Abstract
We have determined the structure of a catalytically inactive D70N variant of the Escherichia coli RusA resolvase bound to a duplex DNA substrate that reveals critical protein–DNA interactions and permits a much clearer understanding of the interaction of the enzyme with a Holliday junction (HJ). The RusA enzyme cleaves HJs, the fourway DNA branchpoints formed by homologous recombination, by introducing symmetrical cuts in the phosphodiester backbone in a Mg2+ dependent reaction. Although, RusA shows a high level of selectivity for DNA junctions, preferring to bind fourway junctions over other substrates in vitro, it has also been shown to have appreciable affinity for duplex DNA. However, RusA does not show DNA cleavage activity with duplex substrates. Our structure suggests the possible basis for structural selectivity as well as sources of the sequence specificity observed for DNA cleavage by RusA.
INTRODUCTION
Homologous DNA recombination is a source of genetic diversity and an important repair mechanism for cellular survival and normal growth in both prokaryotes and eukaryotes. Its common and key component is the DNA intermediate known as the Holliday junction (HJ) that comprises a crossover point with four arms of duplex DNA. Formation of this junction from two duplex substrates can lead to exchange of DNA and the ability to move important cellular machinery, such as the replication fork apparatus away from sites of damage (1–4). Eventual resolution of a HJ back to duplex products is a critical step in the recombination process. It is carried out by structure specific endonucleases called resolvases that recognize the HJ and cut two DNA strands of like polarity located symmetrically at the crossover. The subsequent formation of patched or spliced DNA products depends upon the orientation of the resolvase on the junction and thus which pair of strands is cleaved (2,5). A range of different proteins interact with HJs and many protein–HJ complexes have been investigated but just three systems have been structurally determined: those with the branchpoint migration protein RuvA and with the site-specific recombinases Cre and Flp (6–10). The structure we present here of Escherichia coli RusA and a DNA duplex is the first DNA complex with a HJ resolvase.
The resolvase enzyme RusA is found in many bacteria including E.coli where it is encoded by the defective prophage gene rusA that originates from the DLP12 prophage of E.coli K-12. In E.coli, the normally quiescent rusA is expressed when an IS2 or IS10 insertion upstream of the coding sequence activates its transcription. The RusA thus expressed acts as a suppressor of mutations in the ruv genes encoding the RuvABC system involved in the processing of HJ intermediates formed during homologous recombination (11,12). The suppressor activity of RusA in vivo is coupled to the presence of the RecG helicase that also binds HJs and forked DNA substrates, such as those found at stalled replication forks where it is believed to exhibit its primary role in their repair and restart (13,14). However, RusA does not interact directly with RecG or RecG-bound DNA and may require RecG activity solely to establish suitable HJ substrates (11,12). The ability of the RusA enzyme to function independently of other recombination proteins in the selective resolution of HJs has seen it used to probe eukaryotic genomes and to confirm the DNA structures targeted by RuvAB and RecG (15–18). Functionally, RusA is analogous to the RuvC resolvase enzyme that acts at the final resolution step to cleave the HJs that are formed during recombination. However, RusA displays no structural similarity to RuvC, although, it is also a Mg2+ cation-dependent, dimeric endonuclease and shares the common model of using clusters of three or four acidic residues to bind its divalent cations (19). Like RuvC but unlike other phage-encoded endonucleases, RusA is selective for binding and cutting HJs and prefers to cleave at particular DNA sequences. However, the degree of selectivity and specificity is lower for RusA. Unlike RuvC, RusA also binds other branched DNA substrates and duplex DNA and has less stringent sequence requirements favouring sites of cleavage 5′ of a CC dinucleotide at the point of crossover of the junction (20–22). Electrophoretic mobility assays of the binding of RusA to HJs or immobile fourway junction points coupled with restriction endonuclease cleavage of arms of the DNA substrates have shown that it binds to the central portion of the junction where it induces a conformational change in the junction from a stacked-X form typically found for free junctions under conditions resembling those in vivo where there are appreciable levels of Mg2+. These assays suggest that RusA perhaps organizes the junction into an approximately tetrahedral conformation and also that subsequent to formation of an initial complex further RusA dimers are loaded onto the arms of the DNA substrates (20,23). More detailed mutational analyses of specific residues within RusA have confirmed the identity of those residues (D70, D72, K76 and D90) critical for endonuclease activity (24,25). The aspartate residues are believed to be involved in Mg2+ binding and the lysine residue is proposed to have a role in orienting an attacking nucleophile during catalysis. Further analysis has suggested a major role in DNA binding for R69 and a more minor role for N73 in catalysis. The structure determined for the free RusA enzyme was consistent with these interpretations (19,24,25).
We have carried out a series of experiments to determine the structure of a catalytically inactive D70N variant of RusA both with and without DNA substrates. The ultimate aim of this work is to determine the structure of a RusA–HJ complex to enable a fuller understanding of the basis for structural selectivity and sequence specificity by this enzyme. Thus far, we have determined the high resolution crystal structure of the enzyme variant and a medium resolution structure of a complex with a piece of duplex DNA that highlights key features in the protein–DNA interaction and a possible mechanism for the observed sequence specificity.
MATERIALS AND METHODS
Protein expression and purification were carried out as described previously (19,26) except for the substitution of a phospho-cellufine column kindly donated by Chisso Corp. at the second step of the protocol. Crystallization trials were performed by the hanging drop vapour diffusion approach. Trials on the free enzyme were carried out at 17°C using the D70N protein at 5 mg/ml. The best crystals were grown from 18% PEG 3350, 0.2 M Na Tartrate in Tris–HCl buffer at pH 8.0. They belong to space group C2221 and have one monomer in the asymmetric unit with the biologically active dimer generated by crystal symmetry. Data from these crystals were collected at 100 K using an ADSC detector to a resolution of 1.2 Å on station ID 23 of the European Synchrotron Radiation Facility (ESRF). Data were processed using MOSFLM (27) and scaled and merged with SCALA (28). The protein phases were then calculated following molecular replacement with MOLREP (29) using a monomer of wild type (wt) RusA as a search model. For the DNA complex trials, DNA was purchased as high-performance liquid chromatography (HPLC) purified from Sigma GenoSys. Trials were set down using both protein and DNA at a concentration of 200 μM with a 1:1.2 ratio of protein to DNA. Crystals grew from 20% PEG 3350, 0.2 M NaF. They belong to space group P21 and have two dimers and two DNA duplexes in the asymmetric unit. Data were collected at 100 K using a Mar345 image plate detector mounted on a Rigaku MM007 generator to a resolution of 3.1 Å. The data were processed using the HKL suite (30). An initial phase set was determined following molecular replacement with MOLREP (29) using the D70N structure with the flexible loops removed as a search model. After initial rigid body refinement of the protein moiety of the complex with REFMAC, the course of the DNA backbone could be clearly seen in difference electron density maps and duplex B-form DNA was added to the model. With both the free enzyme and the DNA complex, model building was carried out using TURBO-FRODO (31) and COOT (32) and stereochemistry was monitored using PROCHECK (33). Data collection and refinement statistics are presented in Table 1. All figures have been produced using PyMOL (http://www.pymol.org).
Table 1.
The data collection and refinement statistics for the free D70N and D70N–DNA complex structures
| RusA D70N | RusA-D70N–DNA complex | |
|---|---|---|
| Data collection | ||
| Space group | C2221 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 60.2, 84.7, 57.3 | 64.7, 59.5, 90.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 101.7, 90 |
| Resolution (Å) | 1.2 | 3.1 |
| Rmerge | 0.051 (0.082) | 0.071 (0.372) |
| I/σI | 17.6 (6.0) | 18.8 (3.2) |
| Completeness (%) | 83.4 (45.0) | 99.9 (100) |
| Redundancy | 3.1(1.5) | 5.1(5.1) |
| Refinement | ||
| Resolution (Å) | 35–1.2 | 44–3.1 |
| No. reflections | 46 447 | 12 485 |
| Rwork/Rfree | 19.4/20.0 | 24.7/28.9 |
| No. atoms | ||
| Protein | 971 | 3655 |
| DNA | 874 | |
| Water | 175 | 2 |
| B-factors | ||
| Protein | 13.2 | 92.6 |
| DNA | 92.6 | |
| Water | 31.2 | 92.6 |
| Rmsd | ||
| Bond lengths (Å) | 0.010 | 0.004 |
| Bond angles (°) | 1.42 | 0.922 |
Figures in parentheses refer to values in the outer resolution shell. In the DNA complex structure the following residues are missing or have been truncated (indicated with *): M1, N15*, H20*, N21*, R66*, R67*, R68*, K101*, N119 and E120 from monomer A, M1, R19*, H20*, N21*, R22*, R24*, K43*, M46*, R66*, R68*, K76*, K101*, E116*, N119 and E120 from monomer B, M1, N2*, L8*, R16*, R19*, N21, R22, G23, R24, T25, H26, N37*, I41*, K43*, L47*, K56*, R67*, R68*, R69*, K76*, K84*, K101*, E116*, N119 and E120 from monomer C and M1, S13*, R16*, R19*, H20, N21, R22, G23, R24, R40*, I42*, K43*, R66*, R67*, R68*, K76*, K106*, R109*, N119 and E120 from monomer D. There are no residues missing or truncated in the structure of the free enzyme.
RESULTS
Structure of the D70N variant
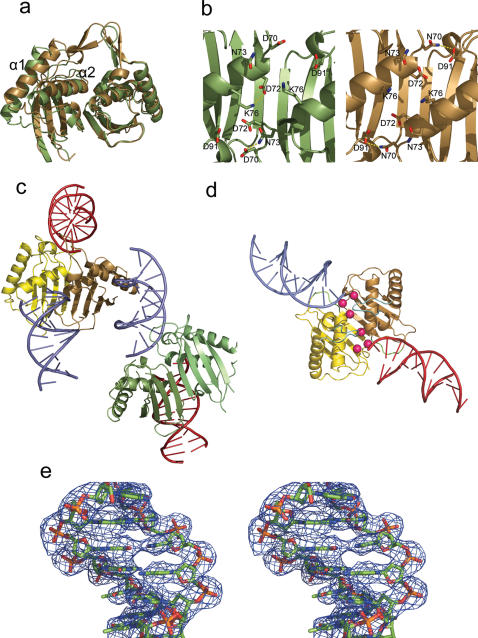
One of the key residues identified as essential for junction resolution is D70 (24). This residue is believed to be involved in binding Mg2+ cations that are required for the endonuclease activity of RusA. Replacement of the aspartate with asparagine gives an 80-fold reduction in junction resolution activity but the variant still binds junctions with affinities comparable or better than wt enzyme (24). Experiments in vitro on the stability of a junction complex with the D70N variant have shown it to have a longer half-life than that of the wt enzyme (24), when there is Mg2+ present. Further advantages of working with this catalytically inactive variant were improvements in both gene expression and protein stability. The variant enzyme was crystallized in a number of forms of which the best diffracted to 1.2 Å resolution with a monomer in the asymmetric unit in space group C2221. A full structure determination and refinement was carried out including an analysis of the solvent structure (see Table 1). As expected, the overall structure was as observed for the wt enzyme in which a monomer is composed from a four-stranded mixed β-sheet flanked on one side by two α-helices. The biological dimer of the D70N variant is formed by application of crystal symmetry. Unlike the wt enzyme, the loop formed by residues 15–28 in the variant is very clearly observed in the electron density and adopts an extended conformation similar to that seen only poorly in monomer A of the wt enzyme, with maximum differences in α-carbon positions of 4 Å. Crystal packing contacts are the source of loop stabilization rather than direct effects of the mutation.
Upon dimer formation in the D70N variant, the interface formed between the β-sheets in each monomer is maintained relative to wt enzyme but the helices alter their relative positions. The relative movement of helix α1 is quite substantial with equivalent alpha carbon positions having moved by ∼6 Å resulting in a closer approach of their N-termini (Figure 1a). Helix α2 also shifts but less markedly with mean changes of ∼2.5 Å. The net effect is that the residues forming the cation binding site in the variant (D70/N70, D72 and D91) are brought closer together such that hydrogen bonds are formed between the side chains of N70 at the site of mutation in one subunit and D91 in the other and between R68 and D90. This is linked to the reduction in charge repulsion arising from the D70N mutation and indicates a degree of flexibility in the dimer interface that might also arise when magnesium cations are bound to wt enzyme. Further detailed differences between wt and variant lie in the proposed cation binding sites (Figure 1b) where in one active site of the wt enzyme the sidechains of K76 and D72 form a contact that is lost through adoption of different sidechain rotamers in the variant and a hydrogen bond contact at the other active site between the sidechains of N73 and D70/N70 is lost in the variant.
Figure 1.
Structure of E.coli RusA D70N and its DNA complex. (a) Overlay of wt RusA (green) onto D70N variant (brown) showing marked shift in position of helices α1 and α2. (b) Regions around catalytically critical residues in wt and D70N variant, including mutation site showing backbone shift and conformational changes of residues D72 and K76. Hydrogen bonds are shown in yellow. (c) Crystal packing of RusA–DNA complex with asymmetric unit content shown with protein dimers in yellow/brown and light green/dark green and associated DNA in light blue and symmetry related duplexes shown in red. (d) Dimer of RusA with two duplexes bound. The locations of the catalytically critical aspartate residues are highlighted with pink spheres, arginine-rich DNA binding regions (residues 66–69) are in green and flexible loop regions (residues 15–28) are in cyan. (e) Stereo representation of electron density from the final 2Fo–Fc map contoured at 1σ around a DNA duplex.
The combined effect of these changes resulting from the loss of charge at residue 70 is to produce a stable, symmetric dimer that may be unable to bind Mg2+ appropriately for catalytic activity. We have repeatedly failed to observe cation binding in either cation cocrystallization or crystal soaking experiments with the wt and D70N variant although assays of junction binding with wt and D70N in the presence or absence of Mg2+ do suggest a cation-dependent effect upon the presumed non-specific binding of the enzyme to duplex DNA in the arms of a junction (24).
RusA–DNA complex
We have used the D70N variant of RusA in attempts to obtain a protein–DNA complex because of its stability and its reported high affinity for junctions (24) whilst being catalytically inactive and thus unable to degrade HJ DNA samples. Crystallization screens using a range of DNA constructs with RusA D70N aimed at producing HJ complexes gave crystals in a variety of forms. The best results from those investigated so far involved the use of a DNA oligonucleotide with the base sequence 5′-CCGGTACCGGT-3′. The sequence of this oligonucleotide is identical to that used in crystallization experiments that yielded the structure of free HJ (34) apart from an additional 3′ T. Upon annealing, however, the construct can also obviously form a duplex with a central region of 10 bp and an overhanging 3′ T on both ends. Data collected from a crystal of this complex indicated that it belonged to space group P21 and was shown to have two RusA dimers and two DNA duplexes in the asymmetric unit (see Figure 1c and Table 1).
Structure of D70N in complex with duplex DNA
Each of the two dimers of RusA D70N in the asymmetric unit binds one of the pieces of duplex DNA and a symmetry related copy of the other (Figure 1c). The overall structure of RusA D70N has altered slightly upon DNA binding relative to that seen for the free enzyme. The major difference lies in the flexible loop regions formed by residues 15–28 noted above in the comparison with wt enzyme which have a modified conformation in one monomer of one dimer (dimerAB) and are not observed for the other dimer (dimerCD) in the DNA complex. These differences are driven by packing considerations related to other proteins in the crystal lattice rather than direct interactions with bound DNA. Overall there are 25 residues missing and the sidechains of another 47 residues have been truncated (see Table 1). The relative separation of the α1 and α2 helices in the dimers noted above is intermediate in the DNA complex structure between that seen for the free D70N and wt enzyme. The only other notable difference occurs in the arginine-rich regions formed by residues 65–69 in dimer CD. Here contacts with the DNA play a role and a root mean square deviation (rmsd) of 0.72 Å is seen compared to free enzyme. The rmsd values for a comparison of the alpha carbon positions excluding the flexible loops of the free D70N enzyme dimer and the two dimers in complex with the DNA are 0.8 Å (dimerAB) and 1.4 Å (dimerCD). An rmsd value of 1.3 Å is obtained for a comparison of the two dimers in the asymmetric unit of the DNA complex. The DNA in the structure is approximately B-form, with rmsd values compared to ideal B-form DNA for the two pieces of duplex DNA, termed DNA X and DNA Y, of 0.99 Å (X) and 0.89 Å (Y).
Protein–DNA interaction
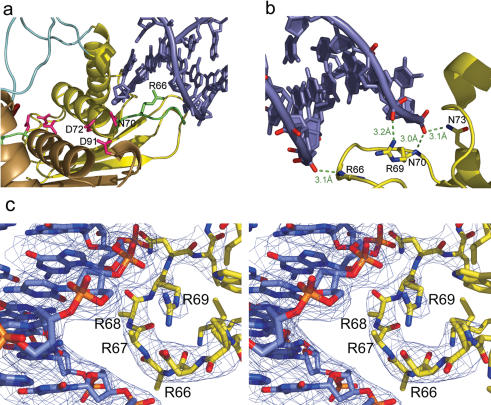
Via the symmetry equivalence, a DNA duplex can be considered bound to each monomer of RusA (Figure 1d). Binding to monomers A and B is very similar but in the case of monomers C and D, the terminal base pair is closer to the dimer dyad axis by ∼3.4 and 6.8 Å i.e. via additional 1 and 2 bp, respectively. In spite of these differences, the chief source of contacts between the protein and DNA in all monomers in our structure is made by the arginine-rich regions formed by residues 65–69 (Figures 1d, 2a and b) that show a strong conservation of positively charged residues in sequence alignments (19). Mutagenesis studies of the highly conserved residue R69 have shown its importance in DNA binding by RusA (25). However, despite the close proximity of the DNA backbone and the positively charged sidechains of the four arginine residues found in this region, only the sidechains of R69 in subunits A, B and D, R67 in subunit B and R66 in subunit C are ordered and show interpretable electron density.
Figure 2.
DNA binding interactions. (a) Insertion of arginine-rich binding loop highlighted in green into DNA minor groove and relative location of DNA backbone to active site residues highlighted in pink. (b) Conserved interactions within the DNA binding region between main-chain amides and phosphate groups either side of the minor groove and the ion-pair interaction between residue R69 and the DNA phosphate backbone. (c) Stereo representation of electron density from the final 2Fo–Fc map contoured at 1σ around residues 65–70 and associated DNA.
There are three conserved interactions formed with the duplex DNA by the arginine-rich region, which anchor the DNA via its minor groove to the protein (Figure 2b). Allowing for uncertainties in interaction distances because of the medium resolution (3.1 Å) nature of the structure, there are a number of likely interactions that can be inferred. Hydrogen bonds seem to be formed between the main-chain amides of R66 and N70 to oxygens of phosphate groups on either side of the minor groove. The well-conserved R69 probably forms an ion-pair with a phosphate of the DNA backbone in three of four subunits. These interactions are conserved despite the variation in binding of the DNA to each of the subunits. There is also a notable contact likely between the sidechain of N73 and a DNA phosphate oxygen, which is conserved in three of the four subunits. Earlier biochemical data have shown that N73 is required for optimal resolution of a HJ (25). Residues 13 to 16 in the helical turn preceding the flexible loop also seem to form interactions with the DNA bases and phosphate backbone. As a result of these sets of interactions, the phosphate backbone of the DNA is positioned close to the active site residues believed to bind the catalytically essential Mg2+ cation.
DISCUSSION
We have shown that the structure of the catalytically inactive D70N variant is very similar to that of wt RusA. The overall fold remains unchanged, but there are changes in the relative orientations of the α-helices in the dimer and in sidechain conformations in the active site region. The loss of charge on D70 leads to perturbation of local structure and alteration of the hydrogen-bonding network and hence the nature of the Mg2+ binding site. The RusA D70N variant in our DNA complex shows little conformational change when bound to duplex DNA relative to its free form.
Cation binding site
We have been unable to observe directly the divalent cation binding by RusA in any crystal form obtained. Our D70N structure suggests that Mg2+ binding might be completely abolished in this variant. It has been shown (20,23) that RusA will bind HJs in the absence of divalent cations and the manner in which the arms of a fourway junction are organized is also independent of the presence of Mg2+. However, data from assays of both wt and D70N binding to HJ constructs do indicate a Mg2+ effect (24), interpreted as the elimination of binding to the DNA duplex arms of a junction following RusA binding at the crossover point when 10 mM Mg2+ is present. This effect could arise from Mg2+ binding to sites remote from the active site that inhibit the binding of additional RusA dimers perhaps through disruption of favourable interactions. Alternatively, the catalytic inability of the D70N variant might arise from a reorganization of the active site such that cations still bind there but do not promote efficient catalysis yet still induce a conformational change in RusA that inhibits its binding to DNA duplex arms.
DNA binding
In our DNA complex structure we observe the extent of bound duplex DNA on the surface of RusA varies between the four copies of the monomer in the asymmetric unit but a constant feature of the interaction is the use of the arginine-rich loop (residues 66–69) inserted into the minor groove of the DNA. In addition, the DNA interaction of the highly conserved N73 and the residues around S13 indicate a role in aligning the DNA optimally for cleavage. Superposition of the monomers and associated duplex DNA does show that the phosphate backbone and helical axis of the DNA are maintained in the same orientation relative to the protein surface and supports the suggestion that we are observing a real mode of RusA binding to DNA rather than a crystallographic artefact. The consistent features of these interactions have given us the confidence to build a model for the interaction of RusA with its natural HJ substrate.
RusA–HJ complex model
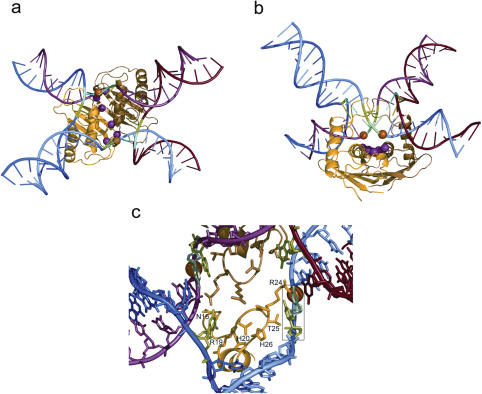
Using the copy of monomer D and its bound DNA duplex, we generated a second copy of the duplex by a 2-fold rotation around the protein dimer axis and created two of the arms of a HJ. As suggested previously (19), it is not possible to bind duplex DNA across the centre of RusA because of the presence of the flexible loops (residues 19–27) and the placement of the other two arms of a bound HJ is also restricted. Solution studies of the binding of RusA to synthetic HJs (20,23) have suggested that a RusA-bound HJ adopts an approximate tetrahedral conformation that is intermediate between a stacked-X and a flat, open 4-fold symmetric structure. Using these features as a guide, we placed two more 2-fold related duplex arms of a junction close to the RusA dimer and attempted to adjust their orientation and helical rotation to enable formation of bonds with the first two bound duplex arms. An immediate finding was that a single unpaired base per strand had to be incorporated in the model to enable a stereochemically reasonable link to be made that avoided unwanted steric clashes with the flexible loops. As a result, the centre of the junction in our model is opened up such that a hole of diameter of ∼20 Å is created that is large enough to accommodate the flexible loop regions. The opening up of the centre of a HJ to accommodate the binding of a resolvase has also been proposed for Schizosaccharomyces pombe Ydc2 (35) and Bacillus subtilis RecU (36). In these cases the junction is proposed to adopt an approximately flat, 4-fold symmetric conformation that creates a hole at the centre although it does not require unpairing of bases and permits the insertion of loops on the dimer interface that form a ‘pin’ structure in Ydc2 and a ‘stalk’ region in RecU. In our model, the four duplex arms of the DNA adopt a distorted tetrahedral arrangement (Figure 3) that still complements the 2-fold symmetry of the protein dimer. There are fewer contacts with the two purely modelled arms of the junction but possible contacts might be formed with the DNA phosphate backbone by the sidechains of R19, H20, H26 and R24 and the main-chain around residue S28 and A29 at the C-terminus of helix α1.
Figure 3.
Model of RusA in complex with a DNA HJ. (a) View down the 2-fold axis of the protein dimer with the locations of the DNA scissile bonds and catalytically critical residues marked with orange and pink spheres, respectively, and the cytidines from the CC dinucleotide sequence recognized during sequence specific cleavage highlighted in green (unpaired base) and cyan. The partner guanosine of the unpaired base is also shown in green. (b) View perpendicular to that in (a). (c) Central region of HJ model at the crossover point with protein residues implicated in DNA sequence recognition labelled and the CC dinucleotide sequence boxed. The colour scheme is as in (a).
The resultant HJ model resembles the junction in a state where a single base pair from each of two duplex arms related across the dimer axis has broken apart and the bases are moving to the other pair of duplex arms i.e. the junction is undergoing branch migration. In vitro data on junction cleavage by RusA has shown a preference for a mobile junction capable of branch migration to enable resolution (20) although RusA can cleave static junctions with appropriately positioned recognition sequences (23).
Our model suggests that the likely scissile bonds lie 1 bp removed from the crossover point in the duplex arms of the DNA that are bound more tightly by the protein i.e. via the arginine-rich loop as seen in our experimental structure. Interactions with conserved R35 and K84 as well as bound metal cations might direct the path of the single-stranded portion of the phosphate backbone in our model. Combined with the experimentally observed contacts around S13 and N73, they could help to position the scissile bond for catalysis.
Sequence specificity
Although, earlier work had suggested a strong preference for mobile junctions capable of branch migration (20), HJ cleavage assays using RusA and a static junction showed strong cleavage when a CC dinucleotide sequence was positioned at the point of crossover (23). Cleavage occurred on the 5′ side of this sequence as also observed when using mobile junctions (21,23). This is consistent with our model if RusA is able to access and stabilize a junction in which the first C–G base pair has been broken and hence the centre of the junction opened up to permit insertion of the flexible loops. The DNA sequence specific recognition required for optimal cleavage by RusA occurs directly at the crossover point. Attention is thus focussed upon possible recognition of the unpaired bases by residues N15, R16 and R19 on one side and R24 and T25 on the other. The intact second C–G base pair might also be recognized via N15 (Figure 3c).
Binding to other DNA constructs
Our observation of the binding of RusA to the ends of pieces of duplex DNA agrees well with the solution studies on the protein. In HJ binding assays with increasing RusA concentration there are progressively larger species formed up to a total of five (20). These species could correspond to selective binding to the centre of the junction followed by loading onto the ends of each arm of the HJ with the final form corresponding to RusA-bound at the centre of the HJ and on the end of each arm. Binding assays using RusA and a cruciform DNA species with two long (25 bp) arms and two short arms (6 bp) showed the presence of only three species (20) and would correspond to binding to the centre of the junction point and the longer arms. Our complex structure and HJ-bound model would suggest that arm lengths of at least 10 bp are required in order to enable additional RusA dimers to be loaded following initial binding at the centre of a junction and is consistent with the solution studies. Assays with a three-way junction also suggested the formation of three species, which likely seem to correspond to binding of RusA to the ends of each arm (20). Binding to a 50 bp duplex DNA or a 50 nt duplex DNA with a central ‘bubble’ created by three mismatched base pair each produced a single retarded species (20). The affinity for the ‘bubble’ DNA substrate was much higher and nearly comparable to that for a HJ. This might suggest more specific binding at the site of the bubble. Our model would predict that approximately four unpaired bases might be required for optimal binding with the flexible loops of RusA fully inserted and interactions formed analogous to those seen in our complex. Destabilization of the duplex DNA by incorporation of the three mismatch bases might allow RusA to induce extended base unpairing that was compensated for by interactions with the protein. However, only one protein–DNA species is observed with the ordinary or mismatch ‘bubble’ duplex rather than two or three species, respectively, that might be expected for binding the ends and the ‘bubble’ site. This suggests that perhaps favourable interactions between multiple, DNA-bound RusA dimers found with 3- and 4-way junction points cannot be formed with 50 bp long duplex and that the interaction with the bubble distorts the DNA in a manner that disfavours additional dimer binding at the ends.
CONCLUSION
Our structure of the RusA D70N variant bound to duplex DNA is the first for a resolvase–DNA complex and has allowed us to construct a plausible model for the RusA–HJ complex formed during the latter stages of homologous DNA recombination. We have been able to provide clearer explanations for many of the observed properties of the enzyme in vitro and insight into its mode of target structure selection and sequence specific cleavage. These insights form a basis for further investigation of the action of the enzyme but also highlight the need for a full structure of the RusA–HJ complex that might help to confirm the hypotheses and explain some of the remaining uncertainties. In particular, it is not clear what the structural consequences are of simultaneous binding of a divalent cation and DNA substrate nor precisely which residues are involved in recognizing key features to facilitate sequence specific cleavage. Given that RusA has already been used as a biotechnological tool to probe eukaryotic genomes because of its ability to function independently of other recombination machinery and its high level of structure selectivity for HJs, detailed structural information on the basis of sequence recognition might enable engineering of RusA variants with altered specificities that could be of further use as biotechnological probes.
Acknowledgments
The authors wish to thank the staff at the ESRF for data collection support and the Royal Society and BBSRC for funding. Coordinates and structure factors for the free D70N enzyme and the DNA complex have been deposited in the Protein Data Bank with codes 2H8E and 2H8C, respectively. Funding to pay the Open Access publication charges for this article have been waivered by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Allers T., Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 2.Lilley D.M., White M.F. The junction-resolving enzymes. Nature Rev. Mol. Cell. Biol. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- 3.Seigneur M., Bidnenko V., Ehrlich S.D., Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 4.McGlynn P., Lloyd R.G. Recombinational repair and restart of damaged replication forks. Nature Rev. Mol. Cell. Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 5.Sharples G.J. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 2001;39:823–834. doi: 10.1046/j.1365-2958.2001.02284.x. [DOI] [PubMed] [Google Scholar]
- 6.Roe S.M., Barlow T., Brown T., Oram M., Keeley A., Tsaneva I.R., Pearl L.H. Crystal structure of an octameric RuvA-Holliday junction complex. Mol. Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- 7.Ariyoshi M., Nishino T., Iwasaki H., Shinagawa H., Morikawa K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc. Natl Acad. Sci. USA. 2000;97:8257–8262. doi: 10.1073/pnas.140212997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F., Gopaul D.N., van Duyne G.D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y., Narendra U., Iype L.E., Cox M.M., Rice P.A. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell. 2000;6:885–897. [PubMed] [Google Scholar]
- 10.Hargreaves D., Rice D.W., Sedelnikova S.E., Artymiuk P.J., Lloyd R.G., Rafferty J.B. Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nature Str. Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 11.Mahdi A.A., Sharples G.J., Mandal T.N., Lloyd R.G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J. Mol. Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 12.Mandal T.N., Mahdi A.A., Sharples G.J., Lloyd R.G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB and ruvC mutations. J. Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGlynn P., Lloyd R.G. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl Acad. Sci. USA. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGlynn P., Lloyd R.G., Marians K.J. Formation of Holliday junctions by regression of stalled replication forks: RecG stimulates fork regression even when the DNA is negatively supercoiled. Proc. Natl Acad. Sci. USA. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., III, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 16.Doe C.L., Ahn J.S., Dixon J., Whitby M.C. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 17.Doe C.L., Dixon J., Osman F., Whitby M.C. Partial suppression of the fission yeast rqh1(−) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolt E.L., Lloyd R.G. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell. 2002;10:187–198. doi: 10.1016/s1097-2765(02)00560-9. [DOI] [PubMed] [Google Scholar]
- 19.Rafferty J.B., Bolt E.L., Muranova T.A., Sedelnikova S.E., Leonard P., Pasquo A., Baker P.J., Rice D.W., Sharples G.J., Lloyd R.G. The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold. Structure. 2003;11:1557–1567. doi: 10.1016/j.str.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Chan S.N., Vincent S.D., Lloyd R.G. Recognition and manipulation of branched DNA by the RusA Holliday junction resolvase of Escherichia coli. Nucleic Acids Res. 1998;26:1560–1566. doi: 10.1093/nar/26.7.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan S.N., Harris L., Bolt E.L., Whitby M.C., Lloyd R.G. Sequence-specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 22.Sharples G.J., Chan S.C., Mahdi A.A., Whitby M.C., Lloyd R.G. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraud-Panis M.J., Lilley D.M. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J. Mol. Biol. 1998;278:117–133. doi: 10.1006/jmbi.1998.1681. [DOI] [PubMed] [Google Scholar]
- 24.Bolt E.L., Sharples G.J., Lloyd R.G. Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol. 1999;286:403–415. doi: 10.1006/jmbi.1998.2499. [DOI] [PubMed] [Google Scholar]
- 25.Bolt E.L., Sharples G.J., Lloyd R.G. Analysis of conserved basic residues associated with DNA binding (Arg69) and catalysis (Lys76) by the RusA Holliday junction resolvase. J. Mol. Biol. 2000;304:165–176. doi: 10.1006/jmbi.2000.4196. [DOI] [PubMed] [Google Scholar]
- 26.Muranova T.A., Sedelnikova S.E., Leonard P.M., Pasquo A., Bolt E.L., Lloyd R.G., Rafferty J.B. Crystallization of RusA Holliday junction resolvase from Escherichia coli. Acta Crystallogr. D. 2003;59:2262–2264. doi: 10.1107/s0907444903018948. [DOI] [PubMed] [Google Scholar]
- 27.Leslie A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 28.CCP4. The CCP4 suite: programs for protein crystallography. Acta. Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A., Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter C.W., Sweet R.M., editors. Methods in Enzymology. Vol. 276. San Diego.: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Roussel A., Fontecilla-Camps J.C., Cambillau C. XV IUCr Congress Abstracts. 1990. TURBO-FRODO: a new program for protein crystallography and modelling; pp. 66–67. [Google Scholar]
- 32.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Eichman B.F., Vargason J.M., Mooers B.H., Ho P.S. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc. Natl Acad. Sci. USA. 2000;97:3971–3976. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceschini S., Keeley A., McAlister M.S.B., Oram M., Phelan J., Pearl L.H., Tsaneva I.R., Barrett T.E. Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J. 2001;20:6601–6611. doi: 10.1093/emboj/20.23.6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGregor N., Ayora S., Sedelnikova S., Carrasco B., Alonso J.C., Thaw P., Rafferty J. The structure of Bacillus subtilis RecU Holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure. 2005;13:1341–1351. doi: 10.1016/j.str.2005.05.011. [DOI] [PubMed] [Google Scholar]