgCl2, 1 mM ATP, 50 μg/ml BSA and 125 mM KCl at 37°C. MutH endonuclease activity was scored by the appearance of cleaved products (analyzed by 6% polyacrylamide gel electrophoresis).
Size-exclusion chromatography
MutS variants were analyzed by size-exclusion chromatography using a Sephadex 200 10/300 column. The column was calibrated using the following standard proteins (Sigma): thyroglobulin (MW = 669 kDa, RS = 8.5 nm), apoferritin (443 kDa, 6.1 nm), β-amylase (200 kDa, 5.4 nm), alcohol dehydrogenase (150 kDa, 4.5 nm), albumin (66 kDa, 3.55 nm), carbonic anhydrase (29 kDa, 2.35 nm), myoglobin (16.9 kDa, 2.0 nm) and cytochrome c (12.5 kDa, 1.77 nm). Samples (100 μl) containing different concentrations of MutS full-length or CTD were injected into the column equilibrated with 10 mM HEPES–KOH (pH 8.0), 500 mM KCl, 1 mM EDTA, at flow rates of 0.5 ml/min. Elution profiles were monitored at 280 nm.
Circular dichroism
Circular dichroism (CD) spectra were recorded in a Jasco J-710 dichrograph between 185 and 280 nm at 20°C in a cylindrical cuvette of 0.05 cm path length. A baseline was recorded and subtracted after each spectrum. Protein was 20 μM in 10 mM HEPES–KOH (pH 8.0), 0.1 mM EDTA, 50 mM KCl. CDNN Deconvolution software (version 2; Bioinformatik.biochemtech.uni-halle.de/cdnn) was employed for estimation of secondary structure content.
DNA binding
DNA binding of MutS was determined by electrophoretic mobility shift assay (EMSA). The following oligodesoxynucleotides were used: G-strand 5′-TAT TAA TTT CGC GGG CTC GAG AGC TTC ATC CTC TAC GCC GGA, T-strand 5′-TCC GGC GTA GAG GAT GAA GCT TTC GAG CCC GCG AAA TTA ATA and C-strand 5′-TCC GGC GTA GAG GAT GAA GCT CTC GAG CCC GCG AAA TTA ATA. G-strand was labeled at 3′ end using terminal desoxynucleotidyl transferase (Fermentas) following the manufacturers instructions. G/T heteroduplex or G/C homoduplex of 42 bp were prepared by annealing the G- with the T or C-strand, respectively. MutS was incubated with 1 nM of [32P]-labeled 42 bp G/T heteroduplex in binding buffer [20 mM HEPES–KOH (pH 7.5), 125 mM KCl, 5 mM MgCl2, 50 μg/ml BSA and 0.5 mM ADP] in a total volume of 20 μl. After 10 min at 37°C 4 μl of 50% glycerol and 20 mM EDTA solution was added, samples were placed on ice and loaded under voltage onto 4% native polyacrylamide gels (29:1 acrylamide:bisacrylamide) in 40 mM Tris (pH 7.5), 20 mM sodium acetate, 1 mM EDTA. After electrophoresis at 4°C under 11.4 V/cm for 70 min, gels were analyzed using an Instant-Imager (Packard). SigmaPlot was used to fit the data to the following sigmoidal equation:
where Y is the fraction of bound DNA, Y0 is the background signal, Ymax is the maximum fraction of bound DNA, S is the concentration of MutS in monomers, K is the apparent dissociation constant and n is the Hill-coefficient.
Sedimentation equilibrium analysis
For determination of molecular masses in solution, sedimentation equilibrium experiments in the analytical ultracentrifuge (Beckman-Coulter XL-A, UV absorption optics) were exploited. Samples containing MutS-CTD at concentration between 14 and 57 μM in 10 mM HEPES–KOH (pH 7.9), 250 mM KCl were run in two-channel centerpieces filled with 150 μl sample and underlaid with 50 μl FC43 (ABCR, Karlsruhe, Germany) as artificial bottom. Equilibrium runs were performed at both 16 000 and 23 000 r.p.m. at 20°C for at least 32 h. Concentration profiles were measured every hour and equilibrium attainment was assumed when no change in these concentration profiles could be observed over at least 12 h. Buffer absorption was determined after sedimenting the protein for 7 h at 44 000 r.p.m. Apparent molecular masses were calculated from traces averaged over these last 12 h as described elsewhere (27).
Sedimentation velocity analysis
Determination of sedimentation coefficient s20,w were performed at 20°C in 10 mM HEPES–KOH (pH 7.9), 0.1 mM EDTA and 50 or 250 mM KCl at concentration between 30 and 50 μM of protein. Sedimentation profiles were monitored at 280 nm. Sedimentation rate constants were obtained by analyzing the movement of the sedimenting boundary or by fitting a numerical solution of Lamm's differential equation to the concentration profiles using the BPCFIT software package (28).
RESULTS
Since deletion of the C-terminal 53 amino acid residues (CTD) of MutS results in a truncated protein (MutS800) that no longer forms tetramers in the concentration range studied (7,8), we started to investigate the properties of the CTD in more detail.
Secondary structure predictions
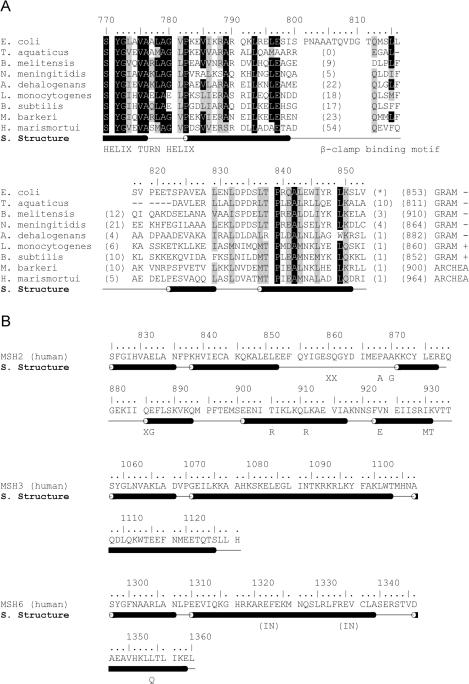
Our secondary structure prediction analysis of the CTD region based on a multiple sequence alignment revealed a bipartite organization of the CTD (Figure 1A). It consists of a less conserved N-terminal region (residues 801–822) with no predicted secondary structure which contains the β sliding clamp binding motif (13) and a conserved C-terminal region (residues 823–853) with a predicted helix–loop–helix motif. Comparison of the MutS-CTD secondary structure prediction from bacteria and eukarya revealed that all homologs contain similar helical content (Figure 1B). Interestingly, several cancer causing mutations are localized in the C-terminus of the human MutS homologs, however, the biochemical consequences of these mutations are still unknown (29).
Figure 1.
Sequence alignment and secondary structure prediction of MutS. (A) C-terminal part of MutS-1 protein sequences from nine different species (accession numbers for these proteins are as follows: E.coli K12 NCBI accession no. 16130640; Haloarcula marismortui, NCBI accession no. 55232253; T.aquaticus, NCBI accession no. 2497995; Neisseria meningitides, NCBI accession no. 15677973; Listeria monocytogenes, NCBI accession no. 47095354; Methanosarcina barkeri, NCBI accession no. 68133332; Bacillus subtilis, NCBI accession no. 1002520; Brucella melitensis, NCBI accession no.17983835; Anaeromyxobacter dehalogenans, NCBI accession no. 66857729) aligned using MUSCLE. Invariant residues are shaded in black whereas similar residues are shaded in gray. Residue numbering and secondary structure predictions are for the E.coli sequence. (B) Secondary structure prediction of the human MSH2, MSH3, and MSH6 C-terminal domains. Cancer causing mutations are annotated with the amino acid exchange, insertion (IN) or deletion (X).
Purification and secondary structure analysis of the MutS-CTD
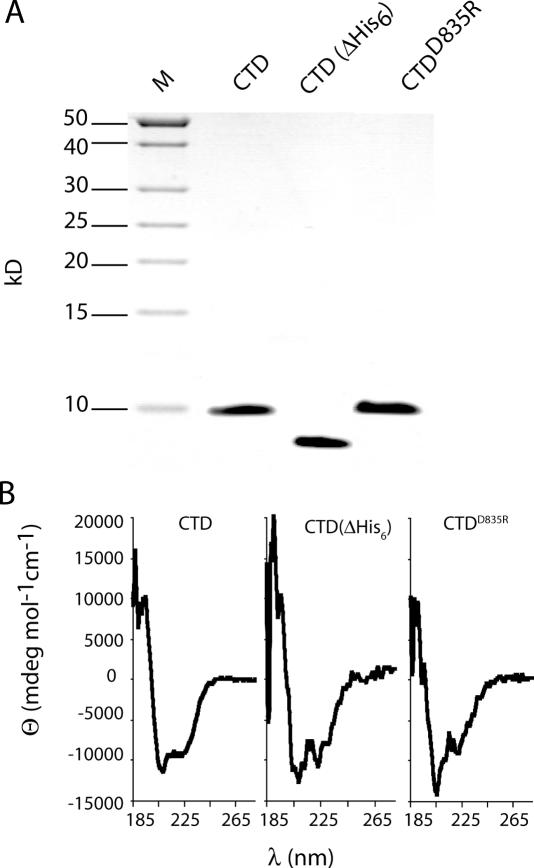
In order to analyze the structure and function of this domain, we created a His6-tagged protein (MutS-CTD) corresponding to residues 801–853 of the E.coli MutS. MutS-CTD was expressed and purified to homogeneity by Ni-NTA affinity and size-exclusion chromatography (Figure 2A). Analysis by electrospray ionization mass spectrometry indicated that the N-terminal methione was missing resulting in a 71 residue protein with a calculated mass of 7.7 kDa (Table 1). Upon treatment with the diaminopeptidase DAPase, the N-terminal His6-tag was removed to give MutS-CTDΔHis6 with a calculated mass of 6.4 kDa (Figure 2A).
Figure 2.
Analysis of purified MutS-CTD variants. (A) Coomassie blue-stained Tris-Tricine 15% polyacrylamide gel is shown. Lane 1, protein markers (M) with sizes indicated on the left; lane 2, MutS-CTD; lane 3, MutS-CTDΔHis6 (His6-tag removed by DAPase; for details see Material and Methods); lane 4, MutS-CTDD835R. (B) CD spectra of MutS-CTD variants. All spectra were recorded with 20 μM proteins in HEPES–KOH 10 mM (pH 8.0), EDTA 0.1 mM and KCl 50 mM at 20°C. Residual molar ellipticity [Θ] of MutS-CTD variants was measured from 190 to 280 nm as described under Materials and Methods. The α-helical content was estimated to be 30% using secondary structure deconvolution program CDNN.
Table 1.
Hydrodynamic properties of MutS-CTD variants
| CTD | CTDΔHis6 | CTDD835R | |
|---|---|---|---|
| Partial specific volume (ml/g)a | 0.727 | 0.738 | 0.728 |
| Sedimentation coefficient s20,w (Svedberg) | 2.1 ± 0.04 | 1.4 ± 0.13 | 1.5 ± 0.04 |
| Stoke radius Rs (nm) | |||
| Calculated from s20,wb | 3.65 | 4.3 | 2.55 |
| Estimated by size-exclusion chromatography | 3.5 | n.d. | 2.3 |
| Molecular weight determined | |||
| By sedimentation equilibrium | 29 800 ± 410c | 25 300 ± 380d | 16 500 ± 370e |
| From sequence (dimer)a | 15405 | 12835 | 15487 |
| From sequence (tetramer)a | 30792 | 25651 | 30956 |
| Oligomeric status | Tetramer | Tetramer | Dimer |
| Perrin factor f/fmin | 1.74 | 2.19 | 1.55 |
aUsing sequence without N-terminal methionine as determined by ESI mass spectrometry.
bUsing SEDNTERP version 1.08 using the vbar method.
cDetermined at 29 μM protein and 23 000 r.p.m.
dDetermined at 50 μM protein and 23 000 r.p.m.
eDetermined at 29 μM protein and 16 000 r.p.m.
Full-length MutS has been shown to form aggregates at elevated concentrations especially in the absence of nucleotide (30). In part the CTD was considered to be responsible for this phenomenon since deletion of this domain allowed crystallization of the protein (5,6). However, MutS-CTD was soluble and homogeneous at concentrations up to 500 μM with no apparent formation of aggregates as judged by size-exclusion chromatography and analytical ultracentrifugation (see below). The CD spectra of MutS-CTD and MutS-CTDΔHis6 revealed that the protein adopts a folded structure with ∼30% α-helical content (Figure 2B) consistent with the secondary structure prediction (Figure 1A).
Quaternary structure analysis
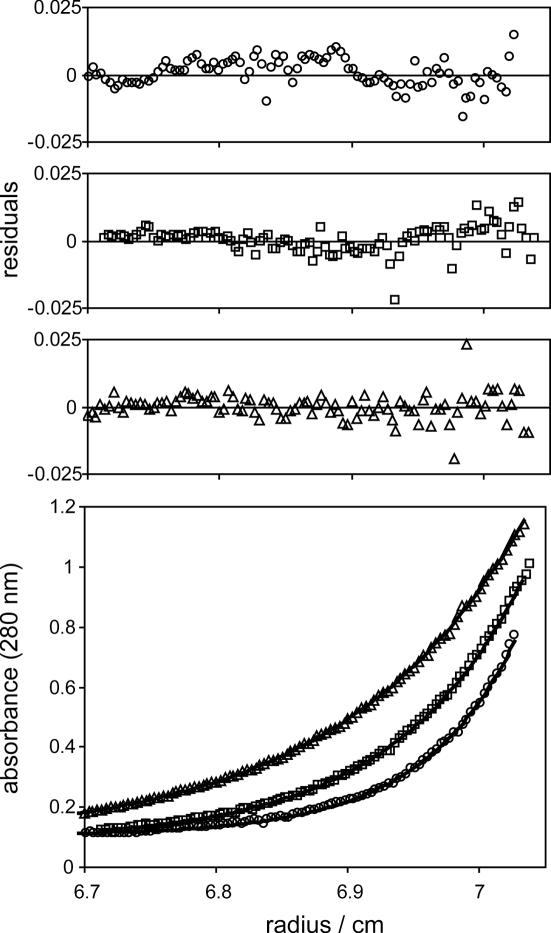
Since the CTD had been shown to be essential for tetramerization of the full-length protein we asked whether the domain on its own is sufficient to form tetramers. Sedimentation equilibrium analysis indicates that MutS-CTD and MutS-CTDΔHis6 form tetramers (Figure 3 and Table 1). Thus, tetramerization is a characteristic feature of this domain and is not influenced by the N-terminal His6-tag. In the concentration range tested (14–60 μM) we did not observe any significant formation of either lower or higher molecular weight species. Moreover, sedimentation velocity analysis revealed sedimentation coefficients s20,w of 2.1 and 1.4 S for MutS-CTD and MutS-CTDΔHis6, respectively. Based on the molecular mass and sedimentation coefficient obtained by the sedimentation analysis, MutS-CTD is a tetramer with highly asymmetric shape (Table 1).
Figure 3.
Sedimentation equilibrium analysis of MutS-CTD. Examples of equilibrium sedimentation profiles of 57 μM (monomer equivalents) MutS-CTD (circles), 50 μM MutS-CTDΔHis6 (squares) and 57 μM MutS-CTDD835R (triangles) run at 23 000 r.p.m. Solid lines are theoretical concentration profiles calculated with partial specific volumes and molar masses given in Table 1. The upper three panels show the residuals for each profile of the bottom panel.
Disruption of the tetramer by D835R mutation
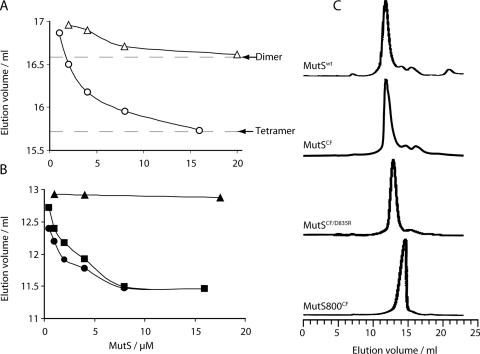
In order to investigate the MutS-CTD in more detail we generated several variants which we analyzed for their ability to form tetramers (L. Manelyte, D. Goldeck and P. Friedhoff, unpublished data). A single point mutant, MutS-CTDCF/D835R abolished tetramerization while still preserving dimerization (Figures 3 and 4 and Table 1). CD analysis of MutS-CTDD835R indicates that the protein is very similar to MutS-CTD with respect to secondary structure content. Therefore, the amino acid exchange did not affect the overall structure of the protein (Figure 2B). Sedimentation equilibrium and velocity experiments revealed that the protein exists in solution as a dimer with asymmetric shape indicated by the frictional ratio of 1.55. Our data imply that the CTD stabilizes dimerization of the protein and allows tetramerization of dimers. Since Asp-835 is located in the acidic loop of the helix–loop–helix motif it is tempting to speculate that tetramerization is mediated via this loop.
Figure 4.
Size-exclusion chromatography analyses of MutS variants. Proteins were analyzed on a Superdex200 column. In the graph is plotted elution volume versus protein concentration at the point of injection for (A) MutS-CTD variants: MutS-CTD (open circles) and MutS-CTDD835R (open triangles) and (B) MutS-variants: MutSwt (squares), MutSCF (circles) and MutSCF/D835R (triangles). Note that MutS-CTD, MutSwt and MutSCF show concentration dependent elution volumes while this is not the case for MutSCF/D835R. Dashed lines indicate the elution volumes of a tetramers and dimers, respectively, based on the hydrodynamic radius obtained for the sedimentation analysis of MutS-CTD and MutS-CTDD835R (see also Table 1). (C) Typical elution profiles for MutS proteins. MutSwt (8 μM), MutSCF (8 μM), MutSCF/D835R (17.4 μM) and MutS800CF (40 μM).
Tetramerization of full-length MutS is important but not essential for in vivo function in mismatch repair
As mentioned above the role of the MutS tetramer in vitro and in vivo is controversially discussed in the literature. This is in part owing to comparing the function of the tetramer-forming full-length protein with a dimer-forming truncated protein missing residues involved not only in tetramerization but also dimerization and probably interaction with other proteins, e.g. the β sliding clamp (13). Consequently, we asked whether the D835R mutation affects the function of the full-length MutS protein. Starting from an active cysteine-free MutS variant (MutSCF), we generated a truncated MutS variant (MutS800CF) and the D835R variant (MutSCF/D835R) which we assayed for MMR activity in vivo (Table 2). MutSwt as well as MutSCF encoded on a multicopy plasmid were able to complement the mutS-mutator phenotype resulting in low mutation frequencies on rifampicin/ampicillin plates compared with the vector control. However, both variant MutS800CF and MutSCF/D835R have impaired MMR activity as indicated by elevated mutation frequencies, ∼5- to 10-fold higher compared with MutSwt and MutSCF, respectively (Table 2).
Table 2.
In vivo MMR activity of MutS variants
| Variant | In vivo activity | |
|---|---|---|
| Mutation frequency (×10−9) | Normalized frequency | |
| Vector (mutS null) | 172 | 156 |
| MutSwt | 1.1 | 1 |
| MutSCF | 0.7 | 0.7 |
| MutSCF/D835R | 7.0 | 6.4 |
| MutS800CF | 7.5 | 6.8 |
| MutS-CTD | 147 | 134 |
For in vivo MMR activity the rpoB mutation assay was used (see Materials and Methods for details). At least three independent experiments were performed for each variant. See Supplementary Table for a larger dataset on determining the number of rifampicin resistant clones.
Further, we purified the proteins and characterized their biophysical and biochemical properties in vitro. Size-exclusion chromatography experiments showed a concentration dependence of elution profiles for both MutSwt and MutSCF in agreement with the observed dimer/tetramer equilibrium for MutSwt (8,31) (Figure 4, Table 3). In contrast to this, MutSCF/D835R did not show any concentration dependence of the elution profiles. An apparent molecular mass of 180 kDa was calculated based on gel-filtration experiments suggesting that the protein exists as a dimer in solution. This agrees with our sedimentation data for the CTDD835R variant being a dimer in solution. Interestingly, we observed concentration dependence in the elution profile for the MutS-CTD which was diminished for MutS-CTDD835R. Thus, we conclude that the D835R amino acid exchange not only abolished tetramerization for the CTD but also for the full-length protein. This enables us for the first time to study a stable dimeric E.coli full-length MutS protein for its function in DNA MMR.
Table 3.
Molecular masses and oligomerization state of MutS variants
| Protein | Calculated mass (kDa) | Size-exclusion chromatography (Mapp/kDa) | AUC (M/kDa) | Oligomeric state | Reference |
|---|---|---|---|---|---|
| MutSwt | 381 | ≈580a | 360 ± 20 | Dimer/Tetramer | (7,8,31) |
| MutSwt | 390 | ≈450a | N.D. | Dimer/Tetramer | this studyd |
| MutSCF | 390 | ≈450a | N.D. | Dimer/Tetramer | this studyd |
| MutS800 | 180 | N.D. | 220 ± 12 | Dimer/Monomer | (7,8) |
| MutS800CF | 183 | 125/175b | N.D. | Dimer/Monomer | this studyd |
| MutSCF/D835R | 190 | ≈180c | N.D. | Dimer | this studyd |
aConcentration dependent in a range of 0.5–10 μM.
bIn the absence or presence of ADP.
cConcentration independent in a range of 0.5–17 μM.
dN-terminal His6-tagged proteins.
Dimer-forming MutS binds to DNA with similar affinity as tetramer-forming MutS
To test whether the reduced in vivo MMR activity of MutSCF/D835R is owing to changed DNA-binding affinity, we measured protein binding to 42 bp G/T heteroduplex DNA using a gel electrophoretic mobility shift assay (Figure 5). MutSCF/D835R was able to bind to DNA with similar affinity as MutSCF (Figure 5A) and MutSwt (data not shown). However, we observed differences in the mobility of the protein–DNA complexes between MutSCF/D835R and MutSCF. At low concentrations MutSCF/D835R forms a fast migrating complex (C1) while MutSCF as well as MutSwt (data not shown) form complexes with lower mobility (C2, Figure 5A). At higher concentration we observe for both proteins the formation of at least a second complex of lower mobility (C2 and C3, respectively). MutSwt has been reported to bind to short DNA oligonucleotides as a tetramer (MutS4–DNA) (7). Hence, complex C2 observed for tetramer-forming MutSCF might correspond with a MutS4–DNA complex while complex C1 observed for dimer-forming MutSCF/D835R to a MutS2–DNA complex. Meanwhile complex C3 could correspond to any higher order complex. In the absence of additional experiments, however, these assignments must be regarded with caution. The DNA binding data do not fit with a simple hyperbolic function (Figure 5B), similar to previously reported (7,32). We evaluated the titration curves with a cooperative binding model yielding Hill-coefficients of 3.6 and 2, while the apparent Kd-values of 42 ± 2 and 45 ± 3 nM for MutSCF and MutSCF/D835R, respectively, are virtual identical.
Figure 5.
DNA binding of MutSCF and MutSCF/D835R variants using EMSA. (A) MutSCF or MutSCF/D835R binding to 42 bp G/T heteroduplex DNA, respectively, was performed in the presence of 0.5 mM ADP. MutS concentrations (monomer equivalents) were 1, 5, 12.5, 25, 50, 65, 80, 100, 300 nM (lanes 2–10 and 11–19). Arrows C1, C2 and C3 indicate different MutS–DNA complexes. Note that complex C1 is not observed for the tetrameric MutSCF meanwhile complex C3 is not observed for the dimeric MutSCF/D835R. (B) Quantitative analysis of DNA binding for MutSCF (circles) and MutSCF/D835R (triangles). Fits to a sigmoidal Hill function (see Materials and Methods) yielded apparent K1/2 = 42 ± 2 nM for MutSCF (Hill coefficient n = 3.6 ± 0.3) and K1/2 = 45 ± 3 nM for MutSCF/D835R (Hill coefficient n = 2 ± 0.2).
Dimer-forming full-length MutSCF/D835R is compromised in mismatch-provoked activation of MutH
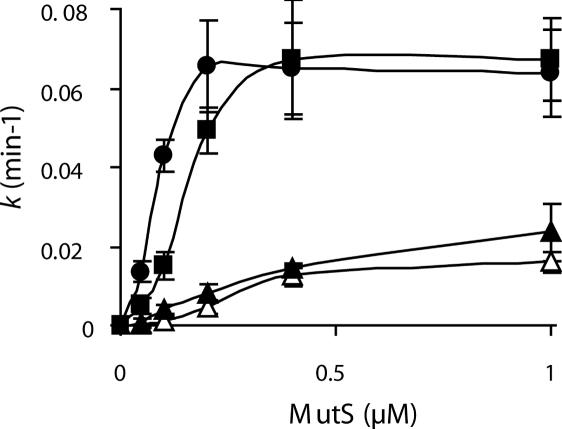
Dimer-forming MutS800 has been reported to be compromised in MutH activation in vitro. However, these results were obtained with a truncated protein missing the CTD that contains several conserved amino acid residues including a β sliding clamp binding motif (7,13). Hence, we have compared the biochemical activity of the dimer-forming full-length MutSCF/D835R and truncated MutS800CF with the tetramer-forming MutSwt and MutSCF in an assay that tests for the initial step of methyl-directed MMR: the mismatch-provoked activation of the MutH GATC endonuclease (33). As reported for MutS800, activation of MutH endonuclease is observed with both dimer-forming variants, however, the level of activation was significantly lower even at elevated MutS concentrations compared with the tetramer-forming variants (Figure 6). Thus, our data support previous observations that the formation of a tetramer is important for the in vivo as well in vitro function of the MutS protein.
Figure 6.
Mismatch-provoked MutH endonuclease activation is less efficient with dimeric MutS variants. Pseudo-first order rate constants k of GATC cleavage of a 484 bp G/T heteroduplex by MutH in a mismatch-provoked MutS- and MutL-dependent assay are plotted against the concentration of MutS protein ‘For details see Materials and Methods’. MutSwt (solid squares), MutSCF (solid circles), MutS800CF (open triangles) and MutSCF/D835R (solid triangles) as indicated.
DISCUSSION
This is the first report on the characterization of the MutS-CTD and of a full-length dimer-forming MutS variant (MutSCF/D835R). MutSCF/D835R shows similar properties as MutS800 both in vivo and in vitro suggesting that the major defects observed for the truncated MutS800 is owing to a lack of tetramerization rather than to missing residues needed for proper function. MutS-CTD can be expressed as a structured soluble protein domain (Figure 2A) with good agreement between predicted (Figure 1A) and experimentally determined secondary structure (Figure 2B). Furthermore, our hydrodynamic analyses using size-exclusion chromatography and analytical ultracentrifugation indicate that MutS-CTD is sufficient for tetramerization (Figure 3 and 4). This corroborates previous results showing that the tetramerization domain of MutS resides within the last 53 amino acids. Similar to full-length MutS, MutS-CTD exists in an equilibrium mixture of dimers and tetramers (Figure 4). Notably, this domain starts to dissociate into monomers at concentrations <200 nM, however, we did not attempt to determine any dissociation constants based on the size-exclusion chromatography data. It has been reported elsewhere that the truncated MutS protein (MutS800) exists as an equilibrium mixture of monomer and dimers that is shifted towards the dimeric form upon nucleotide binding (8). This raises the question whether the observed MMR defect of the MutS800 is owing to a changed monomer/dimer or dimer/tetramer equilibrium. Moreover, deletion of the CTD also removes the ß sliding clamp interaction motif (residues 812–816) although a recent report suggest that deletion of these residues had no effect on either in vivo MMR function or oligomerization in vitro (13).
An important step towards elucidating the function of the CTD and the MutS-tetramer was the generation of a single-point mutant (D835R) that abolishes tetramerization while preserving dimerization of the MutS full-length protein. MutSCF/D835R has a mutator phenotype in vivo similar to the truncated MutS800CF variant (Table 2). DNA binding analysis of MutSCF/D835R and MutSCF clearly demonstrated that both proteins bind to a 42 bp G/T mismatch containing DNA with similar apparent affinities, but qualitatively formed complexes with different electrophoretic mobility (Figure 5). Interestingly, a single hyperbolic function (single binding site mode) did not fit to the data. Proper fitting was obtained using a sigmoidal binding function with Hill coefficient of (n = 2 and 3.6, respectively). It has been previously reported that a simple hyperbolic function (single site binding mode) is insufficient to describe the DNA binding of MutS (7,32). The concentration dependencies of the tetramer-forming MutSCF and dimer-forming MutSCF/D835R support the idea that a more complex binding model is highly probable. Further studies will be required to establish a proper model for the DNA–MutS interaction that must take into account the dimer/tetramer equilibrium of MutS. Our data agree with surface plasmon resonance spectroscopy results, which showed DNA binding of a MutSwt tetramer and a MutS800 dimer, respectively (7). We observed significant differences between the tetramer-forming and the dimer-forming MutS variants in their ability to promote mismatch-provoked activation of MutH (Figure 6). Both, MutS800CF and MutSCF/D835R are impaired in this reaction, which corroborates previous experiments with MutSwt and MutS800 (7).
Our data support a functional role for a tetramer in DNA MMR. In contrast to previous studies we investigated functional differences between dimer- and tetramer-forming MutS variants in the context of the full-length rather than truncated proteins. In many models presented for coupling mismatch recognition by MutS and downstream processes such as strand discrimination by MutH [reviewed in Ref. (2)] only the dimeric form of MutS is discussed. Available experimental evidence including our data presented here strongly suggest that a MutS tetramer should be considered as an active form of MutS which might be important for MutS-induced DNA looping (2). However, since the dimer-forming MutS variant has substantial activity in vitro and in vivo more experiments are warranted that aim to describe mechanistic differences between tetrameric and dimeric MutS. This helps to explain controversial data obtained in assays for mismatch-provoked MutH activation observed under various experimental conditions (2).
Although initial studies on the MutS from T.aquaticus suggested that the protein is predominantly a dimer in solution at concentrations <10 μM even in the presence of the CTD (34), recent data suggest that Taq-MutS can bind to DNA as dimers and tetramers at concentrations between 10 and 20 nM with only one dimer in the tetramer being in contact with the DNA (35). Gel mobility shift experiments using the yeast MSH2-MSH6 heterodimer suggest that an additional MSH2-MSH6 heterodimer binds to the MSH2-MSH6-DNA complex probably via protein–protein interactions (36) thus forming a tetramer on the DNA. However, at present, there is no evidence for heterotetramers in eukaryotic MutS homologs suggesting that a dimeric form of MutS might be sufficient for efficient repair. This is in line with recent findings that in vivo MMR in E.coli depends on the expression level of MutS800 (10). While MMR function is recovered due to increased expression of dimer-forming MutS800 this is not the case for the anti-recombination function (10). It will therefore be interesting to determine whether the dimer-forming full-length MutSCF/D835R is impaired in anti-recombination, thus clarifying the importance of the CTD for this particular function of MutS.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Supplementary Material
Acknowledgments
The excellent technical assistance of Ina Steindorf and David Goldeck is gratefully acknowledged. The authors thank Dr George Silva and Dr Martin Marinus for critical comment and helpful discussion. This work was supported by the Deutsche Forschungsgemeinschaft (Fr-1495/3-2). L.M. and L.G. are members of the Graduiertenkolleg ‘GRK 370-Biochemie von Nukleoproteinkomplexen’. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 2.Iyer R.R., Pluciennik A., Burdett V., Modrich P.L. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 4.Chao E.C., Lipkin S.M. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res. 2006;34:840–852. doi: 10.1093/nar/gkj489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obmolova G., Ban C., Hsieh P., Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 6.Lamers M.H., Perrakis A., Enzlin J.H., Winterwerp H.H., de Wind N., Sixma T.K. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 7.Bjornson K.P., Blackwell L.J., Sage H., Baitinger C., Allen D., Modrich P. Assembly and molecular activities of the MutS tetramer. J. Biol. Chem. 2003;278:34667–34673. doi: 10.1074/jbc.M305513200. [DOI] [PubMed] [Google Scholar]
- 8.Lamers M.H., Georgijevic D., Lebbink J.H., Winterwerp H.H., Agianian B., de Wind N., Sixma T.K. ATP increases the affinity between MutS ATPase domains. Implications for ATP hydrolysis and conformational changes. J. Biol. Chem. 2004;279:43879–43885. doi: 10.1074/jbc.M406380200. [DOI] [PubMed] [Google Scholar]
- 9.Biswas I., Obmolova G., Takahashi M., Herr A., Newman M.A., Yang W., Hsieh P. Disruption of the helix–u-turn–helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol. 2001;305:805–816. doi: 10.1006/jmbi.2000.4367. [DOI] [PubMed] [Google Scholar]
- 10.Calmann M.A., Nowosielska A., Marinus M.G. Separation of mutation avoidance and antirecombination functions in an Escherichia coli mutS mutant. Nucleic Acids Res. 2005;33:1193–1200. doi: 10.1093/nar/gki263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calmann M.A., Nowosielska A., Marinus M.G. The MutS C terminus is essential for mismatch repair activity in vivo. J. Bacteriol. 2005;187:6577–6579. doi: 10.1128/JB.187.18.6577-6579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer N., Reagan M.S., Wu K.J., Canagarajah B., Friedberg E.C. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 13.Lopez de Saro F.J., Marinus M.G., Modrich P., O'Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J. Biol. Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- 14.Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- 17.Kurowski M.A., Bujnicki J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peltomaki P., Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis. Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 20.Cupples C.G., Miller J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng G., Winkler M.E. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of Escherichia coli K-12. BioTechniques. 1995;19:956–965. [PubMed] [Google Scholar]
- 22.Loh T., Murphy K.C., Marinus M.G. Mutational analysis of the MutH protein from Escherichia coli. J. Biol. Chem. 2001;276:12113–12119. doi: 10.1074/jbc.M007935200. [DOI] [PubMed] [Google Scholar]
- 23.Toedt G., Krishnan R., Friedhoff P. Site-specific protein modification to identify the MutL interface of MutH. Nucleic Acids Res. 2003;31:819–825. doi: 10.1093/nar/gkg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giron-Monzon L., Manelyte L., Ahrends R., Kirsch D., Spengler B., Friedhoff P. Mapping protein–protein interactions between MutL and MutH by cross-linking. J. Biol. Chem. 2004;279:49338–49345. doi: 10.1074/jbc.M409307200. [DOI] [PubMed] [Google Scholar]
- 25.Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas E., Pingoud A., Friedhoff P. An efficient method for the preparation of long heteroduplex DNA as substrate for mismatch repair by the Escherichia coli MutHLS system. Biol. Chem. 2002;383:1459–1462. doi: 10.1515/BC.2002.166. [DOI] [PubMed] [Google Scholar]
- 27.Zaremba M., Urbanke C., Halford S.E., Siksnys V. Generation of the BfiI restriction endonuclease from the fusion of a DNA recognition domain to a non-specific nuclease from the phospholipase D superfamily. J. Mol. Biol. 2004;336:81–92. doi: 10.1016/j.jmb.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Kindler B. 1997. PhD Thesis, Universität Hannover, Hannover, Germany. [Google Scholar]
- 29.Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- 30.Nag N., Krishnamoorthy G., Rao B.J. A single mismatch in the DNA induces enhanced aggregation of MutS. Hydrodynamic analyses of the protein–DNA complexes. FEBS J. 2005;272:6228–6243. doi: 10.1111/j.1742-4658.2005.04997.x. [DOI] [PubMed] [Google Scholar]
- 31.Bjornson K.P., Allen D.J., Modrich P. Modulation of MutS ATP hydrolysis by DNA cofactors. Biochemistry. 2000;39:3176–3183. doi: 10.1021/bi992286u. [DOI] [PubMed] [Google Scholar]
- 32.Lebbink J.H., Georgijevic D., Natrajan G., Fish A., Winterwerp H.H., Sixma T.K., de Wind N. Dual role of MutS glutamate 38 in DNA mismatch discrimination and in the authorization of repair. EMBO J. 2006;25:409–419. doi: 10.1038/sj.emboj.7600936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Au K.G., Welsh K., Modrich P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 34.Biswas I., Ban C., Fleming K.G., Qin J., Lary J.W., Yphantis D.A., Yang W., Hsieh P. Oligomerization of a MutS mismatch repair protein from Thermus aquaticus. J. Biol. Chem. 1999;274:23673–23678. doi: 10.1074/jbc.274.33.23673. [DOI] [PubMed] [Google Scholar]
- 35.Wang H., Yang Y., Schofield M.J., Du C., Fridman Y., Lee S.D., Larson E.D., Drummond J.T., Alani E., Hsieh P., et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surtees J.A., Alani E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 2006;360:523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.