Abstract
The eel long interspersed element (LINE) UnaL2 and its partner short interspersed element (SINE) share a conserved 3′ tail that is critical for their retrotransposition. The predicted secondary structure of the conserved 3′ tail of UnaL2 RNA contains a stem region with a putative internal loop. Deletion of the putative internal loop region abolishes UnaL2 mobilization, indicating that this putative internal loop is required for UnaL2 retrotransposition; the exact role of the putative internal loop in retrotransposition, however, has not been elucidated. To establish a structure-based foundation on which to address the issue of the putative internal loop function in retrotransposition, we used NMR to determine the solution structure of a 36 nt RNA derived from the 3′ conserved tail of UnaL2. The region forms a compact structure containing a single bulged cytidine and a U–U mismatch. The bulge and mismatch region have conformational flexibility and molecular dynamics simulation indicate that the entire stem of the 3′ conserved tail RNA can anisotropically fluctuate at the bulge and mismatch region. Our structural and mutational analyses suggest that stem flexibility contributes to UnaL2 function and that the bulged cytidine and the U–U mismatch are required for efficient retrotransposition.
INTRODUCTION
Long interspersed elements (LINEs) and short interspersed elements (SINEs) are mobile genetic elements that transpose through an RNA intermediate. LINEs and SINEs exist in many kinds of eukaryotic genomes where they constitute a significant portion of the host genomic DNA. For example, the haploid human genome contains ∼850 000 LINE copies and 1 500 000 SINE copies, which cover ∼21 and ∼13% of the human genome, respectively (1). In addition, LINEs and SINEs are thought to have a large impact on the complexity and evolution of eukaryotic genomes (2).
LINEs and SINEs are first transcribed into RNA, which is then reverse transcribed into complementary DNA that is subsequently integrated into a new location within the host genome. This ‘copy-and-paste’ mechanism is called retrotransposition and the number of these elements expands by this process. LINEs encode an endonuclease (EN) and a reverse transcriptase (RT), each of which is required for LINE retrotransposition (3–7). The LINE-encoded EN nicks a target site DNA, thereby generating a free 3′-OH group; the LINE-encoded RT then reverse transcribes its own RNA using the 3′-OH as a primer (8,9). This process by which a LINE element is integrated into a host genomic DNA is termed target-primed reverse transcription (TPRT). LINE-encoded proteins should distinguish their own RNA from host mRNAs so that the LINE RNA is selectively reverse transcribed. Some LINE-encoded proteins recognize their respective LINE RNAs through a specific sequence in the 3′-terminal tail (10–12). However, the structural basis by which a LINE protein recognizes a respective LINE RNA has not been elucidated. The mammalian LINE, L1, which recognizes its own RNA through a poly A tail without a specific sequence at the 3′ tail (7,13,14), is the only exception, although the mechanism by which the L1 RT distinguishes its own RNA from endogenous host mRNAs also has not been elucidated.
SINEs differ from LINEs in that they do not encode any protein(s) required for their own retrotransposition. However, many SINEs and LINEs share a common 3′ tail sequence and research has shown that these SINEs utilize this common 3′ tail sequence to exploit the enzymatic machinery of LINEs for retrotransposition (11,15–17). In addition, L1 can also mobilize the mammalian SINEs, Alu, B1 and B2, through the poly (A) tail (18,19). Thus, SINEs are, so to speak, non-autonomous transposable elements that parasitize LINEs.
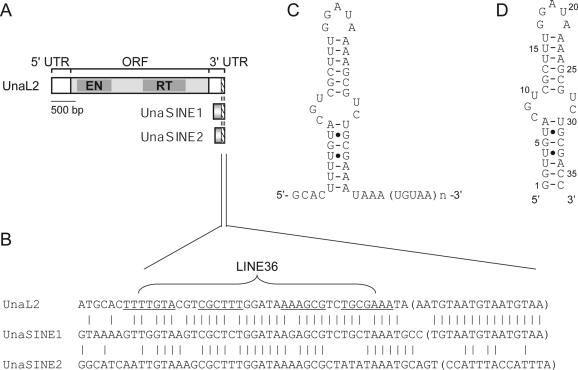
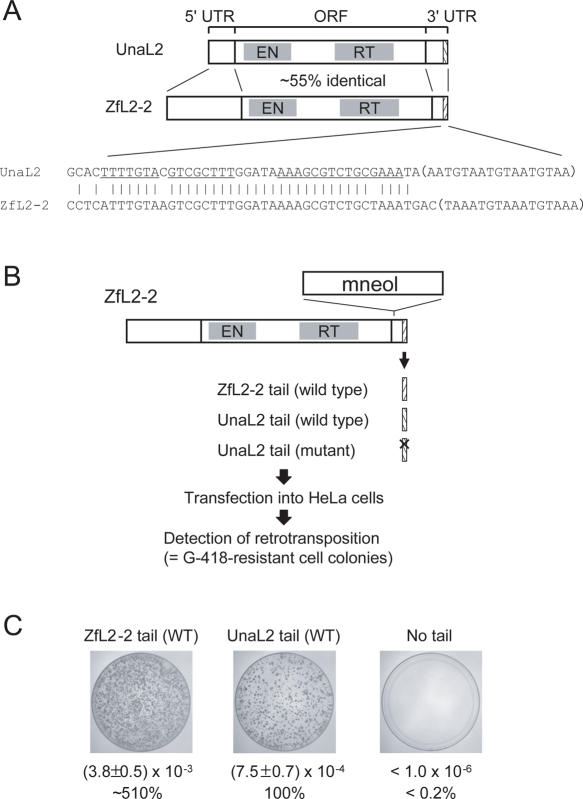
Previously, we isolated one LINE (UnaL2) and two SINEs (UnaSINE1 and UnaSINE2) from the eel genome (11,17). These elements have a conserved 3′ tail of ∼60 bp, the terminus of which has a repeated sequence (Figure 1A and B). Using a retrotransposition assay in HeLa cells, we showed that the 3′ conserved tail of UnaL2 is required for retrotransposition of UnaL2. In addition, an element that we introduced, which contained the 3′ tail of UnaL2, UnaSINE1 or UnaSINE2,could be mobilized efficiently by the UnaL2 retrotransposition machinery in trans (11,17). These results indicated that the 3′ tail of these elements is the only cis element required for retrotransposition and that UnaSINEs are mobilized by UnaL2. These results suggest that the 3′ tails contain a unique sequence specifically recognized by the UnaL2 protein, UnaL2p. The conserved 3′ tail of UnaL2 RNA has two parts, namely the stem–loop region and the 3′-terminal ([UGUAA]n) repeat (usually n = 3), both of which are required—apparently in distinct ways—for UnaL2 retrotransposition (Figure 1C) (11). Reverse transcription of the UnaL2 RNA is initiated from the 3′-terminal repeat and proceeds upstream of UnaL2 RNA through the 3′ conserved region. A template slippage reaction, which is reminiscent of telomere elongation by telomerase, occurs when the reverse transcription of UnaL2 RNA is initiated from the 3′-terminal repeat (11). ‘Repetition’ of the 3′-terminal repeat is probably prerequisite for template slippage, although the role of repetition in retrotransposition has not been elucidated.
Figure 1.
UnaL2, UnaSINE1 and UnaSINE2 from the eel genome. (A) Schematic representation of UnaL2, UnaSINE1 and UnaSINE2. The single open reading frame (ORF) of UnaL2 is indicated by the shaded box. The 5′ and 3′ untranslated regions (UTRs) are also indicated. The 3′ conserved regions are indicated by slash-lined boxes. EN, endonuclease; RT, reverse transcriptase. (B) A sequence alignment of the 3′ conserved regions. The putative stem–loop region of UnaL2 is underlined. The 3′-terminal repeats are indicated by parentheses. The position of LINE36 RNA is indicated. (C) Putative secondary structure of the 3′ conserved region of UnaL2. Watson–Crick base pairs are indicated by horizontal lines and non-Watson–Crick base pairs are indicated by dots. (D) Putative secondary structure of LINE36 RNA.
A part of UnaL2 RNA is predicted to form a stem–loop secondary structure in which the stem is divided into two parts by a putative internal loop (Figure 1C). We previously determined the solution structure of the upper stem–loop region (LINE17 RNA) and confirmed that this region indeed forms a stem–loop (20). The GGAUA loop forms a specific structure in which the uridine is exposed to the solvent and the adenosines are stacked. The second guanosine stacks on the first guanosine and a sharp turn in the phosphodiester backbone occurs between the second guanosine and the adenosine at position 3. Mutational analysis suggested that the particular GGAUA loop structure is requisite for retrotransposition and that UnaL2p specifically recognizes the second guanosine during retrotransposition (20). Although the significance of the stem and putative internal loop structures on retrotransposition has not been thoroughly examined, deletion of the entire putative internal loop abolishes UnaL2 retrotransposition, suggesting that this region is required for the retrotransposition reaction (11).
In the present study, we used NMR techniques with residual dipolar coupling (RDC) restraints to determine the solution structure of a 36 nt RNA, denoted LINE36, that contains nearly the entire stem–loop of UnaL2 RNA including the putative internal loop (Figure 1D). Our results revealed that the putative internal loop region has a compact conformation with a bulged cytidine and a U–U mismatch that separate the upper and lower stems. Although the upper and lower stems are nearly coaxial and thus appear to be a single long stem, molecular dynamics simulation showed that the entire stem fluctuates anisotropically by utilizing the bulged cytidine and U–U mismatch region as a hinge. Mutational analysis indicated that the bulged cytidine and U–U mismatch are required for efficient retrotransposition.
MATERIALS AND METHODS
RNA synthesis, purification and preparation
For structural determination, non-labeled LINE36, stable isotope–labeled [G, C-13C/15N]LINE36 and [A, U-13C/15N]LINE36 were synthesized enzymatically by in vitro transcription with AmpliScribe T7 or T7 Flash transcription kits (Epicentre Technologies Co.) using 13C- and 15N-labeled NTPs (Taiyo Nippon Sanso). Each RNA sample was purified by denaturing PAGE using 30 × 40 cm glass plates (Nihon Eido Co., Ltd.) and the RNAs were recovered from gel slices and salt was removed by ultrafiltration (Centricon YM3, Amicon, Inc.). RNA samples were annealed by heating for 5 min at 95°C followed by snap-cooling on ice. To confirm the formation of the stem–loop structure, we subjected RNAs to native PAGE with a 35mer RNA which forms monomer hairpin as well as duplex dimer. For NMR measurements, RNA samples were dissolved in 10 mM sodium phosphate (pH 6.0) containing 50 mM NaCl. The final concentrations of LINE36, [G, C-13C/15N] LINE36 and [A, U-13C/15N]LINE36 were 0.9, 0.3 and 0.9 mM, respectively. Partial alignment of LINE36 for the RDC measurements was achieved by adding ∼16 mg/ml of Pf1 phage (ASLA Ltd.) to [G, C-13C/15N] or [A, U-13C/15N] LINE36. The solvent of the Pf1 phage sample was exchanged to 10 mM sodium phosphate (pH 6.0) containing 50 mM NaCl by the ultracentrifugation.
NMR measurements
NMR spectra were measured using Bruker DRX-500 and DRX-600 spectrometers. Spectra were recorded at probe temperatures of 4–30°C and NMR data at 20°C were used for structure calculations. The imino proton resonances of G and U residues within RNAs in H2O were distinguished by the 1H-15N HSQC spectra measured with [G, C-13C/15 N]LINE36 and [A, U-13C/15N]LINE36. Exchangeable proton resonances were assigned by NOESY and 15N-edited NOESY in H2O with mixing times of 150 or 200 ms using the jump-and-return scheme (21) or the 3-9-19 pulse (22) for water suppression. Base-pairing schemes were established by 2D HNN-COSY experiments (23). Hydrogen bonding of the U–U mismatch pair was judged from C2 and C4 chemical shifts of the residues obtained from a 2D HNCO experiment (24). HCCH-COSY and HCCH-TOCSY were used to assign sugar spin systems (25), whereas through-backbone assignments were made using 2D HP-COSY (26) and HCP (27). H2 protons of adenosines were assigned using HCCH-TOCSY and 2D HSQC (28). NOE distance restraints from non-exchangeable protons were obtained using NOESY (mixing times of 50, 100, 200 and 400 ms) in D2O (29). Dihedral restraints were obtained from TOCSY (mixing time of 50 ms) and DQF-COSY, as described below. For TOCSY experiments, the modified composite pulse was used to eliminate the ROESY effect with a delay time equal to the 90° pulse (30). Absence of cross peaks between H1′ and H2′ in TOCSY (30) and DQF-COSY experiments was interpreted as the residue being in the C3′-endo conformation (31).
Single-bond 1H-13C RDC values for bases and ribose moieties were measured using non-decoupled 1H-13C HSQC and 1H-13C TROSY spectra (32). Single-bond 13C-13C RDCs for bases and ribose moieties were measured from 1H-13C HSQC experiments with the appropriate selective 13C decoupling.
Structure calculation
A set of 100 structures was calculated using a simulated annealing protocol with the InsightII/Discover package (Accelrys) and NOE distance, dihedral, hydrogen bonding, planarity and chiral restraints were used (Table. 1). NOE intensities between exchangeable protons were interpreted as distances of 2.1–5.0 Å. NOE intensities from non-exchangeable protons were interpreted as distances with a margin of −1.5 to +1.5 Å for the 100 ms NOESY, −1.0 to +2.0 Å for the 200 ms NOESY and −1.0 to +3.0 Å for the 400 ms NOESY. The seven restraints for the absence of NOE cross peaks at 400 ms NOESY were added as distances of 5.0–99.0 Å. The hydrogen bonding restraints were defined as distances of 1.8–2.2 Å; exception was made for U16-A22 (distances of 1.8–5.0 Å), because the imino proton resonances of U16 were broader than other imino proton resonances due to the stem region (20). Information on the anti conformation (residues 1–6, 12–16, 22–26, 31–36), the C3′-endo conformation (residues 1–6, 10–17, 21–36) and the RNA-A conformation for the stem region backbone (residues 2–6, 11–15, 23–27, 31–35) was added to the dihedral torsion restraints. The simulated annealing schedule and force constants were identical to values used previously for RNA (20,33).
Table 1.
NMR restraints and structural statistics.
| Number of experimental restraints | |
| Distance restraints | 411 |
| imino-imino | 16 |
| intra-residue | 198 |
| inter-residue | 190 |
| >5 Å | 7 |
| Dihedral restraints | 172 |
| C3′-endo | 62 |
| RNA-A stems | 110 |
| Hydrogen bonding distance restraints | 62 |
| Residual dipolar coupling | 74 |
| Base planarity restraints | 11 |
| r.m.s. deviations from experimental restraints | |
| Distance (Å) | 0.0387 ± 0.0013 |
| Dihedral (°) | 0.2353 ± 0.1966 |
| r.m.s. deviations from the idealized geometry | |
| Bonds (Å) | 0.0064 ± 0.0001 |
| Angle (°) | 2.5077 ± 0.0115 |
| Impropers (°) | 1.8760 ± 0.1089 |
| Heavy-atoms r.m.s. deviation (Å)a | |
| All | 1.16 |
aAveraged r.m.s. deviations between an average structure and the 10 converged structures were calculated. The converged structures did not contain experimental distance violations of >0.5 Å or dihedral violations of >5°.
Structure refinement with RDC
Xplor-NIH (34) structure calculations with a grid search procedure were used to obtain the optimal Da and R values using low-energy structures calculated by Discover (Accelrys) in the absence of RDCs. The optimized Da values for single-bond 13C–13C RDCs were estimated by multiplying Da for the 1H-13C RDCs by (γCγCr3CH)/(γCγHr3CC) (35). For differences in Pf1 concentrations in the [G, C-13C/15N] and [A, U-13C/15N]LINE36 samples, the deuterium quadrupole splitting of D2O was used to optimize the Da value for the [G, C-13C/15N] sample data (35). The force constant for the single-bond 13C–13C value was also scaled by multiplying the 1H-13C value by (γCγHr3CC)/ (γCγCr3CH) (35). Structures from the Discover calculations were refined with Xplor-NIH using the same distance and dihedral angle restraints with RDCs. Conformational database potentials (36) were used for stems. The 10 final structures that had the lowest total energy were chosen. Experimental RDC data obtained from the well-ordered upper stem region (residues 9–15, 23–29) and lower stem region (residues 2–6, 31–35) were used in the refinement. The susceptibility anisotropy (SANI) force constants increased 0.001–0.5 during the Da and R determination to weight the NOE and dihedral restraints more strongly in the first simulation, as well as geometry constraints. After the optimal Da and R values were determined, final simulation was performed with a higher final SANI force constant of 1.0, according to Leeper and Varani (37). Optimal Da and R parameters for the entire RNA and for the individual upper and lower stem regions were determined to be −32.7/0.11, −32.5/0.12 and −32.8/0.11, respectively. Because the differences between these respective values were small, a single axis system and alignment tensor were used to refine the LINE36 structure.
Molecular dynamics simulation
The averaged structure of LINE36 determined in the present study was used as the starting structure. Molecular dynamics simulation was performed with the SANDER module of the program AMBER, version 7 (38). The model system was built in a periodic box of the TIP3P solvent molecules with a minimum distance of 9 Å from the RNA molecule and we used the LEaP module to add Na+ ions to the LINE36 structure to neutralize the system. Simulation was performed with 100 steps of minimization and 130 ps of solvent equilibration, with a slow warm up from 0 to 298 K over 20 ps (see Supplementary Figure S4). The temperature was set at 298 K for the productive molecular dynamics simulation. The simulation was performed at constant volume and with periodic boundary conditions. The total length of simulation was 8 ns. The simulation was performed on eight processors on a Linux-cluster. Structural analyses of the trajectories from 3 to 8 ns were performed using the CARNAL module.
Retrotransposition assay
The retrotransposition assay was performed as described previously (39). The plasmid pBB4 (39), which contains the entire ZfL2-2 ORF upstream of the mneol cassette (7), was used to make the ZfL2-2 containing the UnaL2 conserved 3′ tail. The 3′ conserved tails of wild type UnaL2 and mutant-stem UnaL2 were PCR amplified and inserted into the BamHI site of pBB4. The resulting plasmids, which contained the 3′ conserved tail of UnaL2 downstream of the mneol cassette, were used in retrotransposition assays. Plasmids pBZ2-5 (39) and pBB4 were used to construct the ZfL2-2 wild type and deletion mutant of the conserved 3′ tail, respectively. As in previous studies (11,17,20,39), we used HeLa-RC (retrotransposition-competent) cells (39) for the retrotransposition assay. HeLa-RC cells (2 × 105 cells/well in 2 ml) were seeded in 6-well dishes and after one day the cells were transfected with 1 μg plasmid DNA and 3 μl FuGENE6 transfection reagent (Roche) according to the manufacturer's instructions. The cells containing the plasmid were selected by hygromycin (200 μg/ml) for 5 days. By comparing the data with cell survival results from negative controls (in which no plasmid was transfected), we estimated that >95% of the transfected cells became hygromycin-resistant (HygR). The HygR cells were trypsinized and reseeded into new 100-mm dishes at densities of ∼1.5 × 105–∼2.5 × 106 cells/dish and grown in 10 ml medium containing 400 μg/ml G-418 antibiotic. After G-418 selection for 12 days, plates were fixed with 100% ethanol and stained with Giemsa solution. G-418R colonies were counted and the retrotransposition frequency (RF) was calculated as the number of G-418R colonies per single HygR cell.
RESULTS
Structure determination
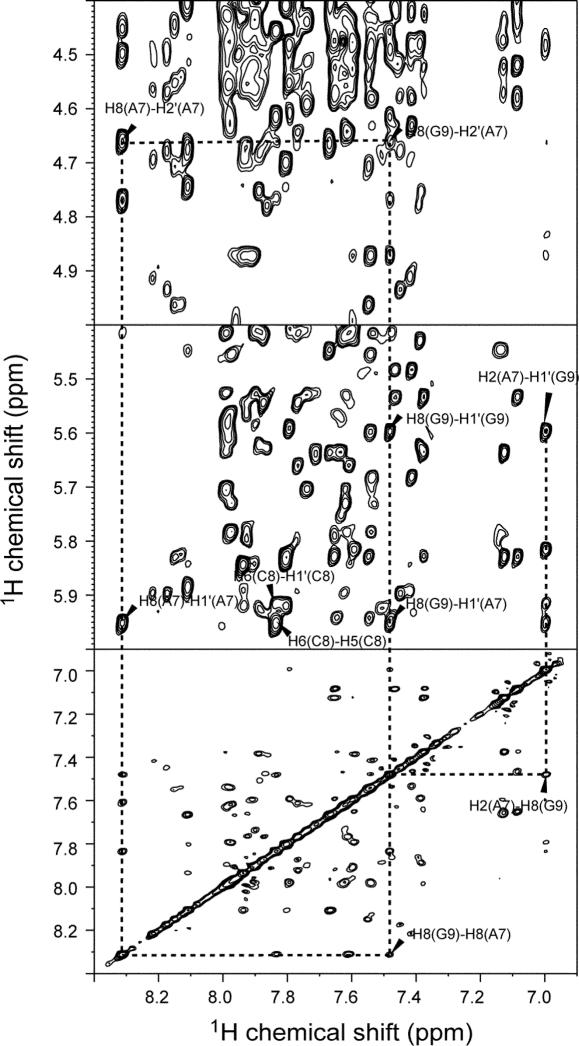
The solution structure of nearly the entire stem–loop of the 3′ conserved region of UnaL2 RNA was determined by utilizing NOE, dihedral and RDC restraints. The NMR signals of LINE36 were assigned using well-established procedures involving heteronuclear methods (40). A part of NMR data is shown in Figure 2. Resonances due to the GGAUA loop of LINE36 were identical to those of LINE17, indicating that the loop structures of the two RNAs (LINE36 and LINE17) are identical (20). Analysis of the TOCSY spectrum indicated that most of the residues adapted the C3′-endo conformation, except for three residues in the GGAUA loop (G18, A19 and U20), two residues in the putative internal loop region (A7 and C8) and two terminal residues (G1 and C36), all of which showed some C2′-endo character. Hydrogen bonds in the Watson–Crick A–U and G–C base pairs were confirmed by HNN-COSY. The NOE connectivities for imino proton resonances of G9, U10, U28 and G27, indicated that G9–C29, U10–U28 and C11–G27 form continuous base pairs in the putative internal loop region. Formation of U10–U28 mismatch was confirmed by a strong NOE between imino protons (see Supplementary Figure S1). The geometry of the U10–U28 mismatch was judged by 13C chemical shifts of the carbonyl C2 and C4 of the uridine residues obtained from HNCO experiments (24). A downfield-shifted C4 signal was observed for U10, whereas a downfield-shifted C2 signal was observed for U28. Thus, the N3 imino proton of U10 forms a hydrogen bond with the O2 carbonyl of U28 and the O4 carbonyl of U10 forms a hydrogen bond with the N3 imino proton of U28. This base pair was categorized as the U–U cis Watson–Crick/Watson–Crick base pair by the standardized nomenclature (41).
Figure 2.
2D NOESY spectrum of LINE36 in D2O. The NOESY spectrum (mixing time = 400 ms) was recorded at 20°C and cross peaks between A7, C8 and G9 in putative internal loop are shown.
During the structure refinement with RDC data, separate grid searches were performed to optimize the Da and R parameters by considering each of the upper and lower stems as a separate unit. In addition, a third simulation was conducted considering the entire molecule as a single rigid unit. Optimal Da and R values for the upper stem, lower stem and entire molecule were determined to be −32.5 and 0.12, −32.8 and 0.11 and −32.7 and 0.11, respectively. Because the differences among the three regions were small, a single axis system and an alignment tensor were used to refine the structure of LINE36, as described below.
A total of 411 distance and 172 dihedral angle restraints were obtained and structures were calculated using restrained molecular dynamics calculations with a simulated annealing protocol (42). Seven restraints for the absence of NOE cross peaks were added as distances of 5.0–99.0 Å (see Supplementary Table S2) to avoid structures containing the closely located proton pairs for which NOE was not observed in the NOESY spectra even with a mixing time of 400 ms. All the protons used in the restraints showed NOEs with other protons, indicating that the rapid relaxation is not the reason for the absence of NOE. Structures calculated by Discover without RDC restraints were well defined, having a heavy-atom r.m.s.d. of 1.55 Å for the 10 lowest energy structures. These structures were refined by Xplor-NIH using 74 RDC restraints along with the distance and dihedral restraints used for the first calculation. Structures calculated with RDC restraints were further refined, having a heavy-atom r.m.s.d. of 1.16 Å for the 10 lowest energy structures (Figure 3), indicating that the calculation with the RDC restraints was successful. The structural statistics are summarized in Table 1. It should be noted that, by using the database potential, the heavy-atoms r.m.s.d. for upper and lower stems were significantly decreased (see Supplementary Table S3). In contrast, the r.m.s.d. for RDC was not changed, indicating that the database potential did not affect the overall structure.
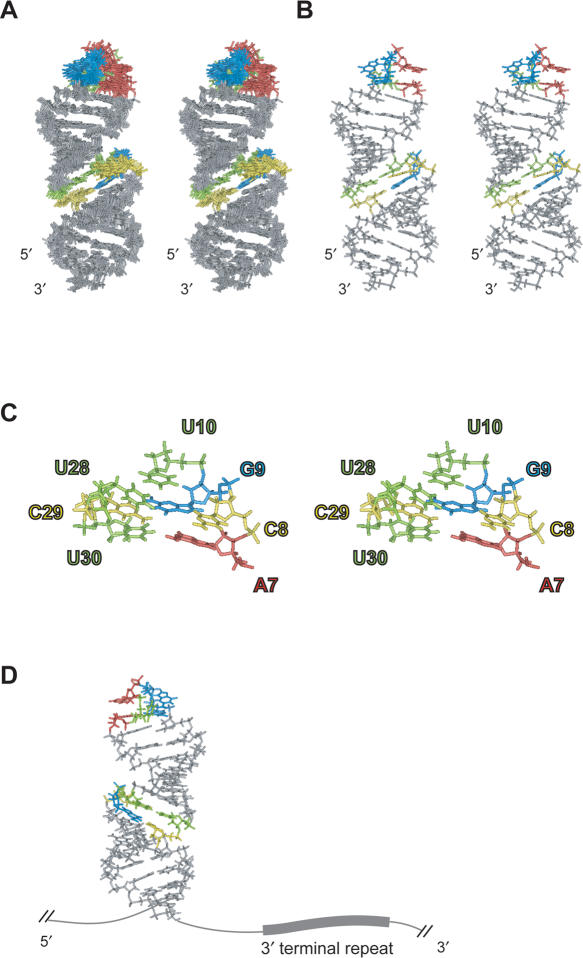
Figure 3.
Solution structures of LINE36. G, A, C and U residues in the GGAUA loop and putative internal loop region are indicated by blue, red yellow and green, respectively. (A) Stereo view of the superposition of the final 10 structures of LINE36. (B) Stereo view of the minimized average structure of LINE36. (C) Stereo view of the bulge region of LINE36 average structure. (D) Location of the second G (blue) in the loop and the 3′-terminal repeat (UGUAA).
Structure and flexibility of LINE36
The solution structure of LINE36 is shown in Figure 3. The entire region looks like a single long stem because the orientations of helical axes are almost the same between the two stems. The second guanosine in the loop, which is probably recognized by UnaL2p (based on mutagenesis studies) and the 3′ end of the stem followed by the UGUAA repeat, which is the initiation site of reverse transcription, may exist on the same side of the stem (Figure 3D); this orientation possibly facilitates UnaL2p binding to the loop to initiate reverse transcription from the UGUAA repeat.
Residues of the putative internal loop region form a compact structure that includes the U10–U28/G9–C29 base pairs and the bulged C8. The A7 and G9 are stacked and this is consistent with the NOE connectivities for H8/H2(A7)-H8(G9), H1′/H2′(A7)-H8(G9) and H2(A7)-H1′(G9). However, the position of the bulged C8 in the calculated structures did not converge to a specific position, indicating that this cytidine is flexible. The imino proton signal of U30 observed at 4°C disappeared at 10°C (data not shown), indicating that the A7–U30 base pair is weak. These observations, namely instability of the A7–U30 base pair, wobbling of sugar puckering at A7 and C8 and conformational heterogeneity of C8 suggest that the putative internal loop region is flexible. This region also contains the U10–U28 mismatch and U–U mismatches generally destabilize a stem, resulting in a decreased Tm. Moreover, the effect of a U–U mismatch on stem stability is greater than that of an A–C or G–A mismatch (43). Thus, the U10–U28 mismatch may also contribute to flexibility in the putative internal loop region.
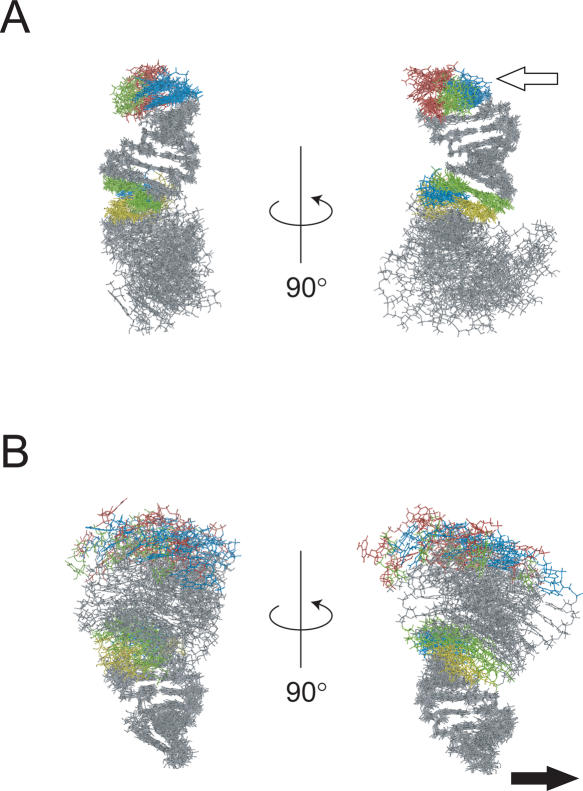
To examine the flexibility of the entire stem of LINE36, we performed a molecular dynamics simulation using the NMR structure. The superposition of the molecular dynamics trajectory of LINE36 is shown in Figure 4. As expected, the conformations of the upper and lower stem regions fluctuate only slightly, whereas the overall structure anisotropically fluctuates by using the putative internal loop region as a hinge. Thus, hereafter, we refer to the putative internal loop region as the ‘hinge’. In Figure 4, the open arrow indicates the putative direction from which UnaL2p recognizes the GGAUA loop and the filled arrow indicates the probable direction of the reverse transcription initiation site (UGUAA repeat). These directions seem to be parallel to the principal component of the hinge motion.
Figure 4.
Superposition of the upper (A) and lower (B) stems of the 10 structures obtained at even intervals by the 3–8 ns molecular dynamics trajectory. G, A, C and U residues in the GGAUA loop and putative internal loop region are indicated by blue, red yellow and green, respectively. The open arrow in panel A indicates the second G in the GGAUA loop of LINE36, which is the UnaL2p recognition site. The filled arrow in panel B indicates the probable location of the 3′-terminal repeat of the UnaL2 RNA.
Requirement of the hinge region for retrotransposition
To examine whether the hinge region of UnaL2 RNA has a role in retrotransposition, we performed a retrotransposition assay in HeLa cells (Figure 5). LINE ZfL2-2, which is a zebrafish homolog of UnaL2, has a 3′ tail that is highly conserved with the 3′ tail of UnaL2 (Figure 5A). As such, the LINE ZfL2-2 protein recognizes the 3′ tail of UnaL2 as well as its own 3′ tail during retrotransposition (Figure 5C). Because the RF of ZfL2-2 (clone ZL15) is ∼30 times higher than that of UnaL2 (clone Aja6-15) (39), we performed retrotransposition assays using LINE ZfL2-2 to analyze a role of the UnaL2 RNA hinge region in retrotransposition (Figure 5B).
Figure 5.
Retrotransposition assay in HeLa cells. (A) Schematics of UnaL2 and ZfL2-2. The single open reading frames (ORFs) of LINEs are indicated by shaded boxes. The 5′ and 3′ untranslated regions (UTRs) are also indicated. The 3′ conserved regions are indicated by slash-lined boxes. EN, endonuclease; RT, reverse transcriptase. A sequence alignment of the 3′ conserved tail of UnaL2 and ZfL2-2 is shown below the schematics. Bases in the upper and lower stem regions of UnaL2 are underlined. The 3′-terminal repeats are indicated by parentheses. (B) Retrotransposition assay procedure. The 3′ tail of UnaL2 and its derivatives were inserted into the 3′ end of ZfL2-2, which contained a retrotransposition detection cassette (mneol) in the 3′ UTR. The ZfL2-2 containing the UnaL2 tail was transfected into HeLa cells and G-418-resistant cell colonies were detected. (C) Results of the retrotransposition assay. Retrotransposition frequencies (RFs) were calculated as described in Materials and Methods. RFs and relative RF values (percentages) compared with the wild type UnaL2 tail are shown. Two different experiments were performed for each construct and average values are shown. Images show each 100 mm plate with G-418-resistant colonies selected from ∼1.5-2.5 × 106 hygromycin-resistant cells. ZfL2-2 tail (WT), ZfL2-2 having the wild type 3′ tail of ZfL2-2. UnaL2 tail (WT), ZfL2-2 having the wild type 3′ tail of UnaL2. No tail, ZfL2-2 having no conserved 3′ tail.
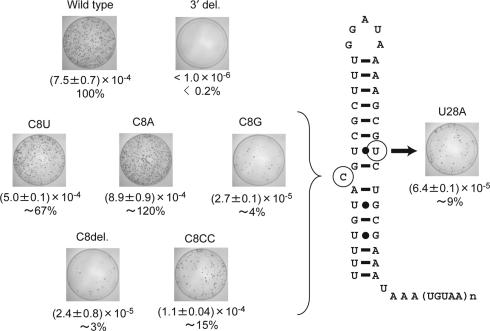
When the bulged C8 was changed to uridine (C8U) or adenosine (C8A), RF values were ∼67 and ∼120%, respectively, relative to wild type (Figure 6). In contrast, substitution of the C8 with guanosine (C8G) severely reduced the RF to ∼4%. These mutations did not alter the secondary structure of the 36mer RNA, except for the C8G mutation, which causes a rearrangement of its secondary structure forming G8–C29/G9–U28 base pairs and a U10 bulge (see Supplementary Figures S1 and S2). Furthermore, deletion of the bulged C8 (C8del) also severely reduced the relative RF to ∼3%. These results indicate that a nonspecific bulged nucleotide (except for G) at position 8 in the stem is requisite for efficient retrotransposition. Addition of another cytidine (C8CC) reduced the RF to ∼15% of wild type, suggesting that the anisotropical feature of the stem flexibility is altered in C8CC and/or that the UnaL2p recognition site, the second guanosine of the GGAUA loop, is oriented differently from the wild type. Substitution of U28 with adenosine (U28A), which presumably forms a U10–A28 base pair instead of the U10–U28 mismatch, decreased the relative RF to ∼10%, suggesting that instability of the U10–U28 mismatch is important for retrotransposition. It should be noted that U28A RNA formed single bulged stem–loop structure similar to LINE36 (see Supplementary Figure S2) and its Tm value was higher than the wild type (see Supplementary Table S1).
Figure 6.
Retrotransposition frequency (RF) in mutants of the putative internal loop region of the UnaL2 3′ tail. The retrotransposition assay was performed as in Figure 5B and 5C. RFs were calculated as described in Materials and Methods. RFs and relative RF values (percentages) for mutants compared with the wild type 3′ tail of UnaL2 are shown. Two different experiments were performed for each construct and average values are shown. Images show each 100 mm plate with G-418-resistant colonies selected from ∼1.5–2.5 × 106 hygromycin-resistant cells. UnaL2 tail (WT), ZfL2-2 having the wild type 3′ tail of UnaL2. No tail, ZfL2-2 having no conserved 3′ tail. C8A, C8U, C8A, C8del, C8CC and U28A: ZfL2-2 constructs having a mutation in the 3′ tail of UnaL2.
DISCUSSION
Evolutionary conservation of the stem structure
We determined the solution structure of the 3′ conserved region of UnaL2 RNA. The hinge region forms a compact structure comprising base pairs U10–U28/G9–C29 and the bulged C8. The two stems separated by the bulged cytidine look like a single long stem. The bulged cytidine along with the U–U mismatch in the hinge region probably confers a high degree of flexibility on the entire stem, allowing it to fluctuate anisotropically.
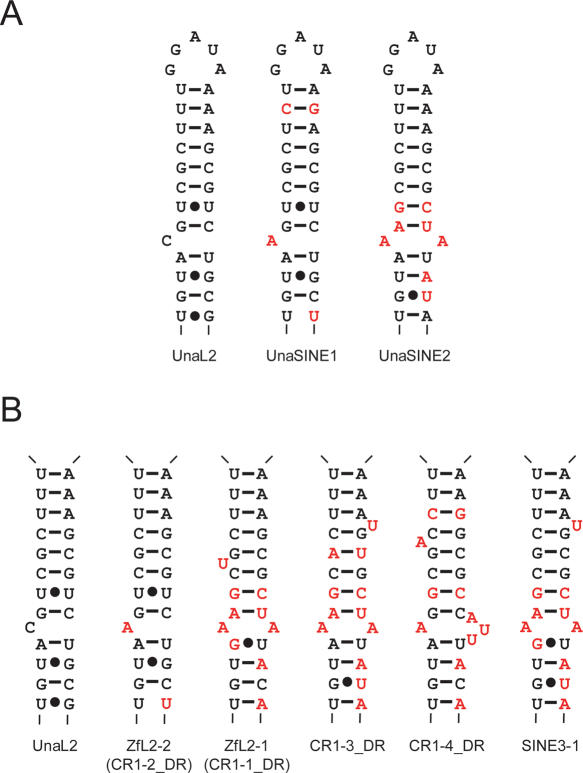
UnaL2 can mobilize UnaSINE1 and UnaSINE2 because the UnaL2 retrotranspositional machinery recognizes their conserved 3′ tails (11,17). Putative secondary structures for the 3′ tail RNA of UnaL2 and UnaSINEs are shown in Figure 7A. The UnaSINE1 structure conserves the bulged nucleotide (A) and the U–U mismatch. The UnaSINE2 structure, on the other hand, lacks the U–U mismatch but instead has a mismatch between two adenosines, once of which is at the same position (position 8) as the bulged nucleotide of UnaL2/UnaSINE1. These adenosines probably impart flexibility to the stem—as in the hinge region of UnaL2—and we speculate that this flexibility is requisite for retrotransposition. To test this point, we predicted secondary structures for the 3′ tail RNA of zebrafish LINEs (and SINE) of the L2 clade (including UnaL2) that have a conserved 3′ tail as UnaL2 by using program Mfold and the sequence alignment with LINE36 (11, 17. 44, 45) (Figure 7B). These elements have similar stem structures, although the loop sequences are highly variable (17). All of these zebrafish elements except for ZfL2-2, which is a zebrafish homolog of UnaL2, have an A–A mismatch-like conformation in the stem. The conservation of this A–A mismatch structure indicates that conformational flexibility in the stem is important for retrotransposition of LINEs/SINEs of the L2 clade.
Figure 7.
Putative secondary structures of conserved 3′ tails. (A) UnaL2, UnaSINE1 and UnaSINE2. (B) Zebrafish LINEs and a SINE, which belong to the L2 clade. All members of the L2 clade have a conserved 3′ tail containing a stem–loop (the loop regions are not shown). Watson–Crick base pairs are indicated by horizontal lines. Non-Watson-Crick base pairs are indicated by dots. Nucleotides that differ from those of UnaL2 are indicated in red. Sequences of the zebrafish elements were obtained from Repbase (47).
Role of the hinge region in UnaL2 retrotransposition
The iron responsive element (IRE), an RNA, contains a stem–loop structure that includes a bulged cytidine, as determined by NMR (46). It has been proposed that the loop structure of IRE RNA makes direct contact with the iron regulatory proteins and that the bulged cytidine functions to orient the hairpin for optimal protein binding (46). Analogously, the bulged cytidine of UnaL2 RNA might allow the stem to bend to achieve optimal binding of UnaL2p to the loop. However, we propose that the function of the bulged cytidine differs between UnaL2 and the IRE, because the stem–loop of UnaL2 has functional aspects other than protein binding (see below). The conserved 3′ tail of UnaL2 RNA is thought to have two different roles in retrotransposition (11). First, the conserved 3′ tail loop structure probably acts as a cis element that is recognized by UnaL2p to form a UnaL2 RNA–protein (RNP) complex (20). Second, a repeated sequence in the conserved 3′ end of the RNA acts as a template to initiate reverse transcription. When the 3′ stem–loop RNA is reverse transcribed, UnaL2p dissociates from the loop RNA and the stem becomes unstructured to facilitate reverse transcription. Previously we found that a slippage reaction occurs during reverse transcription of UnaL2 RNA and we proposed that template slippage is required for dissociation of UnaL2p from the loop RNA (11). UnaL2p repetitively reverse transcribes the UGUAA repeat region during the template slippage reaction and such repetition at the same position should involve repeated conformational changes in the UnaL2 RNP. Flexibility of the stem RNA at the hinge imparts spatial plasticity relative to the loop RNA (binding region) and the UGUAA repeat (reverse transcription initiation region) and this plasticity probably facilitates the conformational change in the UnaL2 RNP for template slippage. On the other hand, the bulged nucleotide and the U–U mismatch in the hinge and the A–A mismatch conformation may promote the transition of the stem RNA from double-stranded to single-stranded by a means of making the stem unstable. There is a bulged nucleotide in the upper stem of many zebrafish LINEs (Figure 7B). These bulges probably also facilitate the conformational transition of the stem RNAs; they do not seem to have a particular role in retrotransposition, however, because they have not been conserved. The dynamic stability of these stem RNAs is an elegant example of the ability of an RNA structure to dictate biological function based on two distinct conformational states.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
We thank Dr A. Pardi for helpful assistance in the measurement of the residual dipolar coupling. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (14035205) and High Technology Research to G.K. and by a Grant-in-Aid to N.O. from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The structure coordinates of LINE36 have been deposited in the RCSB Protein Data Bank (accession code: 2FDT) and the chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank (accession code: 10 018). Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.Xiong Y.E., Eickbush T.H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988;55:235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y., Eickbush T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin F., Maranon C., Olivares M., Alonso C., Lopez M.C. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 7.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 8.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 9.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan D.D., Eickbush T.H. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kajikawa M., Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 12.Osanai M., Takahashi H., Kojima K.K., Hamada M., Fujiwara H. Essential motifs in the 3′ untranslated region required for retrotransposition and the precise start of reverse transcription in non-long-terminal-repeat retrotransposon SART1. Mol. Cell. Biol. 2004;24:7902–7913. doi: 10.1128/MCB.24.18.7902-7913.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 14.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okada N., Hamada M., Ogiwara I., Ohshima K. SINEs and LINEs share common 3′ sequences: a review. Gene. 1997;205:229–243. doi: 10.1016/s0378-1119(97)00409-5. [DOI] [PubMed] [Google Scholar]
- 16.Ogiwara I., Miya M., Ohshima K., Okada N. V-SINEs: a new superfamily of vertebrate SINEs that are widespread in vertebrate genomes and retain a strongly conserved segment within each repetitive unit. Genome Res. 2002;12:316–324. doi: 10.1101/gr.212302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajikawa M., Ichiyanagi K., Tanaka N., Okada N. Isolation and characterization of active LINE and SINEs from the eel. Mol. Biol. Evol. 2005;22:673–682. doi: 10.1093/molbev/msi054. [DOI] [PubMed] [Google Scholar]
- 18.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 19.Dewannieux M., Heidmann T. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 2005;349:241–247. doi: 10.1016/j.jmb.2005.03.068. [DOI] [PubMed] [Google Scholar]
- 20.Baba S., Kajikawa M., Okada N., Kawai G. Solution structure of an RNA stem-loop derived from the 3′ conserved region of eel LINE UnaL2. RNA. 2004;10:1380–1387. doi: 10.1261/rna.7460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plateau P., Gueron M. Exchangeable proton NMR without base-line distorsion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 22.Sklenár V., Piotto M., Lepik R., Saudek V. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102A:241–245. [Google Scholar]
- 23.Dingley A.J., Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 24.Du Z., Yu J., Ulyanov N.B., Andino R., James T.L. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- 25.Pardi A., Nikonowicz E.P. Simple procedure for resonance assignment of the sugar protons in 13C labeled RNA. J. Am. Chem. Soc. 1992;114:9202–9203. [Google Scholar]
- 26.Sklénar V., Miyashiro H., Zon G., Miles H.T., Bax A. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett. 1986;208:94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- 27.Varani G., Aboul-ela F., Allain F., Gubser C.C. Novel three-dimensional 1H-13C-31P triple resonance experiments for sequential backbone correlations in nucleic acids. J. Biomol. NMR. 1995;5:315–320. doi: 10.1007/BF00211759. [DOI] [PubMed] [Google Scholar]
- 28.Legault P., Farmer B.T., II, Mueller L., Pardi A. Through-bond correlation of adenine protons in a 13C-labeled ribozyme. J. Am. Chem. Soc. 1994;116:2203–2204. [Google Scholar]
- 29.Jeener J., Meier B.H., Bachmann P., Ernst R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Phys. Chem. 1979;71:4546–4553. [Google Scholar]
- 30.Griesinger C., Otting G., Wüthrich K., Ernst R.R. Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc. 1988;110:7870–7872. [Google Scholar]
- 31.Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J. Am. Chem. Soc. 1973;95:2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- 32.Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puglisi E.V., Puglisi J.D. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 1998;5:1033–1036. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- 34.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 35.McCallum S.A., Pardi A. Refined Solution Structure of the Iron-responsive Element RNA Using Residual Dipolar Couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- 36.Clore G.M., Kuszewski J. Improving the accuracy of NMR structures of RNA by means of conformational database potentials of mean force as assessed by complete dipolar coupling crossvalidation. J. Am. Chem. Soc. 2003;125:1518–1525. doi: 10.1021/ja028383j. [DOI] [PubMed] [Google Scholar]
- 37.Leeper T.C., Varani G. The structure of an enzyme-activating fragment of human telomerase RNA. RNA. 2005;11:394–403. doi: 10.1261/rna.7222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., DeBolt S., Ferguson D., Seibel G., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 39.Sugano T., Kajikawa M., Okada N. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 40.Varani G., Aboul-ela F., Allain F.H.-T. NMR investigations of RNA structure. Progr. NMR Spectr. 1996;29:51–27. [Google Scholar]
- 41.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilges M., Clore G.M., Gronenborn A.M. Determination of three-dimensional structures of proteins from interproton distance data by dynamical simulated annealing from a random array of atoms. Circumventing problems associated with folding. FEBS Lett. 1988;239:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 43.Meroueh M., Chow C.S. Thermodynamics of RNA hairpins containing single internal mismatch. Nucleic Acids Res. 1999;27:1118–1125. doi: 10.1093/nar/27.4.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Addess K.J., Basilion J.P., Klausner R.D., Rouault T.A., Pardi A. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- 47.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.