Abstract
Z-DNA binding protein 1 (ZBP1) belongs to a family of proteins that contain the Zα domain, which binds specifically to left-handed Z-DNA and Z-RNA. Like all vertebrate proteins in the Zα family, it contains two Zα-like domains and is highly inducible by immunostimulation. Using circular dichroism spectroscopy and electrophoretic mobility shift assays we show that both Zα domains can bind Z-DNA independently and that substrate binding is greatly enhanced when both domains are linked. Full length ZBP1 and a prominent splice variant lacking the first Zα domain (ΔZα) showed strikingly different subcellular localizations. While the full length protein showed a finely punctate cytoplasmatic distribution, ZBP1ΔZα accumulated in large cytoplasmic granules. Mutation of residues important for Z-DNA binding in the first Zα domain resulted in a distribution comparable to that of ZBP1ΔZα. The ZBP1ΔZα granules are distinct from stress granules (SGs) and processing bodies but dynamically interacted with these. Polysome stabilization led to the disassembly of ZBP1ΔZα granules, indicating that mRNA are integral components. Heat shock and arsenite exposure had opposing effects on ZBP1 isoforms: while ZBP1ΔZα granules disassembled, full length ZBP1 accumulated in SGs. Our data link ZBP1 to mRNA sorting and metabolism and indicate distinct roles for ZBP1 isoforms.
INTRODUCTION
The left-handed Z-conformation is an alternative, higher energy form that can be adopted by double-stranded DNA and RNA, preferentially by sequences containing alternating purine–pyrimidine nucleotides (1–3). In Z-DNA and Z-RNA the (desoxy)ribo-phophate backbone forms a left-handed, zigzag helix where the glycosidic bonds between base and sugar alternate between the syn- and anti-conformation and the purines are tilted to the outside. In contrast, in B-DNA and A-RNA the backbone forms a smooth right-handed helix and all nucleotides are in the anti-conformation facing towards the inside of the helix. At the boundary between B-DNA and Z-DNA, a B-Z junction is formed in which a base pair is broken and the bases are extruded on each side (4).
Under physiological conditions, Z-DNA formation is enhanced by negative supercoiling, which is generated by moving RNA-polymerases during transcription (5). Z-DNA can either enhance or repress promoter activity, probably by modulating the local architecture of transcriptional control regions and nucleosome positioning (6–9).
Additionally, Z-DNA and Z-RNA can be bound by Z-DNA binding proteins (ZBPs). These proteins contain one or two copies of a domain, Zα, which can specifically stabilize and induce Z-DNA and Z-RNA. Co-crystallization of Zα domains has revealed that Z-DNA is recognized in a conformation-specific manner (10–12). Identified members of this family in vertebrates are Z-DNA binding protein 1 (ZBP1/DLM-1) (11), the RNA editing enzyme ADAR1 (13,14) and protein kinase containing Z-DNA binding domains (PKZ) (15,16). They contain two structurally related Zα domains, which are tandemly arranged at the amino-terminus of the proteins and are separated by a linker of variable size (15–91 amino acids) (15). Within the context of a protein, the N-terminal of these domains is referred to as Zα, while the C-terminal domain is referred to as Zβ. Vertebrate ZBPs are induced by immunostimulation and are thought to be involved in host defense mechanisms (15,17–19).
To overcome the host response, poxviruses possess a virulence factor, called E3L in vaccinia virus, which contains a single N-terminal Zα domain and a double-stranded RNA binding domain in the C-terminus (13). There is evidence to suggest that the postulated immune function of ZBPs is dependent on their ability to bind Z-DNA/Z-RNA. Single point mutations in the Zα-domain of E3L that prevented Z-DNA binding greatly reduced virus pathogenicity, which was fully restored by substitution of the Zα domain of E3L with that of ADAR1 or ZBP1, although these Zα domains share only ∼25% sequence identity (20). The Zα domain of E3L has been proposed to compete with cellular ZBPs for the binding of target sequences, resulting in inhibition of activation or loss of proper localization of ZBPs (21,22).
ZBP1 was identified as a gene that was up-regulated in mouse tumor stromal cells and it was speculated that it plays a role in the host response against tumors (18). Human ZBP1 is most prominently expressed in lymphatic tissues indicating a role in the modulation of immune functions (22). The ZBP1 gene contains 10 exons and encodes a protein with a predicted molecular mass of 46 kDa, containing two N-terminal Zα domains and a C-terminus of unknown function (22). As a result of alternative splicing and the use of alternative transcriptional start and stop sites, ZBP1 mRNA are very heterogeneous. Exon 2, coding for the first Zα domain, is spliced out in ∼50% of ZBP1 mRNA (22). A similar distribution was observed by analyzing expressed sequence tags (ESTs) of ZBP1 (S. Rothenburg, unpublished data).
In this study, we discovered an association between ZBP1 and stress granules (SGs) and processing bodies (PBs), which are both cytoplasmic aggregates of RNA and proteins. SGs and PBs are distinct granules that play important roles in the regulation of mRNA metabolism and degradation (23–25). SGs are formed in response to environmental stress and consist of stalled 48S translational preinitiation complexes, translation initiation factors and many RNA binding proteins such as 5′–3′ exonuclease 1 XRN1, cytotoxic granule-associated RNA binding protein TIA1 and RasGAP-associated endoribonuclease G3BP (25–27). Although SGs and PBs are distinct structures, they share several components for mRNA sorting and remodelling. Furthermore PBs but not SGs contain components of the decapping enzyme complex including DCP1a and the RNA-induced silencing complex (25). According to the model of Anderson and Kedersha, mRNA can be stored in SGs and can either be exported for translational reinitiation after recovery from cell stress or can be exported to PBs where mRNA can be degraded (25,28). The formation of SGs but not PBs is partly mediated by the phosphorylation of eIF2α following the activation of eIF2α kinases such as double-stranded RNA-activated protein kinase (PKR; activated by interferons and viral RNA) or GCN2 (by amino acid deprivation) (25,29). SG cannot form in the absence of eIF2α phosphorylation (25,26,30).
Here we have characterized the binding of ZBP1 Zα domains to Z-DNA in vitro and show that both, the Zα and Zβ domains, play roles in determining the subcellular localization of ZBP1. Furthermore we show that full length ZBP1 relocates from the cytoplasm to SGs but not to PBs if cells are subjected to environmental stress. In contrast, the isoform lacking Zα is found in granules distinct form SGs and PBs in unstressed cells. The ZBP1ΔZα granules disassemble during stress. The identification of full length ZBP1 as a previously unknown component of SGs and an observed dynamic interaction of ZBP1ΔZα granules with SGs and PBs link ZBP1 to RNA translation and sorting and indicate different roles for ZBP1 isoforms.
MATERIALS AND METHODS
Plasmids
Human leukocyte cDNA was used as the template for PCR using AmpliTaq Gold polymerase (PE Biosystems) with primers hDLM1f (5′-CCG ACT CCT TGC AGC TGC TGT C-3′) and hDLM6r (5′-ACT CCC TGT CAT CTA CTC CTG GCC-3′) as described (22) and cloned into pCR2.1 (Invitrogen). After sequencing clones containing the complete open reading frame (ZBP1 full), or having exon 2 spliced out (ZBP1ΔZα), were chosen as templates for subsequent PCR and cloning. PCRs were performed using vector primer M13R (5′-GGC AGG AAA CAG CTA TGA CC-3′) and hDLM Del2R XbaI (5′-CTG CTC TAG ACC CAC GTG AGG CTG TGC AC-3′). Restriction sites in all primers are underlined. PCR products were subsequently digested with Kpn I and Xba I (New England Biolabs), gel-purified and ligated into pcDNA3.1zeo (Invitrogen, Groningen, Netherlands). The complete open reading frames and cloning sites of each construct reported in this manuscript were sequenced using the dye terminator protocol (PE Applied Biosystems) according to the manufacturer's instructions. Sequence analysis was performed with Lasergene software (DNASTAR).
For native subcellular localization assays ZBP1 full, ZBP1ΔZα, ZBP1e1-5 and ZBP1ΔZαe1-5 were amplified by PCR with hZBP1 1F EcoRI (5′-AGG AAT TCG CCG CCA CCA TGG CCC AGG CTC CTG C-3′) and hZBP1 1R BamHI (5′-CGG GAT CCC CAC CTC CCC ACC AGC TCC-3′) for ZBP1 full and ZBP1ΔZα or hZBP1 2R BamHI (5′-CGG GAT CCT CCC TGG AGA CTG TCT GTC-3′) for ZBPlel-5 and ZBP1ΔZαe1-5. PCR products were cloned into pCR2.1 using TA cloning kit (Invitrogen). The correct plasmids were digested with BamHI and EcoRI, for plasmids containing ZBP1 full and ZBP1ΔZα under limited conditions for EcoRI because of an internal EcoRI restriction site in exon 8 of hZBP1. The fragments were purified and ligated into the multiple cloning site of pEGFP-N1 (Clontech) using T4 DNA Ligase (New England Biolabs). pZBP1full-GFP and pZBP1ΔZα-GFP were used as templates for the construction of pZBP1ΔZβ-GFP and pZBP1ΔZαΔZβ-GFP, respectively, where exon 4 was precisely deleted: the templates were digested with BbsI, which uniquely cuts within exon 4. Subsequently PCRs were performed with forward primer hZBP1ΔZβ MluF (5′-TCC AAC GCG TGG AAG ATT CTG GAA GAA GAG CAA AG-3′), which starts at the beginning of exon 5, and reverse primer hZBP1ΔZβ MluR (5′-CTC TTT GAC GCG TTG TTG GCT GAA CTG AGG GC-3′), which starts at the end of exon 3. PCR products were subsequently MluI (New England Biolabs) digested, gel-purified and ligated. TIA1-GFP and DCP1a-mRFP were kindly provided by Nancy Kedersha (Boston, USA). G3BP-GFP was kindly provided by Jamal Tazi (Montpellier, France). The reading frame of EGFP in pEGFP-N1 was replaced by monomeric red fluorescent protein (mRFP) that was amplified by mRFP1F AgeI (5′-TTG TAA CCG GTG GCC ACC ATG GCC TCC TCC GAG-3′) and mRFP1R NotI (5′-TTA ATC GCG GCC GCT ATT AGG CGC CGG TGG AGT GGC GGC-3′) with indicated enzymes, yielding pmRFP-N1. ZBP1 open reading frames from hZBP1 full-GFP and ZBP1ΔZα-GFP were excised with NheI and BamHI and ligated into pmRFP-N1 yielding hZBP1 full-mRFP and ZBP1ΔZα-mRFP. Site-specific mutations were generated by PCR mutagenesis using KOD Hot start polymerase (Novagen) and primers containing the desired mutation following the Quick Change protocol (Stratagene). Following PCR with 18 cycles the products were digested with DpnI (New England Biolabs) and transformed into ultracompetent XL-10 Escherichia coli (Stratagene).
For two proximate amino acid substitutions in ZαZBP1 two mutagenesis PCRs were performed consecutively. For single mutations in ZBP1Zα PCR with hZBP1 N46D F(5′-CAA GAG GGA GCT CGA CCA AGT CCT CTA CC-3′) and hZBP1 N46D R (5′-GGT AGA GGA CTT GGT CGA GCT CCC TCT TG-3′) or hZBP1 Y50A F (5′-GCT CAA CCA AGT CCT CGC CCG AAT GAA AAA GGA GTT-3′) and hZBP1 Y50A R (5′-CAA CTC CTT TTT CAT TCG GGC GAG GAC TTG GTT GAG C-3′) was performed, followed by a second mutagenesis PCR for double mutation with hZBP1 N46D Y50A F (5′-CAA GAG GGA GCT CGA CCA AGT CCT CGC CC-3′) hZBP1 N46D Y50A R (5′-GGG CGA GGA CTT GGT CGA GCT CCC TCT TG-3′). The substitutions in ZβZBP1 were generated by mutagenesis PCR with hZBP1 N141D Y145A F (5′-ACA GCA AAA GAT GTG GAC CGA GAC TTG GCC AGG ATG AAG AGC AGG-3′) and hZBP1 N141D Y145A R (5′-GGG CGA GGA CTT GGT CGA GCT CCC TCT TG-3′) in one step. For recombinant protein expression cDNA from hZBP1 was PCR amplified and inserted into pET28a (Novagen) with a cutting site for PreScission protease (GE Healthcare) instead of Thrombin (kindly provided by Thomas Schwartz, Cambridge, MA, USA). In detail ZαZBP1, ZβZBP1 and ZαβZBP1 were amplified using primers with NheI and BamHI restriction sites at their termini and cloned into the multiple cloning site of the modified pET28a. For ZαZBP1 the amplified sequence comprises exons 1 and 2: hZBP1 P5F NheI (5′-AAA GGA GCT AGC GCT CCT GCT GAC CCG GGC AG-3′) and hZBP1 P4R BamHI (5′-CGG GAT CCT TAT CAA GGG CTG GAC AAG GCC AGC TC-3′). The ZαβZBP1 construct (comprising exons 1–4) was cloned using hZBP1 P5F NheI as forward primer and hZBP1 P5R BamHI (5′-CGG GAT CCT TAT CAT GGG CGG TAA ATC GTC CAT GC-3′) as reverse primer. For the ZβZBP1 construct, comprising exons 1, 3 and 4, ZBP1ΔZα was used as template for PCR amplification with the same primers as were used for the ZαβZBP1 construct. ZαβADAR1 was constructed essentially as described (31).
Generation and purification of anti-ZBP1 antibodies by genetic immunization
A fragment of the ZBP1 cDNA (exons E1–E4 encoding ZαβZBP1) was amplified by PCR using primers harboring suitable restriction enzyme sites (5′-GAA AGA TCT GCT CCT GCT GAC CCG GGC AG-3′ and 5′-CTT TCC AAG CTT TTG GGC GGT AAA TCG TCC ATG C-3′). The PCR product was cloned into the pME18S mammalian expression vector so that the ZBP1 coding region was fused downstream of the coding region for the CD8 signal peptide followed by a FLAG-tag and upstream of the ART4 GPI-signal sequence (32). Transfection of the pME.CD8Fl_ ZBP1ZαZβ_GPI construct into HEK cells verified that the resulting ZαβZBP1 protein was expressed as a FLAG-tagged-GPI-anchored cell-surface protein. The pME.CD8Fl_ ZBP1ZαZβ_GPI construct was used for genetic immunization of rabbit CR5 essentially as described previously (33). Recombinant ZαβZBP1 was immobilized on Aminolink matrix (Pierce) according to the manufacturer's instructions. This matrix was used for affinity purification of CR5 immune serum, using 100 mM glycine, pH 2.7 for elution of bound antibody.
Immunofluorescence analysis
Cells were fixed for 10 min with 4% para-formaldehyde in phosphate-buffered saline (PBS), permeabilized and blocked (2% BSA, 3% goat serum, 0.5% Nonidet P-40 in PBS) and incubated with affinity-purified anti-hZBP1 antibody for 2 h. After washing three times with high-salt PBS (containing 650 mM NaCl to diminish unspecific binding) and three times with PBS, cells were incubated with secondary R-Phycoerythrin-(RPE) conjugated anti-rabbit F(ab')2 fragment (Dianova) for 2 h and subsequently washed three times with PBS. In order to check the specificity of immunoreactivity of the purified rabbit serum, 10 μg/ml of recombinant ZαβZBP1 was added to cells concomitantly with the antibodies. Nuclei were stained with Hoechst33342 in 1:4000 dilution, incubated for 20 min at 37°C and subsequently washed twice with PBS. Occasionally, small dots in the cytoplasm or outside cells were observed. These dots correspond to transfected plasmid DNA/transfection agent complexes and were never seen in non-transfected control cells. Fluorescence microscopy was performed using Axiovert 200M (Zeiss) with 20× (Plan-Apochromat 20×/0.75), 40× (EC Plan-Neofluar 40×/0.75 M27) or 63× oil objective (Plan-Neofluar 63×/1.25 oil). Photos were taken with AxioCam MR (Zeiss) and analyzed with Axiovision 4.4 SP2. For time-lapse experiments photos were taken every 30 s over a 15 min time period and animations were made with Axiovision 4.4 time-lapse module.
Cell culture and transfection
HeLa cells were kindly provided by Joe Glavy (New York City, USA), Hep2 cells were kindly provided by Matthias Kalitzky (Hamburg, Germany). Cells were grown in D-MEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 μM non-essential amino acids, 1 mM sodium pyruvate and 50 μg/ml Gentamicin at 37°C in 5% CO2 atmosphere. For immunofluorescence experiments tissue cells were seeded 24 h prior to transfection onto 8-well μ-slides or μ-dishes (Ibidi) with 180 μm thin bottoms for short distance microscopy. Cells were transfected with the indicated plasmids using polyethylenimine (jetPEI, Qbiogene) following manufacturer's instructions. Plasmids for double transfections were applied in a 1:1 ratio. In detail, 0.25 μg of each plasmid was used for each transfection in a 250 μl volume. 8–12 h after transfection cells were visualized by fluorescence microscopy, or fixed for subsequent staining. To inhibit nuclear export, 5 μg/ml Leptomycin B (LMB, Calbiochem) was added to HeLa cells 2 h after transfection and retained for 8 h. LMB-treated transfectants were compared to untreated transfectants.
Western blot analysis
HeLa cells were transfected as described above and lysed after 10 h with PBS containing 1% SDS. Total cell lysates were analyzed by SDS–PAGE followed by blotting onto PVDF membrane (Invitrogen). The membrane was blocked with 5% milk powder in TBS and probed with anti-GFP-Antibody (Roche).
Expression of recombinant protein
Proteins were overexpressed in E.coli BL21(DE3) (Novagen). Bacteria were grown at 37°C in Luria Broth Base with 30 μg/ml Kanamycin and 0.4% D-(+)Glucose for catabolite repression and induced between 0.8 and 1.1 A600 nm U with 0.3 mM isopropyl-β-d-thiogalactopyranoside for 4 h. All subsequent steps were performed at 4°C. Cells were spun down at 10 000 g for 10 min and resuspended in BugBuster (Novagen) supplemented with 25 U/ml Benzonase-Nuclease (Novagen), 100 μM 4-(2-aminoethyl)benzenesulphonyl fluoride (MP Biomedicals) and 5 mM β-mercaptoethanol (β-ME). The suspension was rocked for 30 min and centrifuged at 13 000 g for 10 min. The supernatant was incubated with Ni-NTA His-bind resin (Novagen) and washed three times with 12 ml Buffer A [20 mM Tris–HCl (pH 8), 250 mM NaCl, 5 mM β-ME, 7 mM imidazole] and once with 10 ml Buffer B [20 mM Tris–HCl (pH 8), 2 M NaCl, 5 mM β-ME] in a batch. The resin was then transferred into a column and washed with 10 ml Buffer C [20 mM Tris–HCl (pH 8), 250 mM NaCl, 5 mM β-ME, 15 mM imidazole] and finally the His6-tagged protein was eluted with 8 ml Buffer D [20 mM Tris–HCl (pH 8), 250 mM NaCl, 5 mM β-ME, 250 mM imidazole]. Fractions were analyzed by SDS–PAGE followed by Coomassie staining. Fractions with recombinant protein were dialyzed against Buffer E [20 mM HEPES (7.4), 40 mM NaCl, 1 mM DTT] and 1 U PreScission protease (GE Healthcare) per microgram of estimated recombinant protein was added for >12 h in order to remove the N-terminal His6-tag. Recombinant proteins were further purified by cation-exchange chromatography on a HiTrap SP FF column (GE Healthcare). The yield was determined by UV spectrometry at 280 nm. About 10–15 mg purified recombinant protein was obtained from 1 liter of culture.
Circular dichroism measurements
Circular dichroism (CD) spectra were recorded at 24°C on a Jasco J-810 CD spectrometer (Jasco) using a cuvette with optical pathlength of 1 mm. Spectra were recorded in 1 nm-steps as the mean of three measurements with scanning speed of 200 nm/min and were further analyzed with Sigma Plot 9.0 software. The conformational change of the DNA was measured between 220 and 320 nm, and protein spectra were measured between 190 and 320 nm. The measurement was carried out in 10 mM HEPES (7.4), 20 mM KF and 0.1 mM EDTA. When determining the effect of protein on DNA conformation, although the CD signal below 240 nm is dominated by the negative ellipticity of the proteins, no baseline correction was applied because the proteins did not show significant CD signal >245 nm (data not shown). DNA was obtained as ds(poly(dC–dG)∼500) from GE Healthcare and concentration was determined by UV spectrometry at 260 nm. Protein was added to the DNA and incubated for 30 min at 24°C prior to recording.
Electrophoretic mobility shift assay
DNA binding was assayed by native TBE PAGE. (dC–dG)20 oligomers were annealed, FPLC-purified and 5′ end-labeled using polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. A reaction mixture containing 10 mM Tris–HCl (pH 7.8), 20 mM NaCl, 5 mM DTT, 5% glycerol, 100 μg/ml BSA, 600 ng/μl sheared salmon sperm DNA, indicated concentration of recombinant protein and 0.1 pM DNA-probe was incubated for 30 min at 24°C. The mixture was separated on a 6% TBE–Polyacrylamide Gel using 0.5× TBE as running buffer at 10 V/cm. TBE Gels were blotted onto positively charged nylon membranes (Hybond N+, GE Healthcare) and exposed at −80°C to BioMax films (Kodak) with intensifying screen.
RESULTS
The presence of two Zα domains enhances formation of Z-DNA in vitro
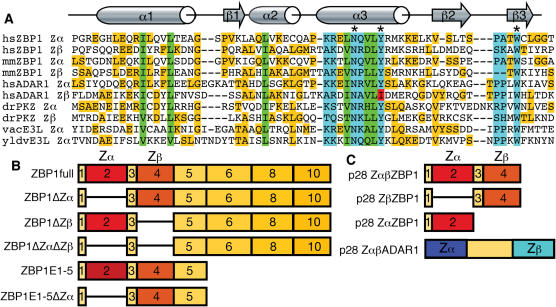
In mammalian ADAR1, the two Zα-like domains vary in DNA binding activity in vitro; ZαADAR1 binds Z-DNA in CD spectroscopy and electrophoretic mobility shift assays (EMSA), while isolated ZβADAR1 has never been shown to bind DNA, although the crystal structure of ZβADAR1 shows that it is folded in a similar way to ZαADAR1 (34). However, together with Zα, Zβ can enhance binding to Z-DNA (31). One reason for the inability to bind the Z-form on its own is the presence of isoleucine at position 335 instead of tyrosine (Figure 1A). The latter is conserved in most Zα domains and is important for hydrogen bonding with Z-DNA as shown in the co-crystal structures of human ZαADAR1, mouse ZαZBP1 and yaba-like virus ZαE3L (10–12). In addition to the conserved tyrosine two further residues, an asparagine and a tryptophan (marked by asterisks in Figure 1A), both important for DNA contact, and critical residues that make up the hydrophobic core are highly conserved in all identified Zα proteins (15) (Figure 1A). The Zβ domain of mammalian ZBP1 lacks the conserved proline adjacent to the β3 sheet (Figure 1A). This residue is important for the shape of the wing and makes van der Waal's contact with the DNA. Therefore it was not clear if ZβZBP1 without an accompanying ZαZBP1 domain, as found in a prominent ZBP1 splice variant, can actually bind the Z-conformation independently and whether the presence of two tandemly arranged Zα domains modulates Z-binding.
Figure 1.
Comparison of Zα domains (A) and schematic representation of constructs used for transfections (B) and protein expression (C). Comparison of Zα domains of human (hs) and mouse (mm) ZBP1, human ADAR1, zebrafish (dr) PKZ, vaccinia virus (vv) E3L and yaba-like disease virus (yldv) E3L is shown (A). The structures of the mouse (mm) ZαZBP1, human (hs) ZαADAR1, yaba-like disease virus (yldv) ZαE3L domains have been determined in complex with Z-DNA. Residues that make contact with Z-DNA, or the analogous residues in other Zα domains, are boxed in light blue. Asterisks mark the conserved asparagine and tyrosine residues that have been mutated in this study in hsZBP1, as well as a conserved tryptophan. Residues that form the hydrophobic core are boxed in green. Residues that are neither DNA contacting nor structural but match the consensus sequence are highlighted in yellow. Isoleucine 335 in ZβADAR1 is highlighted in red. (B) The exon composition of the most prominent ZBP1 splice variants ZBP1full and ZBP1ΔZα as well as that of artificial constructs are shown. Exon 7 is rarely found in mRNA. Exon 9 contains an alternative termination site (22). ZBP1full and ZBP1ΔZα have been expressed as un-tagged or GFP tagged proteins in HeLa cells. ZBP1ΔZβ, ZBP1ΔZαΔZβ, ZBP1E1-5 and ZBP1E1-5ΔZα were expressed as GFP-tagged proteins. Schematic representation of the exon composition of constructs expressed from pET28a (p28) vectors in E.coli are shown in (C).
In order to address the DNA binding characteristics of the two Z-DNA binding domains of human ZBP1, they were recombinantly expressed as His6-tagged proteins in E.coli, either separately (ZαZBP1 or ZβZBP1) or in combination (ZαβZBP1). Human ZαβADAR1, which has been previously shown to bind Z-DNA, was used as control (31). After Ni-NTA purification and subsequent cleavage of the His6-tag by digestion with PreScission protease, proteins were further purified by FPLC (Figure 2). We used CD spectroscopy to monitor the conformational change from B- to Z-DNA as induced by the recombinant proteins. The B-DNA form of poly(dC–dG) can be distinguished from the Z-DNA form by CD due to an inversion of the spectrum between 240 and 300 nm. Figure 3 shows the spectra of poly(dC–dG) either in the absence or presence of different Z-DNA binding domains.
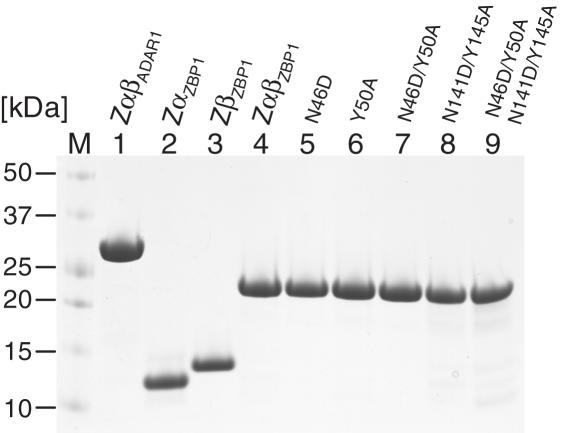
Figure 2.
SDS–PAGE analysis of recombinant proteins used for CD-spectrometry and EMSA. 500 nmol of each purified protein was subjected to SDS–PAGE and visualized by Coomassie Brilliant blue staining: Lane M, molecular mass marker; lane 1: ZαβADAR1; lane 2: ZαZBP1; lane 3: ZβZBP1; lane 4: ZαβZBP1; lane 5: ZαβZBP1N46D; lane 6: ZαβZBP1Y50A; lane 7: ZαβZBP1N46D/Y50A; lane 8: ZαβZBP1N141D/Y145A; lane 9: ZαβZBP1 N46D/Y50A/N141D/Y145A.
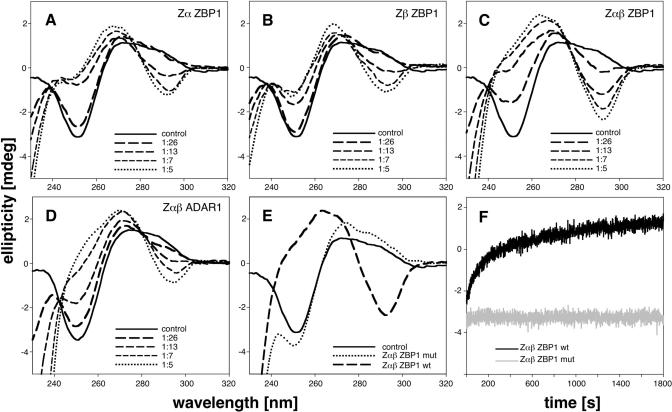
Figure 3.
Conformational change of poly(dC–dG) in the presence of Zα proteins in CD-spectrometry. The CD spectra show the titrations of ZαZBP1 (A), ZβZBP1 (B), ZαβZBP1 (C) and ZαβADAR1 (D). The curves show DNA spectra in the absence (control, B-DNA) and in the presence of protein at protein/basepair molar ratio as labeled. Spectra were measured after 30 min of incubation at 24°C. (E) The CD spectra of wildtype (wt) ZαβZBP1 and quadruple mutant (ZαβZBP1mut) carrying the four amino acid substitutions N46D, Y50A, N141D and Y145A are shown in relation to the control, where no protein was added. (F) Time dependent change of ellipticity at 255 nm wavelength over 30 min ZαβZBP1 and ZαβZBP1mut are shown. Spectra are expressed in absolute values of ellipticity in millidegrees (mdeg).
At low salt concentration and in the absence of protein the DNA adopted the B-conformation, resulting in a CD graph characterized by negative ellipticity at 255 nm and positive ellipticity at 290 nm (Figure 3A–E). Addition of increasing amounts of ZBP domains resulted in inverted CD spectra, similar to DNA at high salt concentration, characterized by positive ellipticity at 255 nm and negative ellipticity at 290 nm. Both Zα domains of human ZBP1 induced a conformational change of double-stranded DNA (Figure 3A and B). If both domains were tethered to one another (ZαβZBP1) Z-DNA was formed more efficiently (Figure 3C) as compared to the single domains. The single Zα domains reached saturation earlier than ZαβZBP1 (compare Figure 3A and B with Figure 3C, respectively). ZαβADAR1 showed a similar spectrum as previously described (Figure 3D) (31). When the spectra are compared at the same molar ratio, ZαβZBP1 showed the highest propensity to induce Z-DNA formation (Figure 3A–D). Time dependent change of ellipticity at 255 nm wavelength in the CD spectra is shown in Figure 3F. Addition of wildtype ZαβZBP1 resulted in a biphasic increase in ellipticity, showing a fast increment for 5 min followed by a slower but progressive increase. Mutation of four amino acids (N46D, Y50A, N141D and Y145A), which are important for Z-DNA binding as deduced from the crystal structures, yielded soluble protein, comparably folded to the wildtype as observed in CD spectra between 190 and 240 nm (data not shown), but with no ability to induce conformational change of DNA (Figure 3E and F). When the conserved asparagines and tyrosines were substituted in only one of the Zα domains, while the other stayed intact, comparably folded, soluble proteins were produced as observed in CD spectra. However, addition of DNA to the mutated proteins resulted in immediate precipitation.
EMSA were previously used to analyze binding of Zα domains of ADAR1 to Z-DNA (31). The binding of Zα proteins to the probe was tested in the presence of a 20 000-fold excess of salmon sperm DNA as competitor (Figure 4A and B). Double-stranded (dC–dG)20 was used as a probe because Zα proteins have been shown to convert it from the B- to the Z-conformation. The proteins were tested at different concentrations and adjusted to the number of Zα domains, i.e. Zαβ was used at half the molar concentration as Zα and Zβ. ZαβZBP1 (Figure 4A, lanes 2–4) shifted the probe at lower concentrations than Zα (Figure 4A, lanes 5–7) and Zβ alone (Figure 4A, lanes 8–10). When Zα and Zβ were mixed together a comparable bandshift was observed as seen when only Zα or Zβ were used (Figure 4A, lanes 11 and 12). A radiolabeled probe with arbitrary sequence of comparable length, without the propensity to adopt Z-conformation was used as control, did not form complexes with Zα proteins (data not shown).
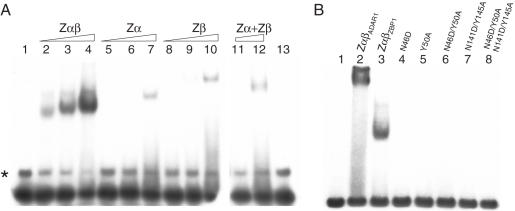
Figure 4.
Binding of Z-DNA by ZBP1 Zα proteins in electrophoretic mobility shift assays. EMSA were performed using 32P-labeled double-stranded (dC–dG)20 as probe in the presence of 20 000-fold access of sheared salmon sperm DNA as unspecific competitor. (A) Molar concentration was adjusted for the number of Zα domains the protein comprises. Zα and Zβ was assayed in twice as high molar concentration as Zαβ. Migration of the free probe is shown in lane 1 and 13. Zαβ (lane 2: 12.5 nM; lane 3: 25 nM; lane 4: 50 nM), Zα (lane 5: 25 nM; lane 6: 50 nM; lane 7: 100 nM), and Zβ (lane 5: 25 nM; lane 6: 50 nM; lane 7: 100 nM) were assayed at indicated concentrations. Reaction mixes shown in lanes 11–13 were prepared at the same time and were run on a separate TBE Gel. Zα and Zβ were mixed together at two concentrations (lane 11: 25 nM of each protein and lane 12: 50 nM of each protein) to see if there is a cooperative effect if both domains are present in different molecules. Sometimes a minor, slower migrating band was observed (asterisk) for the probe, probably due to a different secondary structure of the probe. (B) ZαβADAR1 (lane 2) and ZαβZBP1 (lane 3) form complexes with the radiolabeled probe whereas the indicated ZαβZBP1 mutants at equimolar concentration do not (lanes 4–8). Migration of the free probe is shown in lane 1.
Amino acid residues that are conserved in ZBPs and have been shown to be important for Z-DNA binding were mutated in Zα or Zβ or in both domains of ZαβZBP1. Mutants N46D, Y50A, N46D/Y50A, N141D/Y145A and N46D/Y50A/N141D/Y145A were analyzed for Z-DNA binding at equimolar concentrations. All mutations drastically reduced binding to Z-DNA as compared to the wildtype (Figure 4B). Even single amino acid substitutions abolished shifting of the probe. ZαβADAR1 was used as positive control (Figure 4B, lane 2).
Gene products of ZBP1 splice variants that either contain or lack ZαZBP1 show strikingly different subcellular localizations
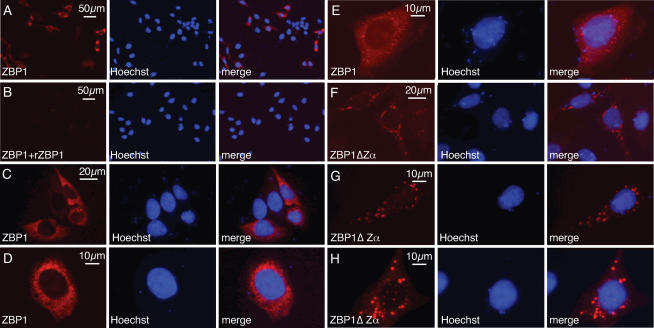
In order to analyze the subcellular localization of ZBP1, human HeLa cells, which do not express endogenous ZBP1, were transiently transfected for 8–12 h with full length human ZBP1 and stained with anti-ZBP1 antibodies generated by genetic immunization. Nuclei were stained with Hoechst33342 (blue, middle panel). Approximately 30% of cells showed strong, mainly cytoplasmic staining for ZBP1 (Figure 5A). Untransfected cells showed no staining. Staining of ZBP1 transfected cells could be completely blocked by the addition of recombinant ZαβZBP1 demonstrating specificity of the antibodies (Figure 5B).
Figure 5.
Different subcellular localization of two major ZBP1 splice variants. HeLa cells transiently transfected with ZBP1 were fixed and stained with polyclonal anti-ZBP1 antibodies and with secondary RPE-conjugated anti-rabbit-Ig antibodies. The left pictures show red fluorescence (RPE) after identical exposure time. The middle panels show nuclear stain using Hoechst33342. Right pictures show merges. In (A) no competitor was present, while in (B) 10 μg/ml of recombinant ZαβZBP1 was added. Anti-ZBP1 antibodies were used to stain for ZBP1 (left panels) in HeLa cells transfected with either full length ZBP1 (C–E) or the splice variant ZBP1ΔZα (F–H). Cells representative of three independent experiments are shown.
The most prominent alternative splice variant of ZBP1 (ΔZα) encodes a protein that differs from full length ZBP1 solely by the absence of ZαZBP1. In order to determine whether the presence of ZαZBP1 influences the subcellular localization of ZBP1, we compared the staining of HeLa cells transfected with constructs containing or lacking Zα. The staining pattern of ZBP1 transfected HeLa cells is shown in Figure 5C–H. The majority (∼80%) of cells transfected with full length ZBP1 showed a finely punctate cytoplasmic staining pattern as shown in Figure 5C and D. The remainder of transfected cells showed small cytoplasmic granules and weak nuclear staining in addition to the punctate staining (Figure 5E). In contrast, cells transfected with ZBP1ΔZα showed a strikingly different staining pattern. In these cells large granules were observed (Figure 5F–H). In the majority of cells (∼70%) the nucleus was not or only faintly stained as exemplified in Figure 5F and G. In other cells few granules in the nucleus and an additional diffuse granular cytoplasmic staining were observed (Figure 5H).
Mutation of the Z-DNA interacting residues N45 and Y50 in ZαZBP1 dramatically affects the subcellular localization of ZBP1
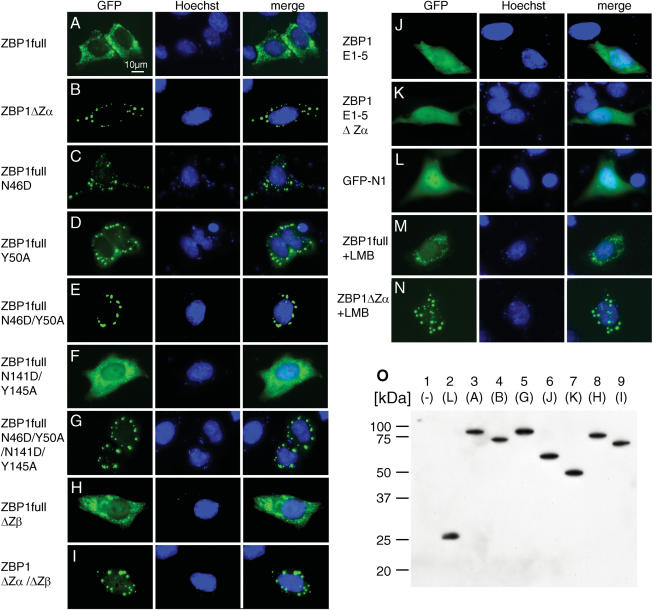
To further analyze the subcellular localization of ZBP1 in living unfixed cells, we cloned the two major splice variants at the N-terminus of GFP in order to generate ZBP1-GFP fusion proteins. The full length ZBP1-GFP fusion protein showed a finely punctate cytoplasmic staining pattern (Figure 6A), while ZBP1ΔZα-GFP was found in large, mostly cytoplasmic granules (Figure 6B). This confirms the results obtained for untagged ZBP1 isoforms detected with antibodies in fixed cells. Additionally, mutations that substitute amino acids important for Z-DNA binding or delete larger parts of the protein were tested in this assay. Asparagine 46 and tyrosine 50 are important for binding of Z-DNA as observed in the co-crystal structure of mouse ZBP1 in complex with Z-DNA (11). Mutation of either of these residues led to a dramatically changed subcellular localization, from a finely punctate distribution as seen in wild type ZBP1 to large granules. In this respect the N46D (Figure 6C) substitution had a stronger effect than the Y50A (Figure 6D) substitution. Mutation of both residues resulted in the formation of large cytoplasmic granules (Figure 6E) that were indistinguishable to those formed by ZBP1ΔZα (Figure 6B). Mutation of the corresponding residues in Zβ (N141D/Y145A) had no such effect. This mutant displayed a finely punctate cytoplasmic fluorescence pattern similar to that observed for wild type ZBP1 and additionally showed nuclear accumulation (Figure 6F). Substitution of all four residues, in both Zα and Zβ, resulted in a comparable pattern to that observed with ZBP1ΔZα (Figure 6G). Precise deletion of exon 4, which encodes the complete Zβ domain, resulted in a comparable fluorescence pattern to that observed for the N141D/Y145A mutant. This deletion led to a finely punctate cytoplasmic fluorescence pattern with some nuclear accumulation (Figure 6H). When exon 4 was deleted from the ZBP1ΔZα splice variant, large cytoplasmic granules were observed (Figure 6I). Deletion of the C-terminal part from wild type ZBP1 (Figure 6J) and ZBP1ΔZα (Figure 6K) resulted in a homogenous distribution within the cell, which was comparable to that of GFP alone (Figure 6L). No granularity or punctate staining was observed. Western blot analysis of whole cell lysates of transfected cells showed no significant level of degradation with an antibody against GFP (Figure 6O).
Figure 6.
Correlation of subcellular localization of ZBP1 with the capability of the first Zα domain to bind the Z-conformation. HeLa cells were transiently transfected with the indicated ZBP1-EGFP expression plasmids for 8–12 h. Subcellular localization was analyzed by fluorescence microscopy (left panels). Nuclei were stained with Hoechst33342 (middle panel). In (A) and (B) localization of the naturally occurring full length ZBP1 and ZBP1ΔZα, respectively, tagged with GFP is shown. Localization is shown for GFP-tagged full length ZBP1 with the indicated single (C and D), double (E and F) or quadruple (G) amino acid substitutions at residues important for Z-DNA binding. (H and I) show the localization of full length ZBP1 and ZBP1ΔZα proteins, respectively, after the precise deletion of exon 4, which encodes the entire second Zα domain. The localization of fusion proteins after deletion of the complete C-terminus (exons 6–10) is shown for ZBP1 (J) and ZBP1ΔZα (K). (L) shows the distribution of GFP alone within the cell. Two hours after transfection with full length ZBP1-GFP (M) and ZBP1ΔZα-GFP (N) constructs, cells were treated with Leptomycin B, an inhibitor of nuclear export, for 8 h prior to microscopy. Cells representative of at least three independent experiments are shown. (O) Western blot analysis shows that ZBP1-GFP fusion proteins were intact and not significantly degraded 10 h after transfection. Total cell lysates of transfected cells were subjected to SDS–PAGE and recombinant proteins were detected with an antibody directed against GFP. In lane 1 lysate of mock-transfected cells is shown as control. Lane 2 shows lysate of GFP transfected cells. The letters written in parenthesis for lanes 3–9 refer to the different fusion proteins that are shown in this figure as fluorescence pictures (A–K).
Because weak nuclear staining was observed in some cells transfected with wild type ZBP1, we tested whether nuclear export might play a role in directing the subcellular location of ZBP1. To this end we treated HeLa cells 2 h after transfection with LMB, an inhibitor of nuclear export. An accumulation of fluorescence in the nucleus was observed for full length ZBP1 (Figure 6M), which was comparable to that observed with ZBP1ΔZβ (Figure 6H). ZBP1ΔZα-GFP transfectants treated with LMB showed cytoplasmatic granules (Figure 6N) comparable to untreated transfectants (Figure 6B). Additionally, weak nuclear accumulation was observed (Figure 6N).
Association of ZBP1-containing cytosolic granules with stress granules and processing bodies
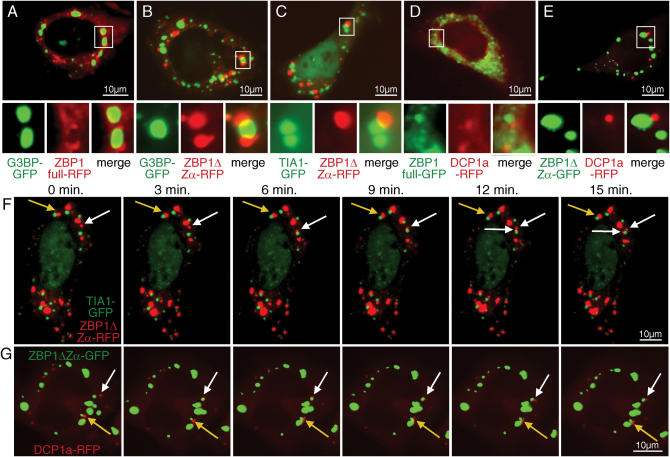
The observed granules, especially those formed by ZBP1ΔZβ, are reminiscent of SGs. SGs are cytoplasmatic foci that are assembled during environmental stress and are thought to modify mRNA turnover and mRNA accessibility to the translational machinery (25,35). To determine whether ZBP1 might be a component of SGs, we generated fusion proteins of full length ZBP1 and ZBP1ΔZα with mRFP and performed co-transfection with GFP-tagged G3BP and TIA1, two markers for SGs whose overexpression has been described to induce the formation of SGs (25,27). Fluorescence pattern of RFP-tagged ZBP1 was comparable to that of untagged and GFP-tagged proteins, showing a finely punctate pattern for full length ZBP1-RFP (Figure 7A) and large granules for ZBP1ΔZα-RFP (Figure 7B and C). The ZBP1ΔZα granules did not co-localize with granules marked by G3BP-GFP and TIA1-GFP (Figure 7B and C). However some of the ZBP1ΔZα granules appear to be closely associated with G3BP-GFP and TIA1-GFP marked granules and even display overlapping at contact areas. Granules marked by G3BP-GFP are surrounded by full length ZBP1-RFP, which appears to be concentrated around G3BP-marked granules (Figure 5A).
Figure 7.
Association and dynamics of interaction of ZBP1 granules with SGs and PBs. HeLa cells were co-transfected with full length ZBP1-RFP (A), ZBP1ΔZα-RFP (B and C), full length ZBP1-GFP (D) and ZBP1ΔZα-GFP (E) plus SG markers G3BP-GFP (A and B), TIA1-GFP (C) or PB marker DCP1a-RFP (D and E). For each photo the indicated sections are shown in better resolution below for green, red and merged fluorescence. The dynamics of interaction between ZBP1 granules with SGs and PBs are shown for co-transfections with ZBP1ΔZα-RFP and TIA1-GFP (F and Video 1), ZBP1ΔZα-RFP and G3BP-GFP (Video 2) or ZBP1ΔZα-GFP and DCP1a-RFP (G and Videos 3 and 4). Cells were observed over a period of 15 min with pictures taken every 30 s. Pictures taken at 3 min intervals are shown in F and G. White arrows indicate ZBP1 granules that were unbound at the beginning of the observation period and later attached to SGs (F) or PBs (G). Yellow arrows indicate ZBP1 granules that stayed attached to SGs (F) or PBs (G) over the complete observation period. Cells representative of at least three independent experiments are shown.
Another form of granules important for RNA turnover are PBs, which can interact with SGs (35). Co-transfection of RFP-tagged PB marker DCP1a with GFP-tagged ZBP1, revealed an association with some DCP1a granules with full length ZBP1 (Figure 7D) or with the large granules formed by ZBP1ΔZα (Figure 7E).
We investigated the dynamics of the interactions between ZBP1ΔZα granules and SGs or PBs over time taking photos of individual cells every 30 s over a 15 min time period. Some SGs as marked by TIA1-GFP (Figure 7F and Video 1) and G3BP-GFP (Video 2) were tethered to ZBP1ΔZα granules over the complete observation interval (e.g. Figure 7F, yellow arrow), while other interactions between these two granules appeared to be more intermittent or temporary. The white arrow in Figure 7F marks a single ZBP1ΔZα granule that was unbound at the beginning of the observation period (Figure 7F, 0–3 min), later attached to a TIA1-marked SG (Figure 7F, 6–9 min) and subsequently split into two granules (Figure 7F, 12–15 min). In addition to fission ZBP1ΔZα granules also displayed fusion of two or more granules into a single larger granule (as observed in Figure 7G middle right). Similarly to SGs, some DCP1a-marked PBs stayed attached to ZBP1 granules over the observed interval (Figure 7G, yellow arrow), while others only temporarily interacted (Videos 3 and 4). The white arrow in Figure 7G marks a PB that was unbound at the beginning of the observation period, attached to a ZBP1ΔZα granule after 2 min and stayed tethered to it until the end of the observation interval. Generally, PBs appeared to be more motile than SGs or ZBP1ΔZα granules, which showed a comparable degree of motility.
Stress-induced accumulation of full length ZBP1 into stress granules and disassembly of ZBP1ΔZα granules
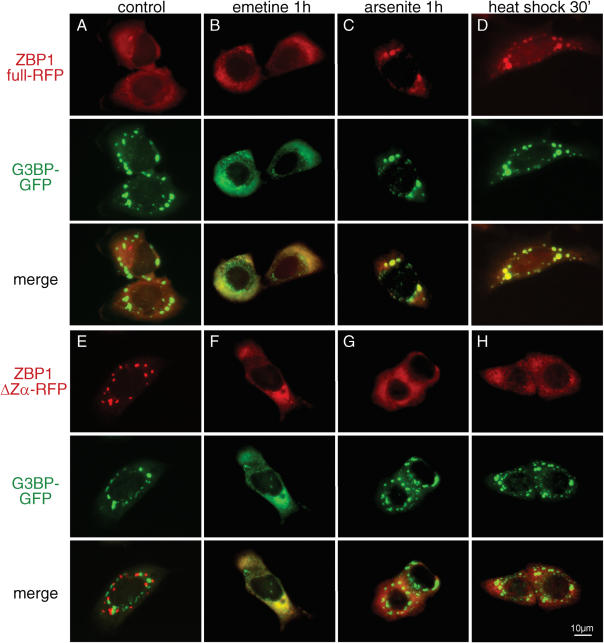
We investigated whether the localization of the two major ZBP1 isoforms can be influenced by stimuli that modify the assembly of SGs. To this end we co-transfected HeLa cells with G3BP-GFP and either ZBP1-RFP or ZBP1ΔZα-RFP and treated cells with arsenite for 1 h or subjected them to heat shock for 30 min, two stimuli that promote SG assembly, or treated cells with emetine, a drug that blocks the disassembly of polysomes and leads to the disintegration of SGs (25,36). Treatment of cells with emetine resulted in the disassembly of SGs marked by G3BP (Figure 8B and F) as well as a relocation of ZBP1ΔZα from granules to the cytoplasm (Figure 8F). It had no effect on the localization of the full length ZBP1. Treatment with arsenite (Figure 8C) or exposure to heat shock (Figue 8D) lead to the relocation of full length ZBP1 into granules which co-localized with the SG marker G3BP. Heat shock was more effective in relocating ZBP1 into the SGs than arsenite treatment. Exposure to arsenite (Figure 8G) or heat shock (Figure 8H) had an opposing effect on ZBP1ΔZα, leading to the disassembly of granules, while G3BP granules were not effected. Comparable results were obtained when cells were co-transfected with TIA1-GFP and when only ZBP1 was transfected (data not shown). The granules formed by full length ZBP1-GFP after heat shock and aresenite treatment did not co-localize with Dcp1a-RFP (data not shown).
Figure 8.
Relocation of full length ZBP1 and ZBP1ΔZα upon arsenite treatment and heat shock. HeLa cells were co-transfected with G3BP-GFP and full ZBP1-RFP (A–D) or ZBP1ΔZα (E–H). After 7 h cells were incubated with 100 μg/ml emetine (B and F) or 500 μM arsenite for 1 h or exposed to heat shock at 44°C for 30 min (D and H). Upper panels show RFP fluorescence, middle panels GFP fluorescence and lower panels the merged views. Cells representative of at least three independent experiments are shown. Supplementary Data.
Online supplemental material
Video 1 shows the dynamic interaction of ZBP1ΔZα granules (red) with TIA1 granules (green) in transiently transfected HeLa cells. Video 2 shows the interaction of ZBP1ΔZα granules (red) with G3BP granules (green). Videos 3 and 4 show the dynamic interaction of ZBP1ΔZα-GFP granules (green) and DCP1a-RFP granules (red). Photos were taken every 30 s over a 15 min time period and time lapse animations were made with 4 pictures per s.
DISCUSSION
The results presented in this paper provide insights into the function of the two Z-DNA binding domains and different isoforms of human ZBP1 and demonstrate association and interaction of ZBP1 isoforms with SGs.
All proteins that can specifically bind to left-handed Z-DNA and Z-RNA known to date contain Zα domains (15). Although the structure of all crystallized Zα domains is remarkably similar, not all of them can actually bind the Z-form by their own (10–12,34). Using CD spectroscopy we have shown that ZαZBP1 (encoded by exon 2) and ZβZBP1 (encoded by exon 4) can bind to left-handed Z-DNA independently of each other, resulting in comparable CD spectra of DNA. This confirms a recent report where both domains, when tested independently, converted DNA from the B- to the Z-form (37). In addition to this finding this report shows that when both Zα domains are tethered to one another, as in the full length protein, the capacity to convert DNA from the B-form to the Z-form is greatly enhanced. Consistently, the results of EMSA experiments show that formation of DNA–protein complexes is greatly augmented when Zαβ is present in one protein molecule as compared to the situation when Zα or Zβ were tested separately or when Zα or Zβ are mixed together.
The complexes of Zα proteins and DNA exhibited different levels of mobility. The larger protein Zαβ (19 kDa) forms complexes which migrate faster and thus appear to be smaller than the complexes formed by the single Zα (9.5 kDa) or Zβ (10.9 kDa) domains. This might be caused by the binding of more protein molecules to the probe if the protein is smaller, leading to the formation of larger complexes.
The importance of two intact Zα domains in one molecule for Z-DNA binding is further supported by the results of our mutagenesis experiments. Mutation of conserved amino acids that contact Z-DNA as seen in the crystal structures (N46, Y50, N141, Y145) resulted in a dramatically reduced capacity to bind Z-DNA. Interestingly, Z-DNA binding of Zαβ was strongly affected even when these amino acids were substituted in only one of the Zα domains.
The results of our in vitro Z-DNA binding experiments led us to hypothesize that the two major isoforms of ZBP1 might show different subcellular distribution. Indeed, following transfection into HeLa cells, full length ZBP1 and ZBP1ΔZα displayed strikingly different subcellular localizations. While both isoforms displayed a predominant cytoplasmic localization, full length ZBP1 showed a diffuse finely punctate distribution, whereas ZBP1ΔZα localized in large granules. Importantly, the finely punctate distribution of ZBP1 correlates with its ability to bind the Z-conformation, since single or double mutations of the Z-DNA contacting residues in ZαZBP1 (N46D and Y50A) resulted in a distribution similar to that of ZBP1ΔZα.
No such effect was observed with analogous mutations of ZβZBP1, even if the complete domain was deleted. It appears that ZαZBP1 exerts a dominant effect over ZβZBP1 with respect to the localization. This result could not be expected from the in vitro experiments, in which both Zα and Zβ appeared to bind Z-DNA with comparable efficiency. However, deletion of Zβ or mutation of its presumptive Z-DNA contacting residues N141 and Y145 resulted in accumulation of protein in the nucleus, indicating that a functional ZβZBP1 is involved in nuclear export of ZBP1. Importance of nuclear export is further supported by nuclear accumulation of full length ZBP1 after treatment with LMB, an inhibitor of nuclear export.
C-terminal ZBP1 deletion mutants (lacking exons 6–10), irrespective of the presence or absence of ZαZBP1, showed a homogenous distribution within the cell, including the nucleus, similar to that of GFP. The C-terminal truncated ZBP1E1-5 and ZBP1E1-5ΔZα fused to GFP have a predicted molecular mass of 52 and 44 kDa, respectively. They appear to be too large to pass the nuclear pore by diffusion, where the cut-off is believed to be between 20 and 40 kDa (38). This indicates that there might be another nuclear export signal apart from ZβZBP1 in the C-terminus of ZBP1. Western blot analysis showed that all ZBP1-EGFP fusion proteins were intact, demonstrating that the different observed fluorescence patterns were not a consequence of protein degradation.
The resemblance of the ZBP1-harboring granules with SGs and PBs led us to investigate whether ZBP1 might be actually a component of these. SGs and PBs are RNA granules which play important roles in mRNA turnover. Translationally silenced mRNA can be packed into these granules and either be degraded or stored and later exported. It is believed that SGs function as triage centers that sort, remodel and export mRNA either for subsequent decay (e.g. export to PBs) or for reinitiation of translation (25,28,35).
It has been shown that SG formation can be promoted by the overexpression of SG proteins G3BP and TIA1 (27,39). Co-transfection experiments showed that G3BP- and TIA1-marked granules are distinct from the ZBP1ΔZα marked granules. However, a close spatial association with overlapping contact areas between SGs and ZBP1ΔZα granules was often observed. A similar association was seen with ZBP1ΔZα granules and some PBs marked by the decapping enzyme DCP1a. Time lapse microscopy showed that these interactions are dynamic. Some ZBP1ΔZα granules were tethered to SGs or PBs over the complete observation period, while some only associated for a limited time. Upon contact with one another, SGs and ZBP1ΔZα granules often appeared to deform and reshape. They share some characteristic features, which include interaction with PBs, fusion and fission (25).
The granules induced by the overexpression of SG protein G3BP are thought to be genuine SGs because they contain typical SG proteins such as PABP1, eIF4G, TIA1 and TIA1/R (27,39). The finding that full length ZBP1 can actually localize into G3BP- and TIA1-marked SGs only after cells have been subjected to environmental stress, i.e. heat shock and arsenite exposure, indicates that G3BP- and TIA1-containing granules in unstressed transfected cells contain ‘stress-independent’ components that are assembled following the overexpression of these proteins. Other proteins such as ZBP1 enter these ‘pre-formed’ granules only after cells have been subjected to environmental stress (stress-dependent).
Inhibitors of translational elongation, such as emetine and cyclophosphamide, impede the disassembly of polysomes and dissolve SGs and PBs. Since mRNA shuttle between SGs and polysomes, this leads to a shift in the equilibrium of mRNAs towards the polysome fraction, resulting in the disintegration of SGs, because mRNA are integral components of SGs and PBs (24,36,40). Interestingly, treatment of cells with emetine led to the disassembly of both SGs and ZBP1ΔZα granules, indicating that mRNA may also be an integral component of the latter. It will be interesting to determine other components of ZBP1ΔZα granules and analyze whether proteins and RNA, which are found in SGs and PBs, are also present in ZBP1ΔZα granules. Remarkably, exposure of cells to arsenite and heat shock dissolved the ZBP1ΔZα granules. The dynamic nature of ZBP1ΔZα granules indicates that these granules do not represent mere aggregates of misfolded proteins. A similar phenomenon has been observed for the AU-rich element binding protein tristetraprolin (TTP). TTP was recruited to SGs after FCCP-induced energy starvation but excluded form SGs in arsenite treated cells. Overexpression of TTP resulted in the formation of SGs, which disassembled after arsenite treatment (41). The exclusion of TTP from SGs was shown to be regulated by the phosphorylation of 14-3-3 proteins via the p38-MAPK/MK2 pathway (41).
Due to the predominant cytoplasmatic localization of ZBP1 it appears that cellular RNA might be a more likely target than cellular DNA. However, as potential Z-DNA forming sequences are found in the genomes of some viruses, e.g. simian virus 40, adenoviruses (6), herpes simplex virus and molluscum contagiosum virus (S. Rothenburg, unpublished), it seems also possible that a major target for ZBP1 is viral Z-DNA or Z-RNA. Indeed ZBP1 is strongly induced after immunostimulation by interferons (18) and its expression is tightly associated with inhibition of Hepatitis B virus replication in a mouse model (42). ZBP1 was further reported to be strongly induced after serum starvation (43). A recent report demonstrated a strong induction of ZBP1 mRNA in mouse embryonic fibroblasts by transfected double-stranded DNA that was dependent on the protein kinase TBK1, a key regulator of the interferon response (44). This indicates that ZBP1 participates in antiviral responses triggered by viral DNA independent of other sensors for nucleic acid like Toll-like receptors and the helicase RIG-1 (44).
Additionally or alternatively, the natural targets for ZBP1, ADAR1, PKZ and E3L Zα domains might be structured RNA that resemble the Z-conformation or slightly differ from it. The latter view is supported by the finding that ZαZBP1 is more important in determining the subcellular localization than ZβZBP1 despite comparable behavior of both domains in in vitro assays for the induction and binding of Z-DNA. This implies that ZαZBP1 might recognize the physiological target(s) better than ZβZBP1.
In this study we demonstrate that ZBP1 isoforms show different subcellular localizations, controlled by its first Z-DNA binding domain. While full length ZBP1 shows a cytosolic finely punctate distribution, the ZBP1ΔZα isoform as well as the mutants N46D and Y50A localize in large granules. ZBP1ΔZα granules are distinct from but interact dynamically with granules formed by SG and PB proteins. Furthermore, we identified full length ZBP1 as a previously unknown component of SGs induced by heat shock and arsenite treatment. The inducible redistribution of ZBP1 into SGs on one hand, and the association and dynamic interaction of ZBP1ΔZα with granules containing G3BP, TIA1 and DCP1a on the other hand, support the notion that ZBP1 isoforms play distinct roles in the regulation of mRNA metabolism and/or in the cellular defense system against viruses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Friedrich Haag, Alan Hinnebusch, Nancy Kedersha, Ky Lowenhaupt, Alexander Rich, Thomas Schwartz and the members of the Rothenburg and Koch-Nolte laboratories for helpful discussions, Nancy Kedersha and Ky Lowenhaupt for carefully reading the manuscript and Kristina Reutlinger and Sandra Gehendges for technical assistance. We are grateful to Nancy Kedersha, Thomas Schwartz and Jamal Tazi for providing expression plasmids and Eckhard Maldelkow and Martin von Bergen for sharing the CD spectrometer. This work was supported by a graduation stipend from the Werner-Otto Foundation to N.D. and a grant from the Novartis Foundation for Therapeutic Research to S.R. Funding to pay the Open Access publication charges for this article was provided by the University Hospital Eppendorf.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rich A., Nordheim A., Wang A.H. The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 2.Rich A., Zhang S. Timeline: Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 3.Popenda M., Milecki J., Adamiak R.W. High salt solution structure of a left-handed RNA double helix. Nucleic Acids Res. 2004;32:4044–4054. doi: 10.1093/nar/gkh736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha S.C., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 5.Wölfl S., Wittig B., Dorbic T., Rich A. Identification of processes that influence negative supercoiling in the human c-myc gene. Biochim. Biophys. Acta. 1997;1352:213–221. doi: 10.1016/s0167-4781(97)00015-8. [DOI] [PubMed] [Google Scholar]
- 6.Nordheim A., Rich A. Negatively supercoiled simian virus 40 DNA contains Z-DNA segments within transcriptional enhancer sequences. Nature. 1983;303:674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- 7.Rothenburg S., Koch-Nolte F., Haag F. DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles. Immunol. Rev. 2001;184:286–298. doi: 10.1034/j.1600-065x.2001.1840125.x. [DOI] [PubMed] [Google Scholar]
- 8.Rothenburg S., Koch-Nolte F., Rich A., Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H., Mulholland N., Fu H., Zhao K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz T., Rould M.A., Lowenhaupt K., Herbert A., Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nature Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 12.Ha S.C., Lokanath N.K., Van Quyen D., Wu C.A., Lowenhaupt K., Rich A., Kim Y.G., Kim K.K. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zalpha bound to DNA. Proc. Natl Acad. Sci. USA. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert A., Alfken J., Kim Y.G., Mian I.S., Nishikura K., Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown B.A., 2nd, Lowenhaupt K., Wilbert C.M., Hanlon E.B., Rich A. The zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl Acad. Sci. USA. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothenburg S., Deigendesch N., Dittmar K., Koch-Nolte F., Haag F., Lowenhaupt K., Rich A. A PKR-like eukaryotic initiation factor 2alpha kinase from zebrafish contains Z-DNA binding domains instead of dsRNA binding domains. Proc. Natl Acad. Sci. USA. 2005;102:1602–1607. doi: 10.1073/pnas.0408714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu C.Y., Zhang Y.B., Huang G.P., Zhang Q.Y., Gui J.F. Molecular cloning and characterisation of a fish PKR-like gene from cultured CAB cells induced by UV-inactivated virus. Fish Shellfish Immunol. 2004;17:353–366. doi: 10.1016/j.fsi.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.George C.X., Samuel C.E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl Acad. Sci. USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y., Comella N., Tognazzi K., Brown L.F., Dvorak H.F., Kocher O. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene. 1999;240:157–163. doi: 10.1016/s0378-1119(99)00419-9. [DOI] [PubMed] [Google Scholar]
- 19.Koeris M., Funke L., Shrestha J., Rich A., Maas S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 2005;33:5362–5370. doi: 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.G., Muralinath M., Brandt T., Pearcy M., Hauns K., Lowenhaupt K., Jacobs B.L., Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl Acad. Sci. USA. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Wolff K.C., Jacobs B.L., Samuel C.E. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology. 2001;289:378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- 22.Rothenburg S., Schwartz T., Koch-Nolte F., Haag F. Complex regulation of the human gene for the Z-DNA binding protein DLM-1. Nucleic Acids Res. 2002;30:993–1000. doi: 10.1093/nar/30.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell. Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell. Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Anderson P., Kedersha N. Stressful initiations. J. Cell. Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 29.McInerney G.M., Kedersha N.L., Kaufman R.J., Anderson P., Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 2005;16:3753–8763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell. Biol. 1999;147:1431–1342. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz T., Lowenhaupt K., Kim Y.G., Li L., Brown B.A., 2nd, Herbert A., Rich A. Proteolytic dissection of Zab, the Z-DNA-binding domain of human ADAR1. J. Biol. Chem. 1999;274:2899–2906. doi: 10.1074/jbc.274.5.2899. [DOI] [PubMed] [Google Scholar]
- 32.Parusel I., Kahl S., Braasch F., Glowacki G., Halverson G.R., Reid M.E., Schawalder A., Ortolan E., Funaro A., Malavasi F., et al. A panel of monoclonal antibodies recognizing GPI-anchored ADP-ribosyltransferase ART4, the carrier of the Dombrock blood group antigens. Cell. Immunol. 2005;236:59–65. doi: 10.1016/j.cellimm.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Koch-Nolte F., Duffy T., Nissen M., Kahl S., Killeen N., Ablamunits V., Haag F., Leiter E.H. A new monoclonal antibody detects a developmentally regulated mouse ecto-ADP-ribosyltransferase on T cells: subset distribution, inbred strain variation, and modulation upon T cell activation. J. Immunol. 1999;163:6014–6022. [PubMed] [Google Scholar]
- 34.Athanasiadis A., Placido D., Maas S., Brown B.A., 2nd, Lowenhaupt K., Rich A. The crystal structure of the Zbeta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005;351:496–507. doi: 10.1016/j.jmb.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 35.Anderson P., Kedersha N. RNA granules. J. Cell. Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell. Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha S.C., Van Quyen D., Hwang H.Y., Oh D.B., Brown B.A., 2nd, Lee S.M., Park H.J., Ahn J.H., Kim K.K., Kim Y.G. Biochemical characterization and preliminary X-ray crystallographic study of the domains of human ZBP1 bound to left-handed Z-DNA. Biochim. Biophys. Acta. 2006;1764:320–323. doi: 10.1016/j.bbapap.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz T.U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W.F., Blackwell T.K., Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland S.F., Vega R.G., Muller R., Evans C.F., Hilbush B., Guidotti L.G., Sutcliffe J.G., Schultz P.G., Chisari F.V. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J. Virol. 2003;77:1227–1236. doi: 10.1128/JVI.77.2.1227-1236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein A., Guhl E., Zollinger R., Tzeng Y.J., Wessel R., Hummel M., Graessmann M., Graessmann A. Gene expression profiling: cell cycle deregulation and aneuploidy do not cause breast cancer formation in WAP-SVT/t transgenic animals. J. Mol. Med. 2005;83:362–376. doi: 10.1007/s00109-004-0625-1. [DOI] [PubMed] [Google Scholar]
- 44.Ishii K.J., Coban C., Kato H., Takahashi K., Torii Y., Takeshita F., Ludwig H., Sutter G., Suzuki K., Hemmi H., et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.