Abstract
L-DNA is the perfect mirror-image form of the naturally occurring d-conformation of DNA. Therefore, L-DNA duplexes have the same physical characteristics in terms of solubility, duplex stability and selectivity as D-DNA but form a left-helical double-helix. Because of its chiral difference, L-DNA does not bind to its naturally occurring D-DNA counterpart, however. We analysed some of the properties that are typical for L-DNA. For all the differences, L-DNA is chemically compatible with the D-form of DNA, so that chimeric molecules can be synthesized. We take advantage of the characteristics of L-DNA toward the establishment of a universal microarray that permits the analysis of different kinds of molecular diagnostic information in a single experiment on a single platform, in various combinations. Typical results for the measurement of transcript level variations, genotypic differences and DNA–protein interactions are presented. However, on the basis of the characteristic features of L-DNA, also other applications of this molecule type are discussed.
INTRODUCTION
For the investigation of structural and functional aspects of nucleic acids, mainly synthetic primer or probe molecules are being used that have the basic chemical structure of naturally occurring DNA or RNA. Although derivatives have been synthesized that exhibit particular characteristics, such as locked nucleic acid (LNA) (1) with its improved duplex stability, the molecules still exhibit the basic conformation of natural nucleic acids. An exception to this is peptide nucleic acid (PNA), which is a synthetic DNA-mimic that is based on an amide rather than sugar-phosphate backbone (2). While PNA behaves similar to normal nucleic acids, it simultaneously has several distinct properties (3) because of its elementary structural difference. As yet, PNA has found only limited use.
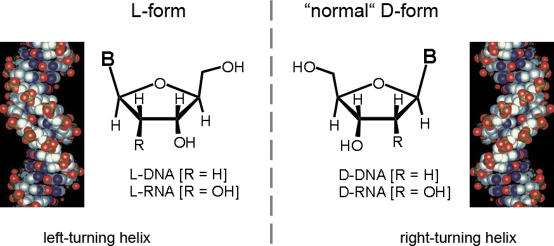
Another artificial molecule, which has attracted even less attention than PNA, is the l-enantiomer of DNA (Figure 1). In principle, L-DNA is identical to the natural d-conformation but for the fact that it is an exact mirror-image of natural DNA and forms a left-turning double-helix upon hybridization to a complementary L-DNA sequence (4–7). L-DNA was examined as a potential antisense reagent but failed to perform adequately (8). Because of its reduced sensitivity to nucleases (9), L-DNA might be an interesting molecule for the creation of aptamer libraries (10). Overall, however, little use has been made of this artificial form of nucleic acid. Only very recently, the use of L-DNA as a tag molecule was demonstrated (11,12).
Figure 1.
Schematic presentation of the structures of the d- and l-enantiomers of nucleic acid.
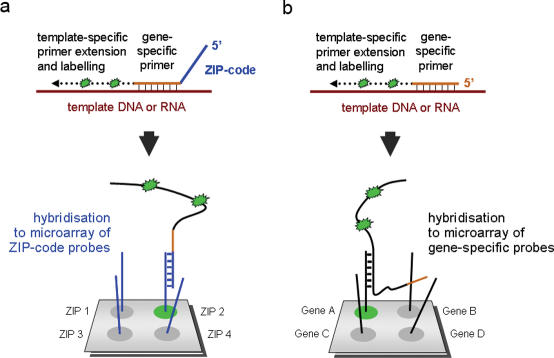
We came across L-DNA when contemplating the design of a microarray platform that is not assaying one specific biological issue at a time—such as transcription profiles, genotypes or protein–DNA interactions, respectively—but would allow a simultaneous analysis of all these and other aspects in a single experiment. Such an array would allow combining different kinds of molecular markers for a more accurate and informative diagnosis, for example. Especially for analyses on samples of limited quantity, simultaneous assaying might become important. For many assays, it would also be advantageous to perform the actual reaction in homogenous solution rather than on a solid support, since the presence of a surface influences many reactions negatively. The establishment of a universal ZIP-code microarray (13,14) could solve these problems. Universal microarrays contain a set of unique and distinct (ZIP-code) oligonucleotides that should not have any complementary sequence in any organism and are made solely for the purpose of addressing with a complementary oligonucleotide a particular location on a microarray (Figure 2). The oligonucleotides should have similar thermodynamic properties so that hybridization can be performed at one experimental condition with identical stringency. Instead of having to produce many different microarrays, a single design can be used for a variety of assays. The actual analysis is carried out with a mixture of probe or primer molecules in homogenous solution. Each oligonucleotide of the mixture is composed of an assay-specific sequence portion that is linked to a distinct, ZIP-code complementary tag-sequence. Only subsequent to the analysis-reaction, the molecules are physically separated by hybridization to the ZIP-code microarray and therefore made available to individual signal scoring. All probe molecules could assay the same kind of information, such as transcript levels for example, or different types of analysis could be combined.
Figure 2.
Basic scheme of a ZIP-code microarray analysis (a). Physical separation of target molecules hybridized to the chip is caused by a specific (ZIP-code) sequence tag. Only one microarray design is needed, irrespective of the kind(s) of assay performed. Standard arrays (b) contain a set of oligonucleotide probes that are designed specifically to fit both the organism analysed and the particular type of experiment performed.
However, the aspect of avoiding tag-sequences that exhibit similarity to any genome is difficult to achieve. Worse, even very short sequence homologies already lead to some cross-hybridization and thus a sequence-dependent accumulation of background signal, if complex samples are hybridized. L-DNA could solve this problem, since its duplexes turn left while natural D-DNA double-helices turn right. Also in terms of stability in an impure environment, L-DNA microarrays could be superior. On this basis, we studied elementary characteristics of L-DNA and describe how this DNA-conformation can be utilized for the generation of a universal microarray platform for a simultaneous analysis of different molecular parameters in a single experiment. However, also other applications of L-DNA are discussed in view of its biophysical and biochemical properties.
MATERIALS AND METHODS
Design of the ZIP-code sequences
Zip-code sequences of 24 nt in length were assembled from a set of 36 tetramer sequence units as described (15). Each tetramer differs from the others by at least 2 nt and is neither palindromic nor complementary to any of the other tetramers. Similarly, each ZIP-code oligonucleotide differs from the others by at least three tetramer units. The result of this design strategy are sequences that have comparable behaviour in terms of hybridization thermodynamics and kinetics while simultaneously maintaining a distinct sequence identity that prevents cross-hybridization (see Supplementary Table S1).
Oligomer synthesis
l-Deoxyphosphoramidites were purchased from ChemGenes Corporation (Wilmington, USA). D-DNA and L-DNA as well as chimeric oligodeoxynucleotides were synthesized at 0.2 μM scale using an ABI 3900 DNA synthesizer and standard synthesis and deprotection protocols. L-DNA and D-DNA ZIP-code molecules with 3′-C6-aminolinker were desalted and used without further purification. Chimeric oligonucleotides that consist of an L-DNA or D-DNA tag sequence, which is complementary to a chip-bound ZIP-code oligonucleotide, and a gene-specific (D-DNA) primer portion were purified by reverse phase HPLC and desalted. All sequences were analysed by electro-spray ionization mass spectrometry. PNA oligomer synthesis was performed as described in detail elsewhere (16).
Tm measurements in solution
HPLC-purified RNA sequences were purchased from IDT (Coralville, USA). To determine the duplex stability, thermal denaturation experiments were performed in a Varian-100 UV-spectrophotometer with solutions of 2 μM oligonucleotide each in 100 mM sodium cacodylate, 100 mM NaCl, pH 7.0.
Enzymatic reactions
Exonucleolytic digests
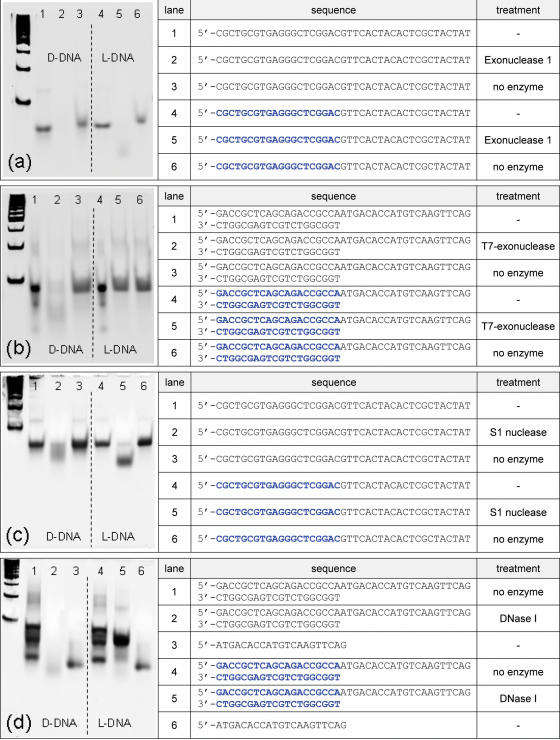
In the experiment, 10 pmol single-stranded D-DNA or chimeric molecules of D- and L-DNA (Figure 5) were incubated with 2 U Exonuclease I (New England Biolabs, Ipswich, USA) in the buffer provided with the enzyme at 37°C for 10 min. For digests with T7-exonuclease, double-stranded molecules were formed by heating complementary single-stranded molecules and a subsequent incubation at room temperature for 30 min (see Figure 5 for sequences). The T7-exonuclease reaction was performed at 25°C for 30 min using 1 U of enzyme (New England Biolabs). The DNA was separated in an 8% polyacrylamide gel and stained with SybrGold (Molecular Probes/Invitrogen, Karlsruhe, Germany).
Endonucleolytic digests
S1-nuclease digestion of 10 pmol single-stranded D-DNA or D-DNA/L-DNA chimeras (Figure 5) was performed with 1 U S1-nuclease (New England Biolabs) at 37°C for 1 min following the supplier's protocol and stopped by heat inactivation. Prior to separation on a gel, the DNA was desalted by MicroSpin G-25 columns (GE Healthcare, Little Chalfont, UK). The DNase I digests were done on 10 pmol double-stranded molecules (Figure 5) with 1 U DNase I (Sigma-Aldrich, Munich, Germany) for 15 min at room temperature at the conditions recommended by the enzyme's supplier. Gel electrophoresis and staining was done as above.
Polymerase reaction
Polymerase chain reactions were carried out in a volume of 50 μl containing a final concentration of 1.5 mM MgCl2, 3 U of either ProofStart DNA polymerase or Taq DNA polymerase (Qiagen, Hilden, Germany), 0.2 mM dNTPs, 10 μM of each primer, 5× Q solution (Qiagen), 10 ng of template DNA and the reaction buffer supplied with the enzyme. The thermocycler conditions were as follows: after an initial denaturation step of 95°C for 5 min, 30 cycles were performed at 94°C for 1 min, 50°C for 1 min, 72°C for 7 min, Subsequently, there was a final extension step at 72°C for 7 min. The PCR-products were checked by electrophoresis in 1% agarose gels and purified with QIAquick kits (Qiagen).
Preparation of oligonucleotide microarrays
For spotting, the oligonucleotides were dissolved at a concentration of 50 μM in 3× SSC (45 mM sodium citrate, 450 mM NaCl), 1.5 M betaine (N,N,N-trimethylglycine, Sigma-Aldrich) and spotted in several copies onto either amino- or epoxy-coated glass slides (Schott-Nexterion, Jena, Germany) using a MicroGrid II (Genomic Solutions, Huntington, UK) equipped with SMP3 pins (TeleChem International, Sunnyvale, USA). After printing, slides were processed and blocked prior to use according to the manufacturer's protocols. To avoid cross-contamination of L- and D-DNA, the respective oligonucleotides were kept in separate microtiter plates and spotted subsequently.
Transcript profiling
Candida albicans cells were grown in either complete yeast medium (YPD) at 25°C for blastospores or cell culture medium (a-MEM; Invitrogen, Carlsbad, USA) at 37°C for hyphaes. Total RNA was isolated using a bead mill (Retsch, Haan, Germany) for cell disruption and the RNeasy kit (Qiagen). 25 μg RNA were reverse transcribed and labelled with Cy3 or Cy5 (GE Healthcare) using the LabelStar kit (Qiagen). Gene-specific primers attached to ZIP-code sequences made of L-DNA were used for cDNA first-strand synthesis. Control reactions were primed with oligo-Dt (15) and hybridized onto conventional arrays of gene-specific probes (17).
Genotyping reaction
Primer extension reactions for genotyping were done in a volume of 10 μl of 26 mM Tris–HCl, pH 9.5, 6.25% glycerol, 12.5 μM EDTA, 0.5% (v/v) Tween-20, 0.5% (v/v) Nonident P-40, 0.125 mM DTT, 12,5 mM KCl, 6.5 mM MgCl2, 1 mM 2-mercaptoethanol, 3.2 U ThermoSequenase DNA polymerase (GE Healthcare), 5 μM each of TexasRed-labelled ddATP, Cyanine-3-labelled ddCTP, Cyanine-5-labelled ddUTP and Fluorescein-labelled ddGTP (Perkin Elmer, Wellesley, USA), 0.1 μM of each primer and 50 nM of each PCR-product (Supplementary Table S3). The reactions were cycled 50 times at 95°C for 30 sec and 40°C for 1 min.
Protein–DNA interaction
Each reaction of protein–DNA interaction probe contained, in a volume of 30 μl of 10 mM HEPES (pH 7.9), 50 mM KCl, 2.5 mM DTT, 10% glycerol, 0.1 mM EDTA, 0.05 mM Nonident P-40 and 1 μM double-stranded PCR-product with a single-stranded L-DNA ZIP-code tag. The sample was heated to 97°C for 5 min and cooled down slowly to room temperature for 1 h. Then, 20 nM of recombinant NF-kappaB (p50) protein (Promega, Mannheim, Germany) labelled with NHS-PEO4-Biotin (Pierce Biotechnology, Rockford, USA) was added and incubated at 30°C for 20 min. Subsequently, a two-step cross-linking reaction (18) was performed using formaldehyde/DSG (Pierce Biotechnology) according to the manufacturer's recommendations. For detection of the labelled protein, 9.9 μg/ml of ExtrAvidin-Cy3 (Sigma) were added to the reaction.
Hybridization
Prior to separating the molecule mixture by hybridization to a microarray, L-formed internal control target (ICT; 5′-TGCGTGATCGTTCTCACAGCCGAA) was added to a final concentration of 33 nM. Hybridizations were done in 4× SSC, 0.1% SDS at 65°C using LifterSlips (Erie Scientific, Portsmouth, USA). After 16 h, the arrays were washed for 1 min and 5 min at 42°C in 2× SSC, 0.1% SDS, followed by subsequent washes at room temperature (10 min in 0.1× SSC, 0.1% SDS, twice for 2 min and once for 1 min in 0.1× SSC). Just prior to drying, the slides were briefly rinsed in 0.01× SSC.
Signal analysis
Fluorescence signals were detected on a ScanArray 5000 unit (Perkin Elmer) or ArrayWorx scanner (Applied Precision, Issaquah, USA) and analysed with the software packages GenePix (Axon Instruments, Union City, USA) or AIDA ArrayMetrix (raytest, Straubenhardt, Germany). Compatibility of the data produced was achieved by cross-checking results on the different systems.
RESULTS
Chemical synthesis of L-DNA, D-DNA and chimeric molecules
Although the chirality of L- and D-formed DNA is inverse, there was no reason to assume that the chemical synthesis of chimeric molecules should be affected by this structural difference. Nevertheless, the chemical compatibility of the L- and D-deoxyphosphoramidites is a critical factor in the production of chimeric molecules that consist of an L-DNA ZIP-code tag and a D-formed primer sequence. No significant differences in the degree of stepwise condensation—monitored by standard trityl assays—or overall yield were observed in syntheses of various oligonucleotides made of pure L-DNA or D-DNA, respectively. More importantly, the same was true for the synthesis efficacy in chimeric oligonucleotides at the junction of the L- and D-sequence portions (data not shown).
Biophysical parameters
Hybridization specificity
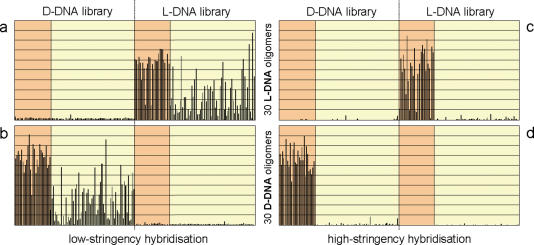
In order to analyse the specificity of the interaction between L-DNA and D-DNA, an identical set of 102 ZIP-code oligonucleotides of 20 nt in length was synthesized in either conformation (see Supplementary Table S1). In addition, we produced 30 fully complementary oligonucleotides in both l- and d-conformation that were labelled at their 5′-terminus with a fluorescence dye. The D- and L-form ZIP-code libraries were spotted next to each other onto microarray surfaces and subjected to hybridizations with the respective set of complementary fluorescently labelled sequences (Figure 3). Even at conditions of low stringency that produced an enormous amount of cross-hybridization of the 30 labelled oligonucleotides at the ZIP-code sequences of the same conformation, no signal above background could be observed at the spots at which the ZIP-code library of the opposite conformation had been placed. As expected, the degree of cross-hybridization within each enantiomeric class decreased with increasing stringency of the hybridization conditions.
Figure 3.
Hybridization specificity of L-DNA and D-DNA. Identical sets of 102 probe oligonucleotides were synthesized in both D- and L-form and spotted next to each other onto microarrays. They were subjected to hybridizations with a set of 30 fluorescently labelled complementary oligonucleotides made of either L-DNA (a and c) or D-DNA (b and d). The raw signal intensities (in arbitrary units) obtained at the 204 spot positions are shown here. Each set of hybridized oligomers is complementary to the probes represented by the orange area. While cross-hybridization occurred within each enantiomeric class at low-stringency (45°C; a and b), there was none between different enantiomers. As expected, cross-hybridization was much reduced at high-stringency conditions (65°C; c and d). Variation in signal intensities also accounts for differences in the amounts of oligomer attached to the support at the various spot positions.
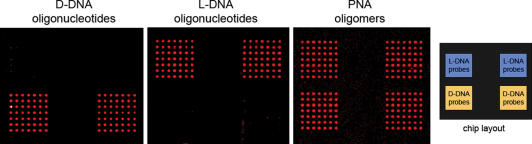
Apart from oligonucleotides, we also synthesized a set of complementary PNA oligomers, each molecule again labelled with a fluorophore. Upon hybridization of the PNA molecules at high-stringency conditions, binding could be observed to both the complementary D- and L-DNA ZIP-code sequences (Figure 4). While D- and L-enantiomeric molecules of complementary sequence did not interact with each other, they both hybridized specifically to complementary PNA sequences.
Figure 4.
PNA binding. DNA probes of identical sequence but different chirality were spotted onto microarrays and hybridized with a set of complementary oligomers that were fluorescently labelled. L-DNA and D-DNA oligomers hybridized only to their respective enantiomeric probe set. PNA oligomers, on the other hand, showed no discrimination between D- and L-form but bound specifically to complementary sequences in either conformation.
Duplex stability
In addition to the array-based analyses, thermal denaturation experiments were performed in solution. There were only insignificant differences in the melting behaviour of pure L- and D-DNA duplexes of identical or inverse sequence (Table 1, duplexes 1–4). The duplex stability of chimeric oligomers, however, comprising of one decamer sequence in pure D- and one decamer stretch in pure L-form which were directly linked by natural 3′–5′-phosphate bridges, dropped considerably (duplex 5). A similar decrease of stability was observed upon the introduction of one tetraethylenglycol (TEG) spacer into a sequence of a 20mer DNA molecule. More spacer molecules decreased the Tm value even further. The influence of TEG spacer molecules on the Tm value was similar irrespective of the chiral nature of the two spaced DNA stretches. The weakest duplex stability was observed with oligonucleotides, which consisted of alternating L-DNA and D-DNA nucleotides (e.g. duplex 6), reducing the melting temperature of a 20mer to about a third of that of the molecules made of either type of DNA only.
Table 1.
Thermal melting temperatures of 20-mer duplexes. D-DNA is shown in black, L-DNA sequences in bold
Biochemical aspects
Exo- and endonucleolytic digestions
We also investigated the activity of nucleases on single-stranded and double-stranded L-DNA molecules. Single-stranded DNA was subjected to a treatment with Escherichia coli exonuclease I, which degrades single-stranded DNA starting from the 3′-terminus. D-DNA was digested entirely. In chimeric molecules, however, only the D-DNA portion was removed, while the L-DNA remained intact (Figure 5a). Incubation of double-stranded DNA with T7-exonuclease, which removes nucleotides from the 5′ end of double-stranded DNA, lead to the degradation of the D-DNA duplex, as expected. However, no digestion of the L-DNA duplex was observed even with an excess of enzyme (Figure 5b).
Figure 5.
Nuclease sensitivity of L-DNA. Single-stranded and double-stranded molecules were digested with commonly used exonucleases and endonucleases. The DNA-sequences are shown. Black letters stand for D-DNA, blue letters indicate L-DNA. While all D-DNA is degraded by all four enzymes used, there is no apparent cleavage of L-DNA with either enzyme. (a) lane 5, the remaining 20mer L-DNA oligomer of the originally 40 nt long molecule is only weakly visible due to smearing effects caused by the presence of the buffer/enzyme cocktail. In (b) lane 5 no digestion of the L-DNA duplex was observed. In panel (c) S1-nuclease digested the D-DNA portion of the chimeric molecule, the L-DNA part, however, remained untouched, lane 5. The three bands in lanes 1 and 4 (d) represent the two single-stranded oligomers and the duplex.
For testing endonucleolytic activity, S1-nuclease was used on single-stranded chimeric oligonucleotides that consisted of an L-DNA and a D-DNA portion. Molecules of identical sequence but made of D-DNA only were used as a control. While the D-DNA portion of either oligomer was degraded, the L-DNA part of the chimeric molecules remained untouched, visible as a distinct band of appropriate size in a gel electrophoresis (Figure 5c). A digest of double-stranded DNA with DNase I resulted in a complete degradation of D-DNA molecules. The L-DNA, however, was not digested; in a chimeric molecule, only the D-formed portion was removed by DNase I (Figure 5d).
Polymerase specificity
The behaviour of chimeric primer molecules in PCR-amplifications using Taq DNA polymerase was studied to see if the L-DNA acted as substrate or blocked the polymerase even within the D-DNA portion of the chimeric primer. As reported earlier (12), L-DNA was no substrate for the polymerase. For a detailed look, the PCR-products were analysed by DNA-sequencing. The incorporation of nucleotides stopped right at the junction of the D- and L-DNA stretches of the chimeric primer, producing a completely double-stranded D-DNA fragment with a single-stranded L-DNA tag.
Application of L-DNA in universal microarray analyses
Internal control probes
In order to control the attachment of the oligonucleotide probes at the array surface, we added to each oligonucleotide spotted to the array an internal control probe (ICP, 5′-TTCGGCTGTGAGAACGATCACGCA), also made of L-DNA. Upon hybridization of a fluorescence-labelled target oligonucleotide of complementary sequence, the amount of attached oligonucleotide could be analysed quantitatively for each individual spot (see Supplementary Figure S1). Also, the accuracy of the gridding and signal homogeneity within the spots could be inspected.
Transcriptional profiling
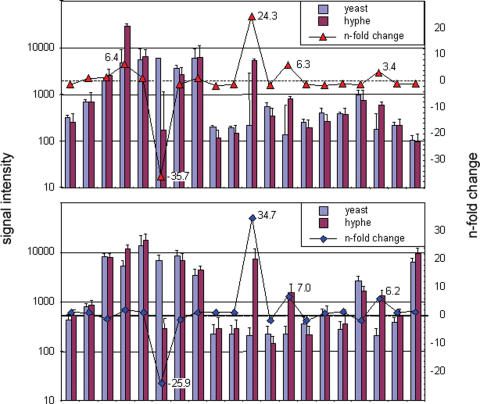
For the validation of an L-DNA microarray for transcriptional profiling, we analysed the transcriptional differences in the well-characterized biological system of hyphal and blastospore growth forms of C.albicans, which are associated with the virulence of this human-pathogenic fungus (17). A mixture of chimeric primers was used for reverse transcription that consisted each of a gene-specific D-DNA portion and a distinct L-DNA ZIP-code. The isolation of total RNA and reverse transcription into cDNA were performed following standard procedures, labelling the cDNA with either the fluorescence dyes Cy5 or Cy3, respectively. Subsequently, the samples were hybridized to ZIP-code arrays. Figure 6, upper panel, presents the signal intensities obtained for a set of 20 genes, which are known to be relevant for the morphological switch from bastospore to hyphal growth (Supplementary Table S2). In comparison, we also performed analyses with conventional oligonucleotides (Figure 6, lower panel). The two sets of results are in agreement as much as one would expect for different experiments. In addition, analysis of the same samples had been performed earlier on a comprehensive microarray made of PCR-products of all C.albicans genes (17). In all cases, data analysis was performed with the data warehouse and analysis software package M-CHiPS, which currently holds some 7800 data sets, following standard protocols (19). In addition, part of the data was confirmed by northern blots (not shown). The results from all these analyses were found to be in agreement, with a degree of variation as known from other comparisons of different analysis platforms.
Figure 6.
Transcriptional profiling on L-DNA microarrays. Typical results obtained for 20 genes of Candida albicans are presented (see Supplementary Table S3), which are known to be relevant for the switch from bastospore to hyphal growth. Both the signal intensities and the variations between the two growth forms are shown. In the upper panel, data from an L-DNA array are presented. The panel below shows the same set of results obtained on arrays made of conventional oligonucleotides.
Typing single nucleotide polymorphisms (SNPs)
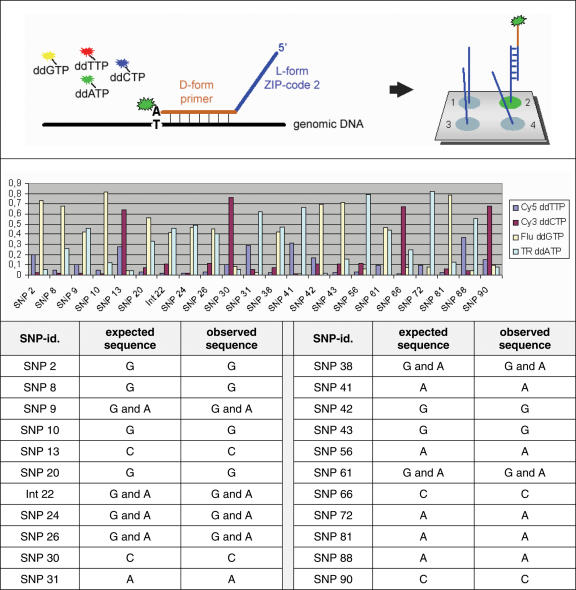
For the genotyping experiments, we used human genomic DNA samples from a case-control study on hay-fever (20). As with the transcript level analysis, the samples had been analysed before by three independent methods, hybridization to in situ synthesized (D-DNA) oligonucleotide arrays, pyro-sequencing and standard DNA-sequencing. DNA regions containing SNPs of 21 candidate genes were PCR-amplified and mixed (for sequences see Supplementary Table S3). As control and for normalization, synthetic fragments were used that were mimicking heterozygous or homozygous fragments. Then, ZIP-code oligonucleotide primers designed to bind adjacent to the bases in question were added and annealed to the DNA-fragments (Figure 7). Dideoxynucleotides that carry a fluorescence label specific for their respective base were incorporated in a polymerase extension reaction, combining the discriminative effect of hybridization with the base-pairing specificity of the polymerase (21). Subsequently, the primers were separated by hybridization to the L-DNA ZIP-code arrays also used for the transcriptional profiling experiments. Figure 7 shows some typical data obtained on a representative patient sample. Base calling was performed as described in detail by Tõnisson et al. (21). The results, including the identification of heterozygous alleles, are in agreement with the data produced by the three other analysis procedures.
Figure 7.
SNP-typing on L-DNA microarrays. Detection of SNPs was done by a primer extension reaction with differently fluorescence-labelled dideoxynucleotides. The basic scheme of the reaction is shown in the upper panel. For one of the analysed patients, the signal intensities obtained for 21 SNPs known to be associated with hay-fever are shown in the second panel. In addition, the result from one artificially synthesized template (Int-22) are presented, which represents a heterozygous polymorphism. In the table below, the expected allele sequences and the actually observed homozygote or heterozygote results are listed. All data could be confirmed by allele-specific hybridization to standard oligonucleotide arrays and gel-based DNA-sequencing.
Protein–DNA interaction
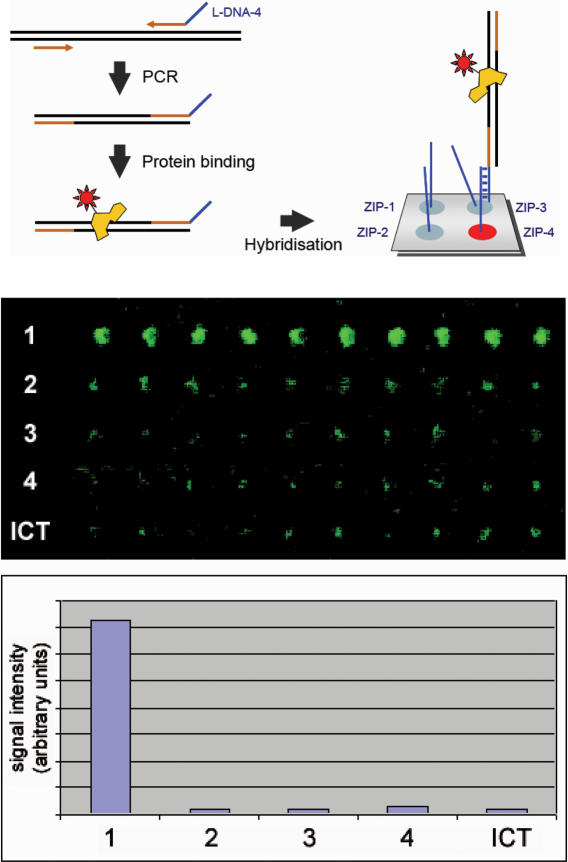
For analysing the interaction of transcription factors and their known or anticipated binding sites, a PCR-amplification with a chimeric ZIP-code primer and a second primer molecule that is made entirely of D-DNA produces a double-stranded DNA-fragment with a single-stranded L-DNA tag (12). Incubation of labelled protein with such molecules will result in binding of the protein to appropriate DNA. Upon a cross-linking reaction for a covalent linkage of protein and DNA, the DNA is hybridized to a ZIP-code array for detection and analysis of protein binding to the various DNA-fragments provided in the experiment. In Figure 8, the result of the binding of purified and labelled proteins to specific double-stranded DNA-sequences with L-DNA tags is shown.
Figure 8.
Detection of protein–DNA interactions. Using chimeric primer molecules, double-stranded DNA molecules with a single-stranded L-DNA tag can be produced. The binding of fluorescently labelled transcription factor NF-kappaB to a mixture of four double-stranded target sequences with L-DNA tags was analysed. Upon incubation and subsequent cross-linking, the DNA was hybridized to an L-DNA ZIP-code microarray. The second panel shows the actual image, each spot being present in 10 copies. Below, the respective signal intensities are shown. Significant binding occurred only to the known target sequence (no. 1). ICT labels the position and signal intensity of a directly labelled control oligonucleotide, which was spotted onto the surface next to the ZIP-code oligonucleotides as a position identifier. By chance, its signal intensity is similar to the background signal at the other spots.
Combination of assays
Combining different kinds of assays on a single chip platform could be advantageous for the application of microarrays as a diagnostic tool. Usually, the number of really informative molecular markers is limited. However, such molecules exist at several molecular levels. We performed a combined analysis of the transcription factor binding assays and SNP-typing (Figure 9). For the former only one label was required, while two differently labelled dideoxynucleotides (ddGTP and ddATP) were used in the latter experiment. As a control for both labelling and hybridization, also the ICT was added to the hybridization mixture. Although detection was done in a single scanning process, data analysis is individual. The occurrence of transcription factor binding could be determined from the variation of signal intensity. Alternatively, the actual signal intensity is irrelevant for the SNP-typing. Base calling depends merely on the ratio of incorporation of the two dideoxynucleotides.
Figure 9.
Combined assay of an analysis of transcription factor binding and SNP-typing. Assays that had been done separately before were performed simultaneously and analysed by a hybridization to a single L-DNA ZIP-code microarray. The top and bottom rows represent Cy3-labelled oligonucleotides, which had been placed to the surface as positional controls. While the mere signal intensity is informative about the interaction of NF-kappaB and DNA, double-labelling had been performed for the SNP-typing. Yellow spots indicate heterozygous samples (Int-22, SNP-9), red signals the presence of a dG in both alleles (SNP-2, SNP-10), while green spots represent homozygous dA sequences. In addition, a Cy5-labelled ICT had been spiked in, hybridizing to a row of ICP oligonucleotides.
DISCUSSION
L-DNA is a versatile but yet mostly ignored form of nucleic acid. Because of its chemical compatibility with the normal D-form of DNA and its identical biophysical behaviour, but for the mirror-image mode, much information can be copied from the vast amount of knowledge on the functioning of D-DNA. Its different chirality, however, offers many possible applications in molecular biology. Here, we demonstrate its utilization toward the establishment of an improved and universally applicable microarray platform. The ZIP-code scheme is not new as such and yet, utilizing L-DNA oligonucleotides for this purpose improves the concept considerably. One important advantage, for example, is the freedom in designing the oligonucleotide sequences, since there is no need to avoid homology to naturally occurring sequences. Therefore, levelling of the dissociation temperatures is simpler to achieve, since the hybridization conditions on an L-DNA microarray can be modified across a wider spectrum of parameters, and many more target molecules can be analysed in parallel. For the lack of any cross-hybridization between L-DNA and D-DNA, also overall background is reduced. In experiments with D-DNA oligonucleotides—also in standard assays—background is produced also via an interaction of target molecules with only a relatively short portion of the probes. Already homologous hexamer sequences can produce considerable background signals, if target is in large excess.
In addition to the assays shown in the result section, there are more that can take advantage of the ZIP-code approach. Since PNA does bind to L-DNA in a sequence-specific manner, for instance, it is possible to synthesize peptides to which during the synthesis—and by the identical chemical process—a PNA-tag is attached. Similar to the transcription factor analysis, the interaction of peptides with analytes could be studied in homogenous solution for purposes such as protein–protein interaction assays or epitope mapping. For detection, the molecules would be separated by the hybridization specificity of the PNA-tag. Another interesting application could be the use of aptamers as ligand binder reagents. Known (D-DNA) aptamer molecules could be synthesized with a unique L-DNA tag attached. Alternatively, new libraries made entirely of L-DNA—including an L-DNA ZIP-code sequence—could be generated.
Since the ZIP-coded assay reaction is performed in solution, sensitivity can be increased by repetition. By cycling a polymerase reaction, for example, more primer molecules will be binding to the template and incorporate label. Therefore, a higher percentage of the molecules hybridizing to the array will carry a label and contribute to the signal produced. Another important element for utilization of microarrays in a routine manner is the aspect of stability. While standard arrays are rather long-living in a dry environment, stability is drastically reduced in the presence of biological samples. The resistance of L-DNA nucleotides to enzymatic degradation had been investigated before using oligonucleotides of mixed L- and D-nucleotides (9). We could demonstrate that this effect is strong in both single-strand and double-strand molecules for both exonucleolytic and endonucleolytic digestion with common enzymes, failing to detect any apparent degradation. The use of L-DNA could therefore allow the positioning of microarrays in a fluidic system through which there is a continuous flow of biological material. Apart from handling advantages, more molecules could be captured in a prolonged incubation in a continuous flow system, thus accumulating signal. This could be important for analysing fermentation or production processes, for example, and fits well with currently ongoing developments toward small-scale lab-on-chip devices.
The production of chimeric molecules could be simplified by linking D-formed primer libraries with L-formed tag libraries enzymatically. Thereby, the ZIP-code libraries could be utilized for all kinds of assays and organisms. Only the D-formed primer had to be synthesized anew. Adding to the 3′-terminus of the L-DNA and the 5′ end of the D-DNA molecules a few common D-formed nucleotides, for instance three each, ligation could be used to link the molecules in presence of a commonly added complementary hexamer. The mixing of the numerous D- and L-formed oligonucleotides could be accomplished robotically. Alternatively, we intend to utilize recent developments in synthetic biology and synthesize the chimeric oligonucleotides on chip surfaces, cleave them off the support, before eluting and using them in subsequent experiments (22,23). We use the system of febit biotech (24) that permits a micromirror-based in situ synthesis of oligonucleotides, similar to the NimbleGen system (25) but for the fact that synthesis takes place in microfluidic channels. Instead of four phosphoramidites, eight are required. The monomers with the respective photolabile groups are available. As a matter of fact, the system already provides connections for eight bottles. Therefore, such synthesis does not require any changes at all. Many different molecules could be synthesized cheaply since in small quantities.
The current costs for L-DNA amidites is around 10 times as high as for standard D-DNA amidites. However, this price is based on small-scale synthesis of L-DNA amidites. Since all the precursors are chiral enantiomers of D-DNA, there is no need to change or re-develop standard synthesis or analysis protocols. The only change is the starting L-ribofuranose instead of the D-ribofurnose. Therefore it is very reasonable to assume that the cost of L-DNA amidite synthesis can reach the range of normal DNA in a short time, once large-scale synthesis schemes are used.
Because of its chiral difference, L-DNA can serve as a useful tool also on normal (D-DNA) microarray platforms and in other areas of molecular biology and biotechnology. One important issue in microarray assays is still the aspect of quantifying the amount of bound analyte in absolute rather than only relative terms. In order to do so, a control for both the amount of probe at the microarray surface as well as the efficiency of the hybridization process is required. This could be achieved by mixing to each probe a small percentage of an L-DNA ICP and spiking the hybridization sample with a known amount of its labelled L-DNA complement (ICT). Neither the probe nor the target will interact with the other probes or the analyte at all, enabling a precise control of microarray quality and hybridization efficiency without any risk of affecting and thus jeopardising the binding of the actual D-formed analyte. Its hybridization will permit the quantification across the entire microarray and between platforms. Even better would be the generation of chimeric probe molecules that consist of a common L-DNA portion for quantification with the ICT and the specific D-portion for binding the actual analyte. This kind of molecule could also be produced on in situ synthesized microarrays.
The apparent lack of enzymatic degradation, the absence of interaction with natural nucleic acids and the ability to produce by standard chemical processes chimeric molecules of L-DNA and D-DNA offer the potential for a number of applications of L-DNA in molecular biotechnology. Especially the combination of the orthogonal hybridization system L-DNA and D-DNA with PNA—with their different structures and distinct patterns of interaction—could be put to good use.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Benjamin Schroeder, Neeme Tõnisson and Michaela Schanne for critical discussions and Gabriele Zelt and Verena Hoerz for technical assistance. We would like to thank Zhaochun Ma for determining the Tm values. This work was financially supported by Applied Biosystems, the German Federal Ministry of Education and Research (BMBF) as part of the NGFN programme, the Deutsche Forschungsgemeinschaft (Graduation Programme 886) and the European Commission as part of the MolTools project. Funding to pay the Open Access publication charges for this article was provided by the above mentioned organisations.
Conflict of interest statement. None declared.
REFERENCES
- 1.Petersen M., Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 1998;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen P.E., Egholm M., Berg R.H., Buchhardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen P.E. Peptiode Nucleic Acids. 2nd edn. Wymondham, UK: Horizon Bioscience; 2004. [Google Scholar]
- 4.Urata H., Shinohara E., Ogura K., Ueda Y., Akagi M. Mirror-image DNA. J. Am. Chem. Soc. 1991;113:8174–8175. [Google Scholar]
- 5.Damha M.J., Giannaris P.A., Marfey P., Reid L.S. Oligonucleotides containing unnatural L-2′-deoxyribose. Tetrahedron Lett. 1991;32:2573–2576. [Google Scholar]
- 6.Urata H., Ogura E., Shinohara K., Ueda Y., Akagi M. Synthesis and properties of mirror-image DNA. Nucleic Acids Res. 1992;20:3325–3332. doi: 10.1093/nar/20.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley G.W. Modeling, synthesis and hybridization properties of (L)-ribonucleic acid. J. Am. Chem. Soc. 1992;114:9731–9736. [Google Scholar]
- 8.Garbesi A., Capobianco M.L., Colonna F.P., Tondelli L., Arcamone F., Manzini G., Hilbers C.W., Aelen J.M.E., Blommers M.J.J. L-DNAs as potential antimessenger oligonucleotides: a reassessment. Nucleic Acids Res. 1993;21:4159–4165. doi: 10.1093/nar/21.18.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damha M.J., Giannaris P.A., Marfey P. Antisense L/D-oligonucleotide chimeras: nuclease stability, base-pairing properties, and activity at directing ribonuclease H. Biochemistry. 1994;33:7877–7885. doi: 10.1021/bi00191a015. [DOI] [PubMed] [Google Scholar]
- 10.Williams K.P., Liu X.H., Schumacher T.N.M., Lin H.Y., Ausiello D.A., Kim P.S., Bartel D.P. Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc. Natl Acad. Sci. USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matysiak S.M., Schroeder B.G., Vinayak R.S., Greenfield L.I. 2003. Heteroconfigurational polynucleotides and methods of use. Patent application. CA2471218AA; US20030198980A1; WO03059929A1; EP1465913A1. [Google Scholar]
- 12.Hayashi G., Hagihara M., Nakatani K. Application of L-DNA as a molecular tag. Nucleic Acids Symp. Ser. 2005;49:261–262. doi: 10.1093/nass/49.1.261. [DOI] [PubMed] [Google Scholar]
- 13.Gerry N.P., Witowski N.E., Day J., Hammer R.P., Barany G., Barany F. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 1999;292:251–262. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- 14.Shoemaker D.D., Lashkari D.A., Morris D., Mittmann M., Davis R.W. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nature Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 15.Favis R, Day1 J.P., Gerry N.P., Phelan C., Narod S., Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol. 2000;18:561–564. doi: 10.1038/75452. [DOI] [PubMed] [Google Scholar]
- 16.Brandt O., Feldner J., Stephan A., Schröder M., Schnölzer M., Arlinghaus H.F., Hoheisel J.D., Jacob A. PNA-microarrays for hybridisation of unlabelled DNA-samples. Nucleic Acids Res. 2003;31:e119. doi: 10.1093/nar/gng120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn K., Urban C., Brunner H., Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as shown by DNA-microarrays. Mol. Microbiol. 2003;47:89–102. doi: 10.1046/j.1365-2958.2003.03300.x. [DOI] [PubMed] [Google Scholar]
- 18.Nowak D.E., Tian B., Brasier A.R. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 19.Fellenberg K., Hauser N.C., Brors B., Hoheisel J.D., Vingron M. Microarray data warehouse allowing for the statistical analysis of experiment annotations. Bioinformatics. 2002;18:423–433. doi: 10.1093/bioinformatics/18.3.423. [DOI] [PubMed] [Google Scholar]
- 20.Nieters A., Linseisen J., Becker N. Association of polymorphisms in Th1, Th2 cytokine genes with hayfever and atopy in a subsample of EPIC-Heidelberg. Clin. Exp. Allergy. 2004;34:346–353. doi: 10.1111/j.1365-2222.2004.01889.x. [DOI] [PubMed] [Google Scholar]
- 21.Tõnisson N., Zernant J., Kurg A., Pavel H., Slavin G., Roomere H., Meiel A., Hainaut P., Metspalu A. Evaluating the arrayed primer extension resequencing assay of TP53 tumor supressor gene. Proc. Natl Acad. Sci. USA. 2002;99:5503–5508. doi: 10.1073/pnas.082100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiler J., Hoheisel J.D. Combining the preparation of oligonucleotide arrays and synthesis of high quality primers. Anal. Biochem. 1996;243:218–227. doi: 10.1006/abio.1996.0509. [DOI] [PubMed] [Google Scholar]
- 23.Tian J., Gong H., Sheng N., Zhou X., Gulari E., Gao X., Church G. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 24.Baum M., Bielau S., Rittner N., Schmid K., Eggelbusch K., Dahms M., Schlauersbach A., Tahedl H., Beier M., Guimil R., et al. Validation of a novel, fully integrated and flexible microarray benchtop facility for gene expression profiling. Nucleic Acids Res. 2003;31:e151. doi: 10.1093/nar/gng151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh-Gasson S., Green R.D., Yue Y., Nelson C., Blattner F., Sussman M.R., Cerrina F. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–978. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.