Abstract
There is an increasing recognition that detailed nucleic acid sequence information will be useful and even required in the diagnosis, treatment and surveillance of many significant pathogens. Because generating detailed information about pathogens leads to significantly larger amounts of data, it is necessary to develop automated analysis methods to reduce analysis time and to standardize identification criteria. This is especially important for multiple pathogen assays designed to reduce assay time and costs. In this paper, we present a successful algorithm for detecting pathogens and reporting the maximum level of detail possible using multi-pathogen resequencing microarrays. The algorithm filters the sequence of base calls from the microarray and finds entries in genetic databases that most closely match. Taxonomic databases are then used to relate these entries to each other so that the microorganism can be identified. Although developed using a resequencing microarray, the approach is applicable to any assay method that produces base call sequence information. The success and continued development of this approach means that a non-expert can now perform unassisted analysis of the results obtained from partial sequence data.
INTRODUCTION
For both surveillance and diagnostic applications, fine-scale pathogen identification and near-neighbor discrimination is important; therefore, an assay that monitors at this very specific level is desirable for many types of samples such as clinical and environmental (1–3). To successfully use any method based on DNA or RNA detection, these assays must be coupled with large databases of nucleic acid sequence information for assay design to ensure that the desired information is provided and for the interpretation of raw data. Several well-established techniques use PCR to amplify individual target pieces of sequenced genomes to provide detection of organisms (4). These methods can roughly be divided into approaches that target individual short sequence lengths or probes (<40 bp) and methods that examine longer probes. The advantage of using short probes is that when the uniqueness of the probe has been assured and unique primers are also selected, this method gives good specificity. This approach is capable of providing fine-scale identification of several genetically close organisms by selecting a sufficient number of probes. However, this can rapidly lead to a very large number of total probes being required to detect all organisms of interest. In addition these selected probes, which in the initial selection process were determined to be unique, are often later found to be less specific as more organisms are sequenced or are less specific under conditions that differ from the original conditions. This is particularly a problem for organisms belonging to a family with a high mutation rate and also for pathogens that have relatively few neighboring pathogens sequenced. In addition, PCR approaches focused on short unique probes are not capable of detecting the presence of new significant mutations nor can they easily resolve base sequence details. Approaches that use longer individual probes avoid many of these issues at the cost of being less specific. This issue means most of these approaches are not suitable for providing the information desired, providing impetus to this work.
High-density resequencing microarrays produce variable length segments, 102–105 bp, of direct sequence information. This target sequence falls in the longer target regime of PCR approaches but rather than being hybridized to a longer less-specific probe on the microarray, many shorter specific probes are placed on the microarray to allow more detailed determinations from the entire PCR amplicon. This also means that the specificity of the primers used can be relaxed. They have been successfully used to detect single nucleotide polymorphism (SNP) and genetic variants from viral, bacterial and eukaryotic genomes (5–12). Their use for SNP detection has clearly established their ability to provide reliable quality sequence information. In most cases, the microarrays were designed to study a limited number of genetically similar target pathogens and for many cases, the detection methods relied only on recognizing hybridization patterns for identification (6,9,10,13,14). Taking advantage of the sequential base resolution capability of resequencing microarrays that is required for SNP detection, resequencing has recently been successfully adapted recently using a different approach for organism identification of multiple bacterial and viral pathogens while allowing for fine detailed discrimination of closely related organisms and tracking mutations within the targeted pathogen (15–16). The new methodology differed from earlier work by using the resolved bases as the query of a similarity search of DNA databases to identify the most likely species and variants that match the base calls from the hybridization observed. The system was capable of testing for 26 pathogens simultaneously and could detect the presence of multiple pathogens. A software program, resequencing pathogen identifier (REPI), was used to simplify data analysis by performing similarity searches of a genetic database using basic local alignment search tool (BLAST) (17). The REPI program used BLAST default settings and would only return sequences that might represent the hybridization if the expect value, a quantity calculated by the BLAST program that indicates the likelihood that the sequence match found would have occurred by random chance in the database, was <10−9. This screened out all cases that had insufficient signal; however, the final determination of what pathogen(s) was detected and to what degree discrimination was possible required manual examination of the returned results. This method successfully allowed fine discrimination of various adenoviruses and strain identifications of Flu A and B samples in agreement with conventional sampling results (15,16). Two important advantages of this approach were that the information was always recovered at the most detailed level possible and that it was capable of still recognizing organisms with recent mutations. This approach also maintained specificity well, as it was not dependent on the uniqueness of a few individual short probes.
Although this analysis method has utility, there are several shortcomings: it is time consuming, not optimized to maximize sensitivity, has complicated results, is suitable only for an expert, and contains redundant or duplicate information. The process was time consuming because only the initial screening was handled automatically while the remaining steps required manual interpretation before the detection analysis was complete. Because a simple criterion (expect value cutoff of 10−9) and non-optimized BLAST parameters were used to consider a pathogen detected, the REPI algorithm provided a list of candidate organisms but did not make a final simple conclusion or relate the results of one prototype sequence to another. Instead a manual process was used to make the final determination, but because the REPI program provided all similar results and the use of public nucleic acid databases containing redundant entries, a large amount of data was presented to a user that was not useful. In addition, with a manual process it was not possible to establish that the algorithm developed was generally applicable for any organism where nucleic acid base resolved sequence information has been provided.
In this paper, we describe a new software expert system, Computer-Implemented Biological Sequence Identifier system 2.0 (CIBSI 2.0), that successfully uses resolved base sequence information from custom designed Affymetrix resequencing microarrays to provide a simple list of organisms that are detected. This algorithm addresses the most important shortcoming of previous methods by incorporating new features to completely automate pathogen identification. We have demonstrated the effectiveness of this algorithm for identification via several examples. The single program is capable of making correct decisions for all 26 pathogens contained on the Respiratory Pathogen Microarray v.1 (RPM v.1), whether detected alone or in combinations, with improved sensitivity. Although the program is currently applied to resequencing microarrays, the methodologies developed remain generally applicable. Only the first portion of the algorithm handles issues specific to microarrays while the remainder deals with sequences that are suitable for use as a query by the BLAST algorithm. In developing the general identification algorithm, we have identified and resolved issues specific to resequencing microarrays that complicate their use. Because the entire decision process for what is detected has been automated, it is straightforward to test whether the rules used to make identifications are rigorous and applicable to any pathogen. With this efficient program, resequencing based assays can provide a competitive method to test simultaneously for many possible pathogens, providing output that can be interpreted by a non-expert.
METHODS
Amplification, hybridization and sequencing determination
The details of the RPM v.1 design and the experimental methods have been discussed in previous work (15,16,18) (Lin et al., submitted for publication). Briefly, the RPM v.1 chip design includes 57 tiled regions allowing resequencing of 29.7 kb of sequences from 27 respiratory pathogens and biothreat agents. These were selected based upon clinical relevance for the population of immediate interest (United States military recruit in training) (19–21). Partial sequences from the genes containing diagnostic regions were tiled for the detection of these pathogens. The experimental microarray data used in the present analysis were obtained using a variety of purified nucleic acid templates and clinical samples culture (throat swabs and nasal washes) using random and multiplexed RT–PCR amplification schemes (for more detail description of amplification methods see Supplementary Data). Resequencing microarrays provide base call resolution by comparing the intensities between a set of four 25mer probes that differ from each other at the same position (13th base). An amplicon or target sequence is represented by numerous overlapping probe sets. GCOS™ software v1.3 (Affymetrix Inc., Santa Clara, CA) was used to align and scan hybridized microarrays to determine the intensity of each probe in every probe set. Base calls were made based on the intensity data of each probe set using GDAS v3.0.2.8 software (Affymetrix Inc.) which used an implementation of the ABACUS algorithm (5). The sequences were represented in FASTA format for later analysis steps.
In this paper, target pathogens are the organisms the assay was specifically designed to detect. The sets of probes that represent reference sequence selected from target pathogen genomes are referred to as a Prototype Sequence or ‘ProSeq’ for brevity. The set of resolved bases that result from hybridization of genomic material to a ProSeq is referred to as the hybridized sequence or ‘HybSeq’. The HybSeq is split into possible subsequences or ‘SubSeqs’.
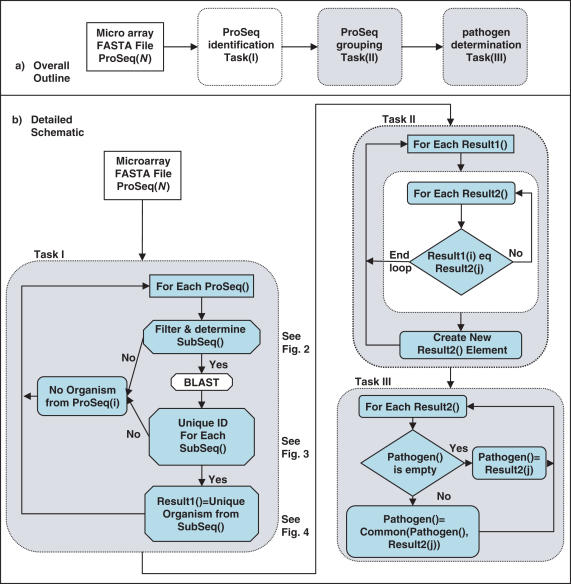
The CIBSI 2.0 program implemented in Perl described in this study handled a hierarchy of three tasks (Figure 1): (I) ProSeq identification; (II) ProSeq grouping; and (III) pathogen determination. The most developed and important portion of the algorithm deals with ProSeq identification Task(I) and is handled in three important subtasks: initial filtering of individual HybSeqs into SubSeqs suitable for sequence similarity comparisons (Figure 2), database querying of individual SubSeqs (Figure 3) and taxonomic comparison of BLAST returns for each SubSeq (Figure 4). The NCBI BLAST and taxonomy databases were used for the queries and images were obtained on February 7, 2006. For the ProSeq grouping Task(II), ProSeqs were compared to determine if they supported the same identified organism. In the pathogen determination Task(III), detected organisms were compared to the list of target pathogens the assay was designed for in order to determine if any were positively detected or were possibly related close genetic near neighbors. The level of discrimination that a particular sample supported was automatically determined.
Figure 1.
Schematic representation of the algorithm representing relationship of three main tasks and logic of subtasks associated with tasks. ProSeq identification Task(I) carries out filtering and subsequence selection, and then determines what database records Subseqs are most similar to. ProSeq grouping Task(II) figures whether prototype sequence identifications support a common organism identification. Pathogen determination Task(III) does final examination and decisions of the detected organism from the microarray data. ProSeq: prototype sequence; SubSeq: subsequences.
Figure 2.
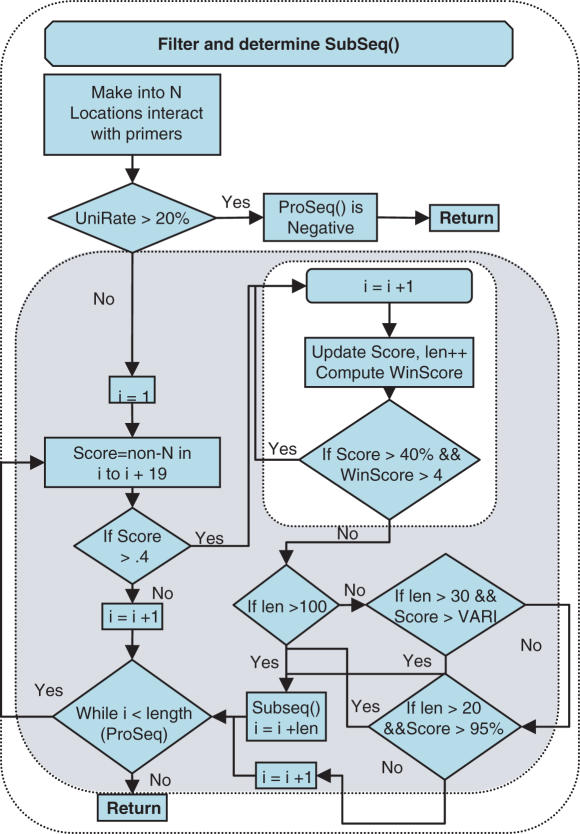
Detail schematic representation of filtering subtask of ProSeq identification Task(I). For each ProSeq, primer regions were masked as N (ambiguous) calls, then UniRate, was calculated from the HybSeq. For ProSeqs, which passed the UniRate requirement, a revised sliding window algorithm attempted to grow a SubSeq that could be used as a query to BLAST. The identity (start location in the ProSeq and length) of a successfully grown SubSeq was placed in a file for batch querying via BLAST. VARI = [(‘SubSeq length’ − 30) * 0.2857 + 70]. Detailed SubSeqs requirement is described in Supplementary Data.
Figure 3.
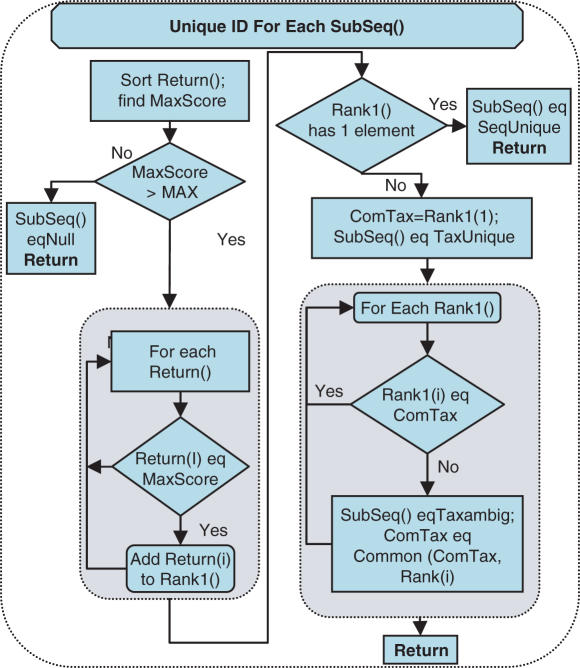
Detailed schematic representation of the second subtask of ProSeq identification Task(I), organism identification for an individual SubSeq. Each SubSeq sent to BLAST returned a list of possible matches contained in a Return array that was sorted through to find best bit score/expect value pair (MaxScore). If the MaxScore was greater than MIN (10−6), all returns that had this best Score were sorted into a new array Rank1. Detailed SubSeqs requirement is described in Supplementary Data.
Figure 4.
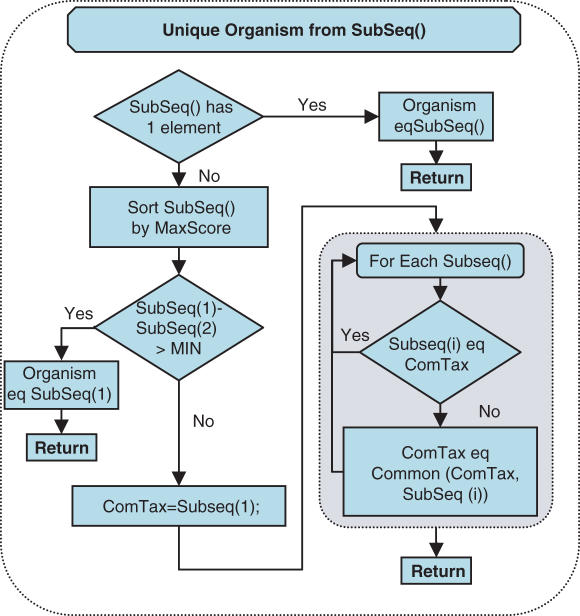
Schematic representation of the third subtask of ProSeq identification Task(I), which determines the organism, determined for a ProSeq based on the results found for its SubSeq. All of the SubSeq of a particular ProSeq are compared to determine the two best scoring SubSeq. When there was a single SubSeq or one scored much better than the other, the ProSeq inherited the properties of that SubSeq. Detailed SubSeqs requirement is described in Supplementary Data.
ProSeq identification Task(I): subtask(I) filtering
An initial filtering algorithm, REPI, was developed previously (16) and the general concepts with revisions were incorporated into the current (automated detection) algorithm used in the CIBSI 2.0 program. Filtering and subsequence selection were used to remove potential biasing caused by reference sequence choice and by other sources (i.e. primers). When PCR amplification was used, microarrays were hybridized in the presence of only primers to determine locations where they resulted in hybridization. Any portions of the ProSeqs that hybridized with the primers were masked as N calls so that the HybSeq did not contain biased information. Normally the primers are designed to be outside the ProSeq region to minimize the interference caused by primers, and so minimize the bases to be masked. There is still the chance that some bases require masking because with the large number of primers used in the multiplex, short stretches of a ProSeq not corresponding to primer locations may still hybridize with the primers. Such regions could be removed from the reference sequences and so not appear on the microarray. However, determining such locations are a difficult and time-consuming task that for most cases is not worth the effort. The first subtask of ProSeq identification Task(I) is noted in Figure 1 and shown schematically in detail in Figure 2. This subtask uses a procedure to examine a HybSeq to find the longest possible subsequence of base calls (SubSeq) that can be submitted as a query to BLAST. It produces a group of SubSeq that contain all portions of a HybSeq that have a chance of producing a limited list of returns from a BLAST query. When a HybSeq has two regions separated by a long stretch of continuous N calls, the relational positioning of the two regions cannot be trusted and so must be sent as separate queries. In addition for shorter subsequences, the number of base calls that must be made is dependent on the length. It was also recognized that for very long sequences a longer WORD size in BLAST may be used. A detailed description of the criteria and process used for each step is contained in Supplementary Data. Upon completion, the algorithm returned to the Task(I) loop and performed the BLAST subtask.
ProSeq identification Task(I): subtask(II) database query
The database query subtask performed a batch similarity search of a database using SubSeq as the queries. The BLAST program used was the NCBI Blastall –p blastn version 2.12 with a defined set of parameters. The masking of low complex regions was performed for the seeding phase to speed up the query; however, low complexity repeats were included in the actual scoring. The entire nucleotide database from NCBI acquired on February 7, 2006 was used as the reference database. (Note that earlier images of the database were used during development but all experiments were rerun with the algorithm as described with the image of the database obtained on this date.) The default gap penalty and nucleotide match score were used. The nucleotide mismatch penalty, –q, parameter was set to −1 rather than the default. The results of any BLAST query with an expect value <0.0001 were returned in tabular format from the blastall program. The information about each return (bit score, expect value, mismatches, length of match) was placed in the Return{hash key}{info} hash using the SubSeq identity as the hash key for further analysis.
ProSeq identification Task(I): subtask(III) and subtask(IV) complete ProSeq identification
The next subtask of ProSeq identification Task(I) carried out was the determination of SubSeq() states and is shown in Figure 3. The BLAST algorithm gives a ranking score which can be reported as accounting for the size of the database (expect value) or not (bit score). The full taxonomic classifications of every return for a SubSeq were retrieved from the NCBI taxonomy database obtained on February 7, 2006. Using the scores and taxonomy relationships it was possible to find a reduced number of returns that had the best match with the HybSeq. These results were summarized by identifying the taxonomic class to which all the returns belonged to, ‘identified organism’, and a parameter that indicated how they are related to each other, ‘organism uniqueness’. A detailed description of the steps is contained in Supplementary Data.
After each SubSeq was examined, the algorithm moved to the next subtask, which was to determine the identified organism of the ProSeq from the SubSeq (Figure 4). The subsequences from the same ProSeq were only allowed to support a single ‘identified organism’ determination. The procedure shown in Figure 4 demonstrates the decision method used to arrive at this determination (detailed description in Supplementary Data). After the subtask covered in Figure 4 was completed, the ProSeq identification Task(I) loop continued until all ProSeqs were examined. A list of ProSeqs that had detected organisms was built up in the Result1 array.
ProSeq grouping Task(II)
After the ProSeq identification Task(I) was completed, ProSeq grouping Task(II) (Figure 1) was used to examine the identified organism values listed in Result1 and grouped them together if they identified the same taxonomic class. Each entry in Result1 was examined and a new entry was created in Result2 if the identified organism did not appear in this list. The entries of Result2 represented the distinct individual organisms identified, but might still contain redundant information. When the ProSeqs were designed to detect the same organism and they all hybridized well, this grouping led to a reduction in redundant information being reported. But, when one ProSeq did not hybridize as well for a variety of possible reasons, multiple entries would appear in Result2 that actually represent hybridization from the same pathogen. This is because there is an alternative cause for the ProSeq hybridizing in this manner. This hybridization could be caused by two different but closely related organisms both being present in a sample and hybridizing to the microarray. Because we have not yet developed methods to distinguish these cases, no further reduction of the list of organisms is made for in ProSeq grouping Task(II) in cases where the level of identification varied on different ProSeq targeted for the same organism.
Pathogen determination Task(III)
Although it was difficult to relate results from separate ProSeqs to each other, it was desired to have a simple final detection decision be made in pathogen determination Task(III). The first task was specifically implemented so that information about what a ProSeq was intended to detect was not considered and the second task only minimal consideration of this was taken into account. This allowed these initial tasks to be capable of recognizing not only just positive and negative identifications of target pathogens but also cases that were indeterminate. In the final task, the algorithm considered whether the identified organisms belonged to the list of organisms the ProSeqs were designed to detect. The task would group organisms from ProSeq grouping Task(II) together that belonged to or were child classes of the taxonomic class of a target organism. The taxonomic class reported was the common taxonomic group of all the organisms. When all the ProSeqs for a pathogen hybridized well, a fine level discrimination was reported. But if one or more ProSeqs hybridized less well, the reported positive target pathogen was only identified at the level of the less detailed level. This is conservative because methods have not yet been developed to clearly discriminate mixtures of very closely related organisms causing different ProSeqs to hybridize from variable hybridization of a single organism on several ProSeqs. The results of all three tasks were reported and a more experienced user can view ProSeq grouping Task(II) results to clarify some cases. Note that organisms identified in ProSeq grouping Task(II) that only belonged to target pathogens were reported as positives. Clear negative ProSeqs were not mentioned in the output. ProSeqs that were indeterminate or that detected close genetic organisms were never reported as positives. These organisms were instead reported as being detected.
RESULTS
A resequencing microarray (RPM v.1) was designed previously for detection and sequence typing of 20 common respiratory and 6 CDC category A biothreat pathogens known to cause febrile respiratory illness based on ProSeqs without relying on predetermined hybridization patterns (15,16,18). Approximately 4000 RPM v.1 experiments performed using different amplification schemes, single and multiple pathogen targets, purified nucleic acids and clinical samples were examined in order to develop the pathogen identification algorithm. Results using this algorithm with clinical samples, identified pathogens and purified nucleic acids are discussed in detail in other works (15,16,18) (Lin et al., submitted for publication). In all cases, the algorithm correctly identified the organism at a species or strain level, depending on the length of the ProSeqs represented on the RPM v.1. Some specific examples will be discussed to illustrate how the algorithm performs under a variety of conditions.
Pathogen identification
Purified Chlamydia pneumoniae nucleic acid samples with 10–1000 genome copies (via method in Lin et al., submitted for publication) were chosen to illustrate how pathogen detection and identification were done when multiple ProSeqs were targeted for the same pathogen. RPM v.1 has three highly conserved ProSeqs selected from the genes encoding for the major outer membrane proteins VD2 and VD4, and the DNA-directed RNA polymerase (rpoB) gene. The HybSeqs from the different samples differed only in the number of unique base calls as shown in Table 1. The percentage of the ProSeq called varied from 80 to 100% except for one case at a concentration of 10 that had only 11% of the rpoB ProSeq producing unique base calls. Because the samples at this concentration are not reproducibly generating the same percentage of base calls, this is probably the detection limit of this ProSeq of the assay. Table 1 listed the determinations made for the SubSeq and at the end of each task for the various samples. The ProSeq from the different cases produced the same number of SubSeqs. These SubSeqs from different samples reported different bit scores for the same top ranked returns from BLAST. In fact VD2 and VD4 produced exactly the same results. The NCBI taxonomy database classified the returns into four distinct groups, which represented the C.pneumoniae taxonomic group and three child strain groups. AE001652, AE002167, AE017159 and BA000008 appeared in the returns of all the ProSeqs for each sample, since they represented database entries of completely sequenced genomes. One rpoB SubSeq produced for its organism uniqueness, SeqUniqu. All other SubSeqs were TaxAmbig as multiple returns from different taxonomic classes were returned. Since the VD2 and VD4 ProSeq each have a single SubSeq, Task(I) assigned the ProSeq the state of the SubSeq. For the rpoB ProSeq, the bit score of one SubSeq was large enough that the algorithm assigned that SubSeq's identification to the ProSeq. Task II of the algorithm grouped all three ProSeqs together since they all had the same identified organism and TaxAmbig was assigned. The result of Task(III) was positive for target pathogen C.pneumoniae and this decision was straightforward as all the ProSeqs agreed with each other and belonged to the same target pathogen taxonomic class. Although the rpoB ProSeq was SeqUniqu, this was not the final conclusion for Task(II) as the ProSeq that was SeqUniqu was not the child taxonomic group and other ProSeq were TaxAmbig. The three recognized strains scored the same, which indicated that the sequence selected for the ProSeqs was very conserved and would not allow discrimination between the strains.
Table 1.
Algorithm decisions for C.pneumoniae at several concentrations for SubSeq, ProSeq identification Task(I), ProSeq grouping Task(II) and pathogen determination Task(III)
| Genome copies | ProSeq | Unique calls (%) | No. of SubSeq | SubSeq organism identification and Uniqueness, Bit score | Task(I) | Task(II) | Task(III) |
|---|---|---|---|---|---|---|---|
| 1000 | VD2 | 89 | 1 | C. pne(G1), TA, 145 | C. pneTA | C. pneTA | POSITIVE C. pne |
| VD4 | 91 | 1 | C. pne(G1), TA, 145 | C. pneTA | |||
| 80 | 2 | C. pne(G2), SU, 307 | C. pneSU | ||||
| rpoB | C. pne(G3), TA, 73 | ||||||
| 100 | VD2 | 100 | 1 | C. pne(G1), TA, 164 | C. pneTA | C. pneTA | POSITIVE C. pne |
| VD4 | 97 | 1 | C. pne(G1), TA, 156 | C. pneTA | |||
| 80 | 2 | C. pne(G2), SU, 343 | |||||
| rpoB | C. pne(G3), TA, 87 | C. pneSU | |||||
| 100 | VD2 | 83 | 1 | C. pne(G1), TA, 136 | C. pneTA | C. pneTA | POSITIVE C. pne |
| VD4 | 91 | 1 | C. pne(G1), TA, 145 | C. pneTA | |||
| 84 | 2 | C. pne(G2), SU, 318 | |||||
| rpoB | C. pne(G3), TA, 82 | C. pneSU | |||||
| 10 | VD2 | 100 | 1 | C. pne(G1), TA, 164 | C. pneTA | C. pneTA | POSITIVE C. pne |
| VD4 | 97 | 1 | C. pne(G1), TA, 156 | C. pneTA | |||
| 90 | 2 | C. pne(G2), SU, 340 | |||||
| rpoB | C. pne(G3), TA, 89 | C. pneSU | |||||
| 10 | VD2 | 100 | 1 | C. pne(G1), TA, 164 | C. pneTA | C. pneTA | POSITIVE C. pne |
| VD4 | 93 | 1 | C. pne(G1), TA, 148 | C. pneTA | |||
| rpoB | 11 | 0 | Null | Null |
Influenza and Human Adenovirus (HAdV) were the only pathogens that had ProSeq selected that would permit detailed strain level discrimination as discussed in previous work (15,16). This previous work using manual analysis found that the microarray results were in excellent agreement with the conventional sequencing results for clinical samples. A few of the results of running the CIBSI 2.0 program using the updated NCBI database on the raw microarray results are presented in Table 2 (the results for all samples used in the previous work are presented in Supplementary Table A). The identified organisms were not identical to the original findings due to the difference in database used and because all ProSeqs were considered rather than only the Flu A and B hemagglutinin. In fact, the conventional sequencing results that were submitted to NCBI from that work were found for every sample to be among the returns with the best score for the hemagglutinin ProSeq (Supplementary Table B). It should be noted that the previous work based its analysis upon only the results of the hemagglutinin ProSeq. For 8 of 13 Influenza A and 3 of 12 Influenza B cases, the results of ProSeq identification Task(I) and ProSeq grouping Task(II) found that the conventional sequencing was the single best return for the hemagglutinin ProSeq. Owing to the large number of isolate sequences in the database for the hemagglutinin gene it was not surprising that in some cases a single unique entry was not found. In each of the remaining five Influenza A samples, the other sequences returned differed by <0.2% from the conventional sequence. The fewer samples with unique isolate identifications for Influenza B were due to an older reference sequence used for the ProSeq, which allowed less hybridization to occur (18). This also meant that when multiple sequences were returned for a sample they represented greater genetic variation, up to 2%. As a result of the current method of making pathogen determination Task(III) level identification, the final organism reported was less specific (H3N2 or Flu B) for every sample than what was reported as possible in ProSeq grouping Task(II). For HAdV samples, the algorithm also reproduced the finer scale discriminations that had been made previously by manual methods (data not shown).
Table 2.
Algorithm decisions for Influenza A clinical sample identified previously using a manual method for SubSeq, ProSeq identification Task(I), ProSeq grouping Task(II) and pathogen determination Task(III)
| Sample name | ProSeq | No. of Sub | SubSeq organism identification and Uniqueness, Bit score | Task(I) | Task(II) | Task(III) |
|---|---|---|---|---|---|---|
| A/Colorado /360/05 | HA3 | 1 | H3N2 TA,1031 | H3N2 TA | (NY) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | A/NewYork/98/04(NY) SU,1570 | (NY) SU | |||
| M | 4 | 2 Flu A TA,69.7 128 | H3N2 TA | |||
| 2 H3N2 TA,125 393 | ||||||
| A/Qatar /2039/05 | HA3 | 1 | A/Qatar/2039/05(QA) SU,1080 | (QA) SU | (QA) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 2 | 2 H3N2 TA,643 919 | H3N2 TA | |||
| M | 4 | 2 H3N2 TA,505 272 | H3N2 TA | |||
| 2 Flu A TA,115 77.7 | ||||||
| A/Guam /362/05 | HA3 | 1 | A/Guam/362/05(GA) SU,1066 | (GA) SU | (GA) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | H3N2 TA,1610 | H3N2 TA | |||
| M | 4 | 2 H3N2 TA,240 397 | H3N2 TA | |||
| 2 Flu A TA,79.2 79.2 | ||||||
| A/Italy /384/05 | HA3 | 1 | A/Italy/384/05(IT) SU, 1017 | (IT) SU | (IT) SU (NY) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | A/NewYork/371/04(NY) SU,1494 | (NY) SU | |||
| M | 3 | 2 H3N2 TA, 461 359 | H3N2 TA | |||
| Flu A TA,74.5 | ||||||
| A/Turkey/2108/05 | HA3 | 1 | A/Turkey/2108/05(TU) SU,952 | (TU) SU | (TU) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | H3N2 TA,1363 | H3N2 TA | |||
| M | 3 | 2 H3N2 TA,412 239 | H3N2 TA | |||
| Flu A TA,76.1 | ||||||
| A/Korea/298/05 | HA3 | 1 | A/Korea/298/05(KO) SU,1011 | (KO) SU | (KO) SU (NY) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 3 | A/NewYork/98/04(NY) SU,243 | (N1) SU | |||
| 2 Flu A TA,110 98.3 | ||||||
| M | 4 | 2 Flu A TA,66.6 76.1 | H3N2 TA | |||
| 2 H3N2 TA,328 255 | ||||||
| A/Japan /1383/05 | HA3 | 1 | A/Japan/1383/05(JA) SU,935 | (JA) SU | (JA) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | H3N2 TA,1369 | H3N2 TA | |||
| M | 5 | 3 Flu A TA,125 114 76.1 | H3N2 TA | |||
| 2 H3N2 TA,175 247 | ||||||
| A/Ecuador /1968/04 | HA3 | 1 | H3N2 TA,1071 | H3N2 TA | H3N2 TA | POSITIVE H3N2 |
| NA2 | 2 | H3N2 TA,1584 109 | H3N2 TA | |||
| M | 4 | 3 Flu A TA,158 164 104 | H3N2 TA | |||
| H3N2 TA,131 | ||||||
| A/Iraq /34/05 | HA3 | 1 | A/Iraq/34/05(IR) SU,1028 | (IR) SU | (IR) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 3 | 2 H3N2 TA,125 1402 | H3N2 TA | |||
| Flu A TA,109 | ||||||
| M | 5 | 3 H3N2 TA,137 350 234 | H3N2 TA | |||
| 2 Flu A TA,131 74.5 | ||||||
| A/Peru /166/05 | HA3 | 1 | A/Peru/166/05(PU) SU,1061 | (PU) SU | (PU) SU H3N2 TA | POSITIVE H3N2 |
| NA2 | 1 | H3N2 TA,1686 | H3N2 TA | |||
| M | 3 | H3N2 TA,508 | H3N2 TA | |||
| Flu A TA,76.1 | ||||||
| A/NewYork/461/2005 SU,247 |
Note: Within a row the first listing of a specific strain was followed by a two-letter abbreviation used in the remaining columns of that row.
The next example of detection for the Mycoplasma pneumoniae pathogen demonstrated a case where there was only a single ProSeq for the target pathogen. A total of 48 test samples were performed using multiplex PCR (via method in Lin et al., submitted for publication) where for 46 of the samples M.pneumoniae organism was spiked into nasal wash with several other pathogens from 100 to 100 000 colony forming units per ml, the remaining 2 samples were purified with nucleic acid from culture stock at a concentration of 1000 genome copies per reaction volume. This ProSeq was also not optimal for fine discrimination because it was selected from a highly conserved region (345 bp) of the cytadhesin P1 gene. In every case taxonomic database entries for M.pneumoniae or its one recognized distinct strain tied for MaxScore (Supplementary Table 3). To better understand these returns, the database sequences were examined and subdivided into three groups of sequences, A, B and C, based on how well they matched the reference sequence used to make the ProSeq. The placement of the database entries into the three groups was determined from a CLUSTAL alignment of the sequences of this gene. This alignment confirmed that the database entries differed significantly more from each other in regions not represented by the ProSeq and contained sufficient variability that would have allowed finer discrimination. Members of Group A exactly matched the ProSeq and could not be distinguished between on the microarray. Similarly, members of group B matched the ProSeq except at the 199th position where the base called was C rather than T. Group C sequences contained a few database entries that were more variable and might be distinguished from other entries within the ProSeq. For the 48 experimental tests of M.pneumoniae, as much as 80% of the ProSeq hybridized for 19 samples, yet only 5 of these samples had an unambiguous base call at the 199th position. When it was unambiguous, it always matched group B sequences. In the cases where an N base call was made at the 199th location, both groups A and B sequences were returned with the same score. Regardless of this, the target pathogen positively identified was M.pneumoniae for every sample tested.
These examples showed how decisions were made independent of whether single or multiple ProSeqs were dedicated to a target pathogen. They also illustrated that the level of discrimination possible was strongly determined by the quality of the selected ProSeq. It is possible that for some pathogens fine level discrimination is not required and the currently tested selections on RPM v.1 would provide satisfactory information. The CIBSI 2.0 algorithm demonstrated its capability to automatically report the maximum level of discrimination that could be supported by the HybSeq information.
Genetic near neighbors
To demonstrate how the algorithm handled closely related genetic species, a sample of a non-targeted pathogen was considered using multiplex PCR (via method in Lin et al., submitted for publication). For Variola major virus, one of the biothreat pathogens on the RPMv.1, the validation runs demonstrated that Variola major virus purified DNA templates of plasmids were always positively identified when detected (Table 3). Table 4 shows the results when purified Vaccinia genomic DNA was spiked into nasal washes and processed at various concentrations using multiplex PCR. The array has two ProSeqs from hemagglutinin (VMVHA, ∼500 bp) and cytokine response modifier B (VMVcrmB, ∼300 bp) genes for Variola major virus detection. The percentage of the ProSeq that hybridizes is sufficient that if hybridization patterns were only considered one might assume that this tile is identifying the presence of its target. This would indicate that reference sequence selected was not the best choice. However, when our algorithm was applied none of the samples is in fact identified as Variola major or minor virus. Vaccinia was always one of the Orthopoxvirus species listed with the highest scores for VMVcrmB ProSeq, but in only seven cases was it uniquely identified as the probable species detected. In only one sample at the lowest concentration and fraction of VMVcrmB hybridizing, did this ProSeq even identify Variola major and minor virus as one among the Orthopoxvirus species that could be the cause of the hybridization. The lower limit of detection for the amplification method used was between this concentration and the one above it for Variola major itself. The VMVHA ProSeq exhibited much lower sensitivity and made identifications of Orthopoxvirus species in only two experiments and Variola major virus was listed as one of the tied best scoring returns. In both cases, VMVcrmB ProSeq specifically identified Vaccinia virus as the best match. The percentage of the hybridized ProSeq correlated with concentration of the sample.
Table 3.
Organism identification and algorithm decisions from Variola Major virus Nucleic Acid templates for SubSeq, ProSeq identification Task(I), ProSeq grouping Task(II) and pathogen determination Task(III)
| Genome copies | ProSeq | Unique calls (%) | No. of SubSeq | SubSeq organism identification and Uniqueness, Bit score | Task(I) | Task(II) | Task(III) |
|---|---|---|---|---|---|---|---|
| 1000 | CRMB | 83.90 | 1 | Variola TA 355 | Vari., TA | Vari., SU | Positive |
| HA | 77.00 | 1 | Variola SU 567 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 80.90 | 1 | Variola TA 342 | Vari., TA | Vari., SU | Positive |
| HA | 75.50 | 1 | Variola SU 554 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 76.40 | 1 | Variola TA 324 | Vari., TA | Vari., SU | Positive |
| HA | 73.30 | 1 | Variola SU 538 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 80.10 | 1 | Variola TA 339 | Vari., TA | Vari., SU | Positive |
| HA | 74.90 | 1 | Variola SU 551 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 81.60 | 1 | Variola TA 345 | Vari., TA | Vari., SU | Positive |
| HA | 76.10 | 1 | Variola SU 561 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 77.90 | 1 | Variola TA 329 | Vari., TA | Vari., SU | Positive |
| HA | 75.50 | 1 | Variola SU 556 | Vari., SU | Vari.,TA | Variola | |
| 1000 | CRMB | 81.60 | 1 | Variola TA 299 | Vari., TA | Vari., TU | Positive |
| HA | 74.90 | 4 | Variola TU 106 84 | Vari., TU | Vari., TA | Variola | |
| Variola TA 103 69.7 | |||||||
| 100 | CRMB | 84.20 | 1 | Variola SU 624 | Vari., SU | Vari., SU | Positive |
| HA | 5.60 | 0 | Null | Null | Variola |
TA, TaxAmbig in this case Variola, Variola major and minor taxonomic classes.
Table 4.
Organism identification and algorithm decisions from Vaccinia sample on Variola Major virus ProSeqs for SubSeq, ProSeq identification Task(I), ProSeq grouping Task(II) and pathogen determination Task(III)
| CFU | ProSeq | Unique calls (%) | # SubSeq | SubSeq organism identification and Uniqueness, Bit score | Task I | Task II | Task III |
|---|---|---|---|---|---|---|---|
| 5 × 107 | CRMB | 77.90 | 2 | Orth.TA 156(H1), Vacc.TU 153(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 29.40 | 1 | Orth.TA 60.2(H3) | Orth., TA | Orth., TA | Vaccinia | |
| 5 × 107 | CRMB | 79.80 | 2 | Orth.TA 164(H1), Vacc.TU 115(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 25.70 | 1 | Orth.TA 66.6(H3) | Orth., TA | Orth., TA | Vaccinia | |
| 1.6 × 107 | CRMB | 79.40 | 2 | Orth.TA 161(H1), Vacc.TU 114(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 14.80 | 0 | Null | Null | Vaccinia | ||
| 1.6 × 107 | CRMB | 77.50 | 2 | Orth.TA 153(H1),109(H4) | Orth., TA | Orth., TA | Detected |
| HA | 24.50 | 0 | Null | Null | Orthopox | ||
| 1.6 × 107 | CRMB | 76.80 | 2 | Orth.TA 155(H1), Vacc.TU 112(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 21.60 | 0 | Null | Null | Vaccinia | ||
| 1.6 × 107 | CRMB | 74.50 | 2 | Orth.TA 152(H1), 106(H5) | Orth., TA | Orth., TA | Detected |
| HA | 17.30 | 0 | Null | Null | Orthopox | ||
| 5 × 106 | CRMB | 77.90 | 2 | Orth.TA 155(H1), Vacc.TU 112(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 25.70 | 0 | Null | Null | Vaccinia | ||
| 5 × 106 | CRMB | 78.30 | 2 | Orth.TA 153(H1), 115(H5) | Orth., TA | Orth., TA | Detected |
| HA | 22.00 | 0 | Null | Null | Orthopox | ||
| 5 × 106 | CRMB | 73.00 | 2 | Orth.TA 150(H1), Vacc.TU 99.9(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 13.00 | 0 | Null | Null | Vaccinia | ||
| 5 × 106 | CRMB | 73.40 | 2 | Orth.TA 153(H1), 115(H5) | Orth., TA | Orth., TA | Detected |
| HA | 7.80 | 0 | Null | Null | Orthopox | ||
| 1.6 × 106 | CRMB | 75.30 | 2 | Orth.TA 158(H1), 107(H5) | Orth., TA | Orth., TA | Detected |
| HA | 8.60 | 0 | Null | Null | Orthopox | ||
| 1.6 × 106 | CRMB | 49.80 | 2 | Orth.TA 60(H1), Vacc.TU 90.3(H2) | Vacc., TU | Vacc., TU | Detected |
| HA | 6.60 | 0 | Null | Null | Vaccinia | ||
| 1.6 × 106 | CRMB | 65.50 | 2 | Orth.TA 136(H1), 91.9(H5) | Orth., TA | Orth., TA | Detected |
| HA | 10.00 | 0 | Null | Null | Orthopox | ||
| 1.6 × 106 | CRMB | 62.90 | 2 | Orth.TA 126(H1), 87.2(H5) | Orth., TA | Orth., TA | Detected |
| HA | 8.20 | 0 | Null | Null | Orthopox | ||
| 5 × 105 | CRMB | 58.40 | 2 | Orth.TA 110(H1), 90.3(H5) | Orth., TA | Orth., TA | Detected |
| HA | 9.00 | 0 | Null | Null | Orthopox | ||
| 5 × 105 | CRMB | 56.20 | 2 | Orth.TA 77.7(H6), 96.7(H5) | Orth., TA | Orth., TA | Detected |
| HA | 8.00 | 0 | Null | Null | Orthopox | ||
| 5 × 105 | CRMB | 49.00 | 1 | Orth.TA 87.2(H5) | Orth., TA | Orth., TA | Detected |
| HA | 9.30 | 0 | Null | Null | Orthopox | ||
| 5 × 105 | CRMB | 44.60 | 1 | Orth.TA 90.3(H5) | Orth., TA | Orth., TA | Detected |
| HA | 7.80 | 0 | Null | Null | Orthopox |
Vacc., Vaccinia; Orth., Orthopox. (H1) Rabbitpox, Buffalopox, Cowpox, Vaccinia, Callithrix jacchus, Taterapox. (H2) Vaccinia. (H3) Vaccinia, Variola (Major and Minor), Cantagalo, Ectromelia, Elephantpox, Aracatuba, Cowpox, Taterapox. (H4) H2 and Cowpox. (H5) H4 and Camelpox. (H6) H1 and Variola, Variola Major, Variola Minor.
Filtering
This example demonstrated the importance of the filtering portion of the algorithm by considering the HybSeqs of the ProSeqs for the H1N1 neuraminidase (NA1) and matrix genes from human Influenza A/Puerto Rico/8/34 (H1N1) strain. Filtering was necessary because sending the HybSeq of a ProSeq to BLAST in a single query can bias the scores against strains that have insertions or deletions relative to the ProSeq, especially when using BLAST parameters that maximized the use of base calls. The sliding window test was the portion of the algorithm that controlled filtering. If filtering were turned off, the entire HybSeq would be used in a single subsequence for two influenza ProSeqs that showed significant hybridization. A/Weiss/43 (H1N1) strain was identified as the most likely strain from the HybSeq of the NA1 ProSeq while the HybSeq of the matrix ProSeq correctly identified A/Puerto Rico/8/34. To better understand the source of biasing, CLUSTAL alignment of the NA1 gene of the two strains and the reference sequence used to make the ProSeq are shown in Figure 5. The two strains showed 95% identity (67 mismatches in 1362 aligned bases); however, there was a stretch of 45 bases inserted in both A/Weiss/43 and the NA1 ProSeq compared to A/Puerto Rico/8/34. With the default filtering on, the NA1 ProSeq was split into five SubSeqs as the algorithm encountered large stretches of no calls. In ProSeq identification Task(I), the algorithm determined that three of the SubSeqs, which were shorter, had an identified organism of H1N1 as several isolates including A/Puerto Rico/8/34 tied with the best score, while the two other SubSeqs had an identified organism of only the A/Puerto Rico/8/34 strain as being the closest match. The organism identified by the NA1 ProSeq was A/Puerto Rico/8/34 because one of the SubSeqs had a much higher score. This ProSeq supported the same strain identification made in the matrix ProSeq. The organism identified was A/Puerto Rico/8/34 because two ProSeqs detected only that organism. The correct target pathogen was detected with filtering while without filtering the target pathogen level of identification would have been Influenza A (H1N1 subtype) because two organisms were detected, A/Puerto Rico/8/34 and A/Weiss/43. Splitting the HybSeq into SubSeqs to remove biasing may reduce the level of identification as occurred for the three of the five SubSeqs in this case. The previous example for Vaccinia was another instance where the wrong species (Camel Pox or Callithrix jacchus) identification would occur if the filtering was not used. Clinical samples in Table 2 showed that HybSeqs split into multiple SubSeqs were capable of very specific identification.
Figure 5.
Alignment of the influenza A NA1 ProSeq and A/Weiss/43, A/PuertoRico/8/34 strains. Raw and filtered hybridization chip results of A/puertoRico/8/34 are also shown. Asterisks indicate perfectly matched sequences.
To illustrate the other filtering performed, when multiplex strategies rather than generic were used for amplification Figure 5 also contains the raw and mask filtered results of this region for the A/Puerto Rico/8/34 sample. It was necessary to perform additional filtering to remove potential biasing caused by the specific primers as described in the methods. In the case shown in Figure 5, a sequence of 18 bases present in the raw result are made N after filtering since they are in a location that interacts with the primers. If these base calls were included in the subsequences constructed, even though the HybSeq would still be split into the same number of SubSeq, the query for the ProSeq would favor an incorrect strain.
DISCUSSION
The algorithm we have developed successfully provided pathogen identification to the maximum level of detail possible (species or strain) depending on the quality of each ProSeq. This identification capability requires minimal input on the identity of the pathogens, making non-expert use feasible. The crucial feature incorporated that allowed complete automation was the use of taxonomic databases, which classify organisms into ordered groups and provide relationships between organism entries, allowing removal of redundancies, comparison of different related prototype sequences and simplification of data presentation. This allows databases, i.e. NCBI, that are redundant and subject to minimal curation but which constantly receive updated and new sequence information to be used with great success. Although we have demonstrated this using only the NCBI databases, other databases or custom made ones could have easily been used, which might improve performance. The algorithm is capable of providing accurate identifications at all analysis levels for pathogens that are less variable or are represented by highly conserved ProSeqs. For more variable or rapidly mutating pathogens, e.g. Influenza A virus, ProSeq identification Task(I) and ProSeq grouping Task(II) still provided accurate detailed identifications, but the pathogen determination Task(III) was unable to report fine scale discrimination. The comparison of the conventionally sequenced Influenza virus gene sequences illustrated that the algorithm is capable of automatically adjusting for updates in databases. The algorithm demonstrated its capability to properly distinguish hybridizations on a ProSeq caused by the specified pathogen from those caused by genetically close (near neighbor) strains and did not make incorrect identifications, eliminating one potential cause of false positives. Filtering the raw hybridization results served to reduce the computation time, accounted for potential primer interference and more importantly reduced potential biasing. This simple integrated algorithm provided sufficient and accurate identification, so that immediate use of the RPM v.1 or similar resequencing arrays and assay is possible. Although not discussed in this paper, the algorithm has successfully detected the presence of simulated multi-infections (Lin et al., submitted for publication). The algorithm as currently developed will detect mixtures when the organisms have sufficient variation; however, detection of a mixture of an organism and its mutation strain in a sample is uncertain in its present phase. In principle it may be possible to detect such mixtures as the resequencing microarray can detect and sequence diploid organisms.
Besides demonstrating the success of the CIBSI 2.0 program, the work involved in developing the algorithm allowed insight into the importance of proper ProSeq selection. The RPM v.1 was the first resequencing array designed specifically for multiple pathogen detection using database similarity searching and served as a prototype for this application. We have demonstrated that a single ProSeq with as few as 100 bp, when designed correctly, can be sufficient to unambiguously identify an organism. However, it is clearly indicated that several longer ProSeqs provide better confirmation and more detailed information of a pathogen. Although the algorithm provides accuracy equivalent to manual analysis for determinations of individual ProSeqs, the current algorithm is only partially successful in integrating information from multiple ProSeqs. The emphasis of the design to this point has been on capabilities that are generally applicable to any pathogen. We are incorporating these insights in our newer more comprehensive resequencing array designs. Improving on level of detail reported in pathogen determination Task(III) will require more information about an individual pathogen and may have to be developed for each specific pathogen or class of pathogens. This information is also required for the algorithm to identify which differences between a sample and database entries represent significant mutations. Future work will involve improving the use of the current taxonomic database or potentially developing a new relational database that is specific to our needs and then incorporating more specific information of target pathogens. The hierarchal design of the data analysis makes it easy to incorporate analysis that build upon the analysis already performed.
We have met with some success in the current version but want to have increased automated discrimination. We have a well-defined path to completing this aim. The use of properly designed resequencing microarrays and this automated detection algorithm provides a way forward to developing assays that can test for multiple organisms simultaneously while providing fine strain level discrimination giving access to information about detailed strain recognition, antibiotic resistance markers and pathogenicity. This is a capability that other approaches cannot currently provide. In addition, since the design of the original 30 kb RPM microarray, the possible sequence content of the current array has increased 10-fold to 300 kb and further increases in array density are still attainable. This, coupled with our identification algorithms, will allow the analysis of partial sequence information from even more organisms for applications such as differential diagnostics for illnesses with multiple potential causes (i.e. febrile respiratory illness), tracking of emergent pathogens, distinction of biological threats from harmless near genetic neighbors in surveillance applications and for tracking the impact of co-infections or super infections. The concept of categorizing and reporting different degrees of identification depending on the quality of samples and set of target sequences is not limited to resequencing microarrays but is more generally applicable to any platform that is capable of returning sequence level calls that can be used to query a reference DNA database. As the trend for assays that test for multiple pathogens increases, automated analysis tools, such as this one, become more crucial for rapid identification in simple formats useful to the non-expert on a day to day basis. The remaining hurdle to using resequencing microarrays as a routine assay method is now clearly the sample processing methods. Further automating these steps is an important area of future research and development.
The program can be obtained free of charge for research purposes by contacting the authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Kate Blaney, Adam Ligler, Carolyn Meador, Paul Xu, Brian Weslowski and the Silent Guardian team without whom the microarray data would not be available. The opinions and assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the US Government. Funding support for this research was from the Office of Naval Research via the Naval Research Laboratory Base program. Funding to pa56y the Open Access publication charges for this article was provided by ONR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Whelen A.C., Persing D.H. The role of nucleic acid amplification and detection in the clinical microbiology laboratory. Annu. Rev. Microbiol. 1996;50:349–373. doi: 10.1146/annurev.micro.50.1.349. [DOI] [PubMed] [Google Scholar]
- 2.McDonough E.A., Barrozo C.P., Russell K.L., Metzgar D. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol. Cell Probes. 2005;19:314–322. doi: 10.1016/j.mcp.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Roth S.B., Jalava J., Ruuskanen O., Ruohola A., Nikkari S. Use of an oligonucleotide array for laboratory diagnosis of bacteria responsible for acute upper respiratory infections. J. Clin. Microbiol. 2004;42:4268–4274. doi: 10.1128/JCM.42.9.4268-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner S.N., Kuczmarski T.A., Vitalis E.A., Slezak T.R. Limitations of TaqMan PCR for detecting divergent viral pathogens illustrated by hepatitis A, B, C, and E viruses and human immunodeficiency virus. J. Clin. Microbiol. 2003;41:2417–2427. doi: 10.1128/JCM.41.6.2417-2427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutler D.J., Zwick M.E., Carrasquillo M.M., Yohn C.T., Tobin K.P., Kashuk C., Mathews D.J., Shah N.A., Eichler E.E., Warrington J.A., et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingeras T.R., Ghandour G., Wang E., Berno A., Small P.M., Drobniewski F., Alland D., Desmond E., Holodniy M., Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 7.Hacia J.G. Resequencing and mutational analysis using oligonucleotide microarrays. Nature Genet. 1999;21:42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 8.Lin B., Vahey M.T., Thach D., Stenger D.A., Pancrazio J.J. Biological threat detection via host gene expression profiling. Clin. Chem. 2003;49:1045–1049. doi: 10.1373/49.7.1045. [DOI] [PubMed] [Google Scholar]
- 9.Wilson W.J., Strout C.L., DeSantis T.Z., Stilwell J.L., Carrano A.V., Andersen G.L. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell Probes. 2002;16:119–127. doi: 10.1006/mcpr.2001.0397. [DOI] [PubMed] [Google Scholar]
- 10.Wilson K.H., Wilson W.J., Radosevich J.L., DeSantis T.Z., Viswanathan V.S., Kuczmarski T.A., Andersen G.L. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 2002;68:2535–2541. doi: 10.1128/AEM.68.5.2535-2541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwick M.E., McAfee F., Cutler D.J., Read T.D., Ravel J., Bowman G.R., Galloway D.R., Mateczun A. Microarray-based resequencing of multiple Bacillus anthracis isolates. Genome Biol. 2005;6:R10. doi: 10.1186/gb-2004-6-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maitra A., Cohen Y., Gillespie S.E., Mambo E., Fukushima N., Hoque M.O., Shah N., Goggins M., Califano J., Sidransky D., et al. The Human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong C.W., Albert T.J., Vega V.B., Norton J.E., Cutler D.J., Richmond T.A., Stanton L.W., Liu E.T., Miller L.D. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulaiman I.M., Liu X., Frace M., Sulaiman N., Olsen-Rasmussen M., Neuhaus E., Rota P.A., Wohlhueter R.M. Evaluation of Affymetrix severe acute respiratory syndrome resequencing GeneChips in characterization of the genomes of two strains of coronavirus infecting humans. Appl. Environ. Microbiol. 2006;72:207–211. doi: 10.1128/AEM.72.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Daum L.T., Vora G.J., Metzgar D., Walter E.A., Canas L.C., Malanoski A.P., Lin B., Stenger D.A. Identifying influenza viruses with resequencing microarrays. Emerg. Infect. Dis. 2006;12:638–646. doi: 10.3201/eid1204.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin B., Wang Z., Vora G.J., Thornton J.A., Schnur J.M., Thach D.C., Blaney K.M., Ligler A.G., Malanoski A.P., Santiago J., et al. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 2006;16:527–535. doi: 10.1101/gr.4337206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Davignon L., Walter E.A., Mueller K.M., Barrozo C.P., Stenger D.A., Lin B. Use of resequencing oligonucleotide microarrays for identification of Streptococcus pyogenes and associated antibiotic resistance determinants. J. Clin. Microbiol. 2005;43:5690–5695. doi: 10.1128/JCM.43.11.5690-5695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolavic-Gray S.A., Binn L.N., Sanchez J.L., Cersovsky S.B., Polyak C.S., Mitchell-Raymundo F., Asher L.V., Vaughn D.W., Feighner B.H., Innis B.L. Large epidemic of adenovirus type 4 infection among military trainees: epidemiological, clinical, and laboratory studies. Clin. Infect. Dis. 2002;35:808–818. doi: 10.1086/342573. [DOI] [PubMed] [Google Scholar]
- 20.Erdman D.D., Xu W., Gerber S.I., Gray G.C., Schnurr D., Kajon A.E., Anderson L.J. Molecular epidemiology of adenovirus type 7 in the United States, 1966–2000. Emerg. Infect. Dis. 2002;8:269–277. doi: 10.3201/eid0803.010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson W.W., Shay D.K., Weintraub E., Brammer L., Cox N., Anderson L.J., Fukuda K. Mortality associated with influenza and respiratory syncytial virus in the United States. J. Am. Med. Asso. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.