Abstract
Cell-free coupled transcription–translation systems with bacterial lysates are widely used to synthesize recombinant proteins in amounts of several mg per ml. By using reporter green fluorescence protein (GFP) we demonstrate that proteins are synthesized with an unsatisfyingly low-active fraction of (50 ± 20)%. One reason is probably the T7 polymerase used, being up to eight times faster than the intrinsic transcriptase and thus breaking the coupling between transcription and translation in bacterial systems. The active fraction of the synthesized protein was improved by using either a slower T7 transcriptase mutant or lowering the incubation temperature to 20°C. A drop of protein synthesis observed after 7 h incubation time was not due to a shortage of nucleotide triphosphates, but rather to a shortage of amino acids. Accordingly, a second addition of amino acids after 10 h during an incubation at 20°C led to synthesis of up to 4 mg/ml of GFP with virtually 100% activity.
INTRODUCTION
Coupled in vitro transcription-translation systems offer a great potential both as an analytical tool and a method for efficient production of recombinant proteins in amounts of several mg per ml. Cell-free protein synthesis provides advantages over conventional in vivo protein expression method. First of all, in vitro systems can direct most of the metabolic resources of the cell extract towards the production of one protein. Although in vivo expression of proteins occurs in concert with numerous physiological activities, cell-free translation takes place without the need to support processes required for cell viability. Second, the lack of cell wall barrier is another advantage, for in an open system there is the opportunity to create an optimal environment for expression of proteins by directly manipulating the reaction conditions. For example, in vitro systems allow for incorporation of isotope-labeled amino acids (15N, 13C) for NMR studies as well as incorporation of unnatural amino acids for protein design. Third, cytotoxic proteins can be produced in cell-free systems.
Depending on the reactors in which the reaction is performed, several types of cell-free systems can be distinguished: batch system, continuous flow (1), semi-continuous system (2) and hollow fiber membrane reactors (3). In the batch system the reaction mix contains all the necessary components for transcription and translation as well as the synthesized products. In the course of our study with the bacterial Escherichia coli system we used both a batch system and semi-continuous reactors (RTS 500 E.coli HY Kit; Roche ProteoMaster). The latter contains two chambers for the reaction and for the feeding mix, respectively, separated by a semi-permeable membrane. The reaction chamber houses the machinery for mRNA and protein production, together with the DNA template. The chamber with the feeding mix is ∼10 vol larger than that of the reaction and supplies nucleotide triphosphates (NTPs) and amino acids, and removes by-products. The final product, the protein, accumulates in the reaction chamber.
The essential component of the system is the cell-free extract containing most of the cellular cytoplasmic compounds necessary for protein synthesis, such as ribosomes, translational factors, tRNA synthetases and tRNAs. Furthermore, usually the RNA polymerase (RNAP) from the T7 bacteriophage is used. The gene of interest, flanked by a T7 RNAP promoter and terminator, is introduced into the system on a plasmid or as linearized double-stranded DNA.
Besides the above-mentioned advantages of the in vitro system, there are certain difficulties to express genes in the prokaryotic-based systems. A major drawback is, as shown here, the unsatisfyingly low activity of the synthesized protein seen for the well-soluble green fluorescence protein (GFP), which ranges between 30 and 70% and impairs therefore subsequent structural and functional studies.
In this work we solve this major in vitro expression problem and report conditions under which high yields of synthesized proteins with up to 100% activity are achieved.
MATERIALS AND METHODS
Mutants of T7 RNAP
Plasmids encoding the T7 RNAP double mutants P266L/I810S and P266L/I810N were obtained from M. Dreyfus and collaborators. Recently, these authors have described a genetic screen that led to the isolation of the P266L/I810S and the I810N T7 RNAPs. The P266L mutation was introduced into the I810N single mutant, yielding the P266L/I810N double mutant, as described in Ref. (4) for the wild-type enzyme. Finally, plasmids encoding His-tagged versions of the P266L/I810S and P266L/I810N polymerases were obtained by inserting the AlwNI–AlwNI fragment, carrying most of the T7 RNAP coding sequence, into the same sites of pBH161 (5). The mutant polymerases were affinity purified as in He et al. (5).
Rapid translation system (RTS 100 E.coli High Yield Kit; Roche)
The preparation followed the protocol of the manufacturer except that a reaction volume of 10 μl was used instead of the suggested 50 μl. Standard incubation temperature was 30°C. Samples were introduced into ProteoMaster instrument (Roche) and incubated according to the assay requirements; 1.5 μl was applied to either the SDS gel or to the native gel (see below).
RTS 500 E.coli HY Kit (Roche)
The preparation and incubation followed the RTS 500 Kit protocol. The reaction solution was loaded into the 1 ml reaction compartment of the supplied reaction device, and the feeding solution into the feeding compartment with care avoiding air bubbles. The filled reaction device was introduced into ProteoMaster instrument and incubated at 30°C if not otherwise indicated. Incubation time was up to 12 h for RTS 100 reactions, and up to 40 h for RTS 500 reactions.
For the assays with mutant T7 polymerase we had to constitute our own S30 system derived from E.coli BL21. The concentrations were adapted to those published by Ref. (6) and are summarized in Table 1. The yield of synthesized GFP in our homemade system was comparable with that of the commercially available one (Roche RTS), but for unknown reasons the active fraction was only half that of the latter.
Table 1.
Final concentrations in the batch cell-free system for protein synthesis
| Component | Final concentrations according to Ref. (6) | Final concentrations in our reaction mix |
|---|---|---|
| HEPES–KOH (pH 8.2) | 57.2 mM | 60 mM |
| Ammonium acetate | 80 mM | 80 mM |
| Potassium glutamate | 200 mM | 230 mM |
| Sodium oxalate | 2.7 mM | 3 mM |
| DTT | 1.76 mM | 2 mM |
| Cycle-AMP | 0.67 mM | 0.7 mM |
| Folinic acid | 34 μg/ml | 35 μg/ml |
| tRNAs | 340 μg/ml | 350 μg/ml |
| NADH | 0.33 mM | 0.35 mM |
| Coenzyme A | 0.27 mM | 0.3 mM |
| ATP | 1.2 mM | 1.5 mM |
| CTP | 0.86 mM | 1 mM |
| GTP | 0.86 mM | 1 mM |
| UTP | 0.86 mM | 1 mM |
| PEG-8000 | 2% (w/v) | 2% (w/v) |
| Methionine | 2 mM | 2 mM |
| 19 Amino acids | 0.5 mM | 2 mM |
| PEP | 33 mM | 35 mM |
| Magnesium acetate | 15 mM | 12 mM |
| T7 RNAP | 30 μg/ml | 100 μg/ml |
| E.coli S30 cell lysate | 4–6 A260 | |
| Plasmid DNA | 4 μg/60 μl | |
| Rifampicin | 10 μg/ml | |
| 3H-Leu | 1.2 μM | |
| 35S-Met | 1.5 μM |
The reaction-mix had a final volume of 1 ml to fit into the reaction chamber of the RTS 500 device with a volume of the feeding chamber of 10 ml (see section about RTS 500 E.coli HY Kit).
Quantification of the GFP amount in SDS–PAGE
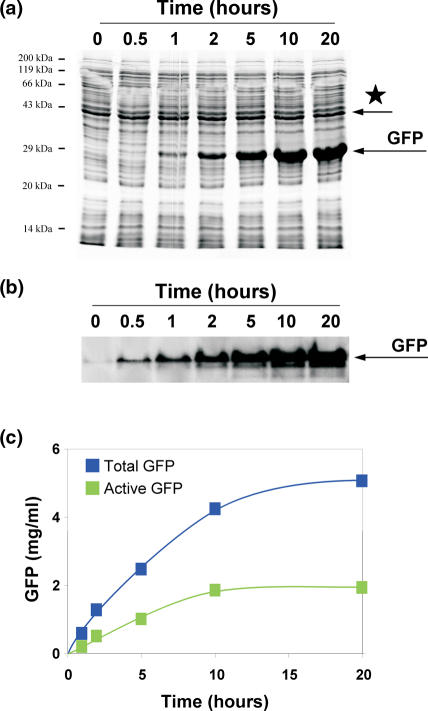
As a standard GFP reporter protein, we used GFP cycle 3. The GFP cycle 3 (GFPcyc3) has three point mutations that allow a fast maturation within 3–4 h, whereas wild-type GFP requires maturation overnight at 4°C (7). GFP was expressed from plasmid DNA for in vitro expression (pIVEX2.2) with T7 promoter and contained a Strep-tag at the N-terminus. The polyacrylamide gels were prepared without SDS addition, the loading and running buffers contained SDS, which is enough for denaturing of the proteins (for buffer components and further details see Bio-rad Application Guide, catalog no. 161-0993). From each 10 μl reaction (RTS 100), 1.5 μl was mixed with 3.5 μl water and 5 μl sample SDS buffer, kept at 95°C for 5 min, cooled down on ice and applied to the 15% PAGE (8). The running conditions for electrophoresis were 75 V for 10 min, 150 V for 3–4 h, which enabled good separation of the GFP protein band from neighboring bands. The SDS gels were stained with Coomassie R-250 (Serva) and scanned by the Personal Densitometer SI (Molecular Dynamic, Sunnyvale, USA). The data were processed using ImageQuant image analysis software, version 5.2 (Molecular Dynamics, Amersham Biosciences). GFP bands were quantified and the total amount of GFP in each lane was determined by comparison with reference bands of known amounts of GFP (catalog no. 11 814 524 001; expressed in E.coli; Roche). The input variations per lane were normalized by scanning of a well-defined band from the S30 pattern (asterisk in Figure 1a), taking into account the respective pixel numbers. Usually amounts of 0.3–0.7 mg/ml of total GFP were produced in 10 h in the RTS 100 reactions (volume 10 μl).
Figure 1.
Quality criteria for the determination of synthesized reporter protein GFP. (a) An aliquot from kinetics of the GFP synthesis in the RTS 500 reaction mix was applied on SDS–PAGE. Asterisk, S30 band was used to normalize input per lane. (b) A sister aliquot was applied to a native PAGE and the fluorescence of GFP was monitored. (c) GFP synthesis in the course of 20 h incubation. Blue squares, total synthesis of GFP (SDS–PAGE); green squares, active GFP (native PAGE).
Fluorometric analysis of GFP
The active GFP present in each reaction sample was calculated by measuring the fluorescence of the GFP at 430–580 nm in the native PAGE (for details see also the above mentioned Bio-rad information). After a maturation period of at least 8 h at 4°C under the conditions of the reaction mixture 1.5 μl from a 10 μl reaction was mixed together with the native loading buffer and directly applied to the 15% PAGE for analysis under the native conditions (9). A longer maturation period of up to 30 h did not improve the active fraction of GFP. Native PAGE, loading and running buffers were prepared without SDS. Conditions for electrophoresis were 75 V for 10 min, 150 V for 2–3 h. The fluorescence was measured directly in the gel with a FluorImager 595 dual-excitation, laser-induced fluorescence scanner (Amersham Biosciences). The images were analyzed using the ImageQuant software. The reference GFP, commercially available and synthesized in vivo, was arbitrarily assigned as 100%, and the relative activity of the newly translated GFP was calculated. On average, the activity of the GFP from the coupled in vitro system was (50 ± 20)% of that of the reference GFP.
Luciferase expression and quantification
The luciferase T7 Control DNA and the luciferase substrate (Steady-Glo® Luciferase Assay System) were purchased from Promega. The expression of luciferase and GFPcyc3 as a control was performed for 6 h at 20 or 30°C in the RTS 100 reaction according to Roche protocol for [35S]Met incorporation in a volume of 25 μl. After incubation, samples of 2 and 1.5 μl from the same tube were taken for measurements of active and total luciferase expression, respectively. The former sample (active luciferase) was mixed with H20M6K150 buffer (20 mM HEPES, 6 mM magnesium acetate, 150 mM potassium acetate, pH 7.6 at 0°C) and then added to a solution containing the luciferase substrate. After 45 min in the dark at room temperature the lumino units were measured with the Centro LB 960 luminometer (Berthold technologies, Germany). The second sample (1.5 μl) was applied to a 15% polyacrylamide protein SDS gel. Electrophoresis was performed for 4 h at 150 V and the gel stained with Coomassie R-250 to monitor the protein separation and then dried on 3 mm Whatman® paper under vacuum at 60°C for 3 h. The dried gel was exposed in a PhosphorImager screen (Fujifilm BAS Cassette 2325) at room temperature for 2 h, scanned in the PhosphorImager STORM 820 (Molecular Dynamics, Amersham Biosciences), and the pixels of luciferase expressions (incorporation of [35S]Met) were quantified using ImageQuant software. The results were normalized to 2 μl of reaction mixture.
Northern blot hybridization (10)
Samples of 10 μl from RTS 100 or RTS 500 were withdrawn at various time intervals as indicated in Figure 4 and extracted with an equal volume of phenol. From the aqueous phase 5 μl of each sample was applied to a 1% agarose gel containing 2% formaldehyde. The running conditions for electrophoresis were as follows: 75 V for 1 h. mRNA was blotted on the Hybond N-plus membrane (Amersham) and the membrane was pre-hybridized in the presence of salmon sperm DNA. Hybridization with the γ-32P-labeled primer 5′-CATCTTCTTTAAAATCAATAC-3′ complementary to a middle sequence of GFP-mRNA was performed in the presence of 50% formamide and 5% dextran sulfate (Sigma) at 42°C overnight. The membrane was washed twice with 2× SSC (0.3 M NaCl and 30 mM Na-citrate) and 0.1% SDS at 65°C for 30 min. Radioactivity was detected in the PhosphorImager system with an exposition time of ∼3–5 h. Data were processed as described above, in vitro transcribed GFP-mRNA served as a reference.
Second addition of a mixture of 20 amino acids
The amino acid mixture was prepared as follows: lyophilized 19 amino acids supplied with the Roche RTS 500 kit were reconstituted by the kit-reconstitution buffer to a volume of 1.5 ml and mixed with the reconstituted methionine solution of 0.9 ml. After 7 h of incubation 2.4 ml was removed from the feeding solution chamber and substituted with this freshly prepared mixture of 20 amino acids.
In case of the RTS 100 we used a reaction volume of 50 μl and added 6 μl amino acid mixture in reconstitution buffer after 8 and 10 h (30 and 20°C incubation temperature, respectively).
RESULTS AND DISCUSSION
GFP as reporter protein and quality criteria for judgment of expression levels
We used the GFP as a reporter. GFP is a fluorescent molecule of 238 amino acids and discovered in the jellyfish Aequorea victoria. The protein has a unique structure consisting of 11 strands β-barrel (β-Can) forming a cylinder with a central and almost coaxial α-helix that forms autocatalytically a fluorophore from the tri-peptide sequence Ser65-Tyr66-Gly67 (11,12). Once folded the fluorescent protein is very stable and, for example, resists heat (up to 65°C) and tolerates a pH up to 11 [for a review see (13)].
The folding and the oxidation process forming the fluorophore is slow, maturation in E.coli requires an overnight incubation at 4°C. Folding of GFP denatured by acidic pH or 6 M guanidine hydrochloride occurs within 1–5 min (14,15) suggesting that most of the maturation time is used for the fluorophore formation. A mutant used here also carries three mutations, namely at positions 100, 154 and 164. The mutations improve the folding efficiency, reduce the maturation times to 3–4 h and improve the yield of stable, active recombinant GFP, not only in E.coli, but also in eukaryotic Chinese hamster ovary (CHO) cells (7,13). The latter observation indicates that the active fraction of GFP can be improved by accelerating the intrinsically slow folding of this protein via mutations. The active fraction of GFP can also be influenced by other factors. They are as follows:
Temperature: wild-type GFP showed a sharply decreased active fraction when synthesized at 37°C, a temperature much higher than that of the cold pacific water habitat of A.victoria (16).
Chaperone concentrations: This was demonstrated previously with proteins essential for the E.coli cell and dependent on GroEL/ES for successful folding (17). When these proteins were overexpressed in E.coli up to 70% were insoluble, whereas upon an ∼5-fold increase of GroEL/ES in vivo, the solubility increased 2- to 3-fold (17). However, a direct involvement of chaperones in the folding of GFP has yet to been shown.
Differences in modes of folding and kinds of chaperones between bacteria and eukaryotes: Here the observation is pertinent that a significantly higher yield of soluble GFP was observed in Saccharomyces cerevisiae (∼90% of the totally synthesized GFP) than in E.coli (∼60%) (18), similar to the amount seen in our experiments. The authors put forward the explanation that this difference reflects the co-translational folding process in eukaryotes, in contrast to the prevailing post-translational folding mode in bacteria (see below). Another non-mutually exclusive explanation is the fact that the different chaperone systems of the eukaryotic cell are superior in supporting the folding of the eukaryotic GFP protein, as compared with the bacterial chaperones. In agreement, Sacchetti et al. (19) suggested that the different expression of GFP variants in E.coli and mammalian cells was caused by different chaperone sets in these organisms.
A different relationship between transcription and translation in bacteria and eukaryotes: In this article we present evidence that the loss of the bacterial coupling of transcription and translation also impairs the output of active and stable GFP.
Since the GFP band hardly overlaps with any of the S30 extract bands in an SDS gel, the total amount of the protein synthesized in vitro can be easily assessed by a densitometry analysis of the GFP band, in comparison with defined amounts of purified reference GFP added to the same gel. Furthermore, one stable S30 band was exploited to normalize the input variations of the S30 reaction mixture per lane (Figure 1a). With GFP we can determine, not only the total yield, but also the active fraction of the synthesized protein by measuring its fluorescence. After the GFP synthesis is finished, its fluorophore has to be folded properly, which is a criterion to detect active GFP by fluorescence using ultraviolet (UV) light excitation. GFP can emit green light even after electrophoresis under non-denaturing conditions (i.e. in the absence of SDS; Figure 1b), which allows an estimation of the amount of active molecules by comparing with a commercially available recombinant GFP reference that was in vivo expressed and assumed to be 100% active (Materials and Methods).
The assumption that the commercially available GFP is 100% active requires a more detailed consideration. GFP synthesized in vivo shows strikingly different active fractions depending on the organism, namely, whether GFP has been synthesized in E.coli or yeast (18). The low-active fraction in E.coli has been explained by the constraints on de novo folding, which is consistent with the assumption that the folding pathways of bacteria are largely post-translational. The authors postulated an ‘interference of GFP with adjacent domains during folding due to the particular topology of the β-barrel GFP structure’. In the same paper evidence was presented that the solubility nicely corresponds to the active fraction of GFP, regardless of whether GFP has been synthesized in yeast or E.coli [Figure 3 in Ref. (18)]. The fact that the commercially available GFP is perfectly soluble therefore justifies our assumption that it has 100% activity. Furthermore, under optimized conditions reported here, the activity of the synthesized GFP could be raised to 100%, but never significantly above this value. This observation adds further credit to the assumption of 100% activity of the reference GFP and indicates that the low-active fraction observed in E.coli is not an intrinsic feature of bacterial systems, but rather might have reasons additional to those mentioned in Ref. (18), namely the unfavorable usage of T7 polymerase routinely employed for inducing gene expression in E.coli (see below).
We can calculate the active fraction of synthesized GFP from both the amount of active GFP and the total amount synthesized (SDS gel). According to this procedure the active fraction was not higher than 30% in the experiment shown in Figure 1c, and (50 ± 20)% on average [see e.g. Ref. (20) and Figure 1b in Ref. (18)]. With the help of these quality criteria we aimed to improve the GFP active fraction up to 100%.
Synchronizing the reactions of transcription and translation
A tight coupling of the transcription and translation processes exists in bacteria: The E.coli RNAP proceeds with a speed of ∼60 nt per second, and a ribosomes initiating translation on the nascent chain of mRNA proceeds with a speed of ∼20 amino acids per second (21) corresponding to ∼20 codons (or equally to 60 nt) per second. It follows that the first ribosome pursues directly the transcriptase causing the tight coupling of transcription and translation and leaving no room for a significant gap between the transcriptase and the following ribosome. Therefore, the nascent mRNA chain cannot form secondary structures and thus complicate or even block translation elongation or transcription via R-loop formation (22). Moreover, the presence of ribosomes also protects the mRNA against endonucleolytic degradation (23).
One of the general problems of in vitro transcription–translation in bacterial cell-free systems is the uncoupling of the naturally coupled processes of transcription and translation. This results from the use of bacteriophage T7 RNAP at 37°C instead of E.coli RNAP (24). The T7 RNAP is five to eight times faster than E.coli RNAP, thus breaking the tight coupling between transcription and translation/ribosome assembly with two unfavorable consequences. (i) Strong secondary RNA structures can form that hinder the path of the translating ribosome over the mRNA probably impairing the co-translational folding and thus the yield of active proteins. (ii) R-loops can be formed, where nascent RNAs emerging from the RNAP exit channel can form heteroduplices with the upstream region of the template DNA strand. These heteroduplices can interfere not only with translation/assembly, but also with the next round of transcription or even with replication [for a review see Ref. (22)]. The result is that only a tiny fraction—within a few percent of the mRNA transcripts—are used for translation (23). Similarly, only a minor fraction of T7 RNAP transcripts of rRNA is used for the assembly of 50S subunits (25). The use of E.coli RNAP together with E.coli promoters is not an easy way to overcome this drawback, since most of the E.coli transcriptases are removed with the membranes, and the addition of the multi-subunit E.coli RNAP is a demanding task in terms of isolation and maintaining activity. Therefore, using the monomeric T7 polymerase with its high processivity is the decision of choice.
A number of slow T7 RNAP mutants have been reported (26,27) and from these mutants we utilized a double-mutant P266L/I810S that has a reduced transcription rate and yet still retains high processivity. The latter mutation slows the transcriptase and the former improves the processivity (4,27). Another double-mutant P266L/I810N used is 2.5 times slower than the WT T7 RNAP (J. Guillerez and M. Dreyfus, unpublished data).
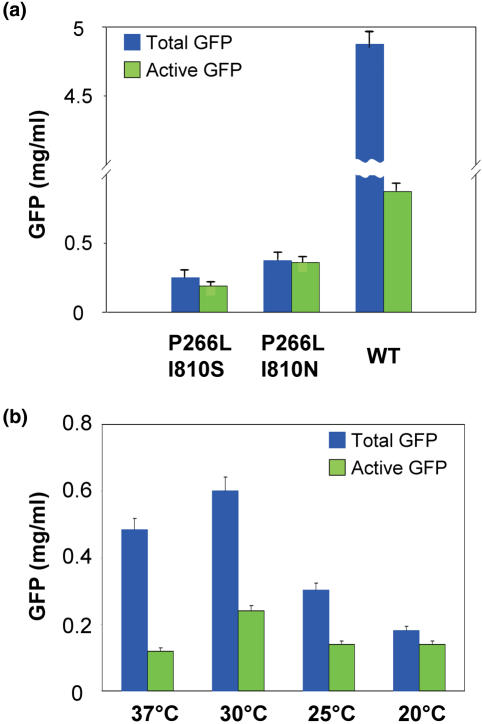
The SDS–PAGE analysis of the GFP production from the transcripts produced by either the double-mutants or by WT T7 RNAPs revealed that the yield of GFP dramatically decreased using the double-mutant variants and is almost undetectable in the case of the P266L/I810S mutant. On the contrary, the native-PAGE analysis of active GFP produced from these transcripts allows an easy detection of the GFP band in all cases, including the P266L/I810S double-mutant (summarized in Figure 2a). In spite of the low yield we can conclude that a reduction in the rate of transcription significantly improves the active fraction of the synthesized protein probably by re-establishing the coupling of transcription and translation.
Figure 2.
Total and active GFP synthesized at various conditions. Blue bars, total GFP synthesis; green bars, active GFP. (a) GFP synthesized after 20 h at 30°C of incubation in the presence of mutant T7 transcriptases and our reaction-mix preparation (semi-continuous system; 1 ml reaction volume; see Materials and Methods; the unusually low active fraction of almost 20% might be due to our preparation procedure of the S30 lysate). (b) GFP synthesis after 12 h in the RTS 100 (batch system; 10 μl reaction volume, wild-type T7 polymerase) at various incubation temperatures.
Another way of slowing down the T7 RNAP is by lowering the incubation temperature. A beneficial effect can be expected, if the reduced temperature slows down the translational rate to a lesser extent than the transcriptional rate, thus also improving the coupling of transcription and translation. In fact, in a similar case lowering the growth temperature from 37 to 25°C improved dramatically the T7-transcribed rRNA fraction assembled into active ribosomes (from 15 to 60%, respectively), which indicated that the rate of T7 RNAP goes down faster than the assembly rate (25).
GFP was synthesized at various temperatures (37, 30, 25 and 20°C; 12 h incubation, batch system) to examine the effect on both total protein synthesis and active GFP. The results are summarized in Figure 2b, from which it is clear that at low temperature in a batch system the total yield is two to three times reduced, but that the active fraction approaches 100% at 20°C. In order to test whether the beneficial effects of lowering the incubation temperature from 30 to 20°C is not restricted to the protein GFP and might be valid for other proteins as well, we performed a control experiment with luciferase. The results (Table 2) show that the specific activity of luciferase (activity per mass unit) is increased 2-fold, similar to the effects seen with GFP in Figures 2b and 5a.
Table 2.
Luciferase expression in a coupled transcription–translation system at 20 and 30°C
| [35S]Met incorporation (pixels) | Luciferase activity (lumino units) | Specific activity (lumino units/ [35S]Met pixels) × 1000 | Relative activity (%) | |
|---|---|---|---|---|
| 20°C | 299 899 ± 10% | 47 175 ± 2% | 157.3 | 100 |
| 30°C | 1 103 493 ± 3% | 94 515 ± 2% | 85.7 | 54 |
[35S]Met incorporation was determined by a PhosphorImager screen as the radioactivity in the luciferase band of a 15% PAGE, and the luciferase activity as described by the manufacturer (Promega). For details see Materials and Methods. GFP expression was determined as a control and showed about the same strong expression as seen in Figure 2b.
The results suggest that the rate of T7 polymerase is more affected by the lower temperature than the translation rate of ribosomes, thus re-establishing the coupling of transcription and translation and causing the beneficial effects observed. Furthermore, lowering the incubation temperature and thus the elongation rate has two beneficial effects on the active fraction. (i) The aggregation of synthesized GFP is prevented thus increasing the active fraction, as has been demonstrated at lower temperatures in vivo (28). The reason is that overexpression of proteins at 37°C in vivo might allow contacts of unfolded proteins, and these contacts of hydrophobic patches of unfolded proteins lead to aggregations and inclusion bodies. (ii) We are dealing with a second effect not seen previously with in vivo studies. Slowing down the elongation rate at 20°C improves the accuracy, since with the higher elongation rates at 30°C we observe a low-active fraction of ∼50% right from the very beginning of kinetic measurements (30 min; Figure 1a–c), where aggregation has not yet occurred.
Increasing the total protein yield
Two additional parameters might influence the yield of the synthesized protein: the stability of mRNA and the availability of building units such as NTP's and amino acids. These parameters are analyzed in this section. The half-lives of bacterial mRNAs at 37°C measure 1 (most labile mRNAs) to 7.5 min [ribosomal proteins mRNA (29)], with 2–3 min on average for most of the mRNAs in E.coli (30).
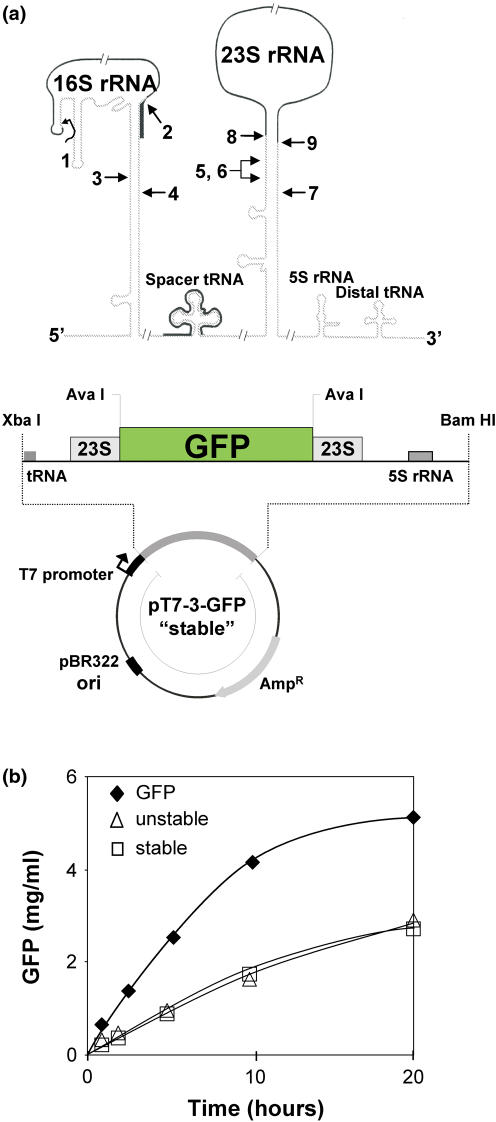
To test whether an increased mRNA stability improves the protein yield, we exploited the enormous stability of the ribosomal precursor RNA due to the long complementary sequences flanking the mature rRNA [(31); upper panel in Figure 3a]. We constructed a GFP-mRNA that is flanked by the highly conserved sequences enclosing the 23S rRNA and forms a strong base-paired stem resulting in a pseudo-circularization of the mRNA similar to that of the precursor rRNA (‘stable’ GFP-mRNA, lower panel in Figure 3a). We expected a prolonged mRNA half-life, since the endo RNase E prefers substrates with unpaired 5′ ends and the exo PNPase and RNase II are specific for single-stranded RNAs (32). As a control we used the same mRNA except that the mutation in the 5′ flanking region disrupts the complementarity and thus pseudo-circularized mRNA fails to be produced (‘unstable’ GFP-mRNA). The latter (in contrast to the former) has been shown to be resistant against in vitro RNase III cleavage, which is specific for secondary structures, thus revealing that no secondary structure has been formed (33). With both constructs, however, we observed levels of GFP production that were two times less than from our usual construct for GFP expression (Figure 3b). Owing to the low yield of GFP synthesis we did not pursue further the question whether indeed the ‘stable’ construct provided a more stable mRNA.
Figure 3.
Various mRNA constructs in the coupled transcription–translation system. (a) Upper panel, stability elements of rRNA genes. Schematic representation of an rrn operon and major processing steps of the 16S and 23S rRNA. The drawing is not to scale. Primary processing cleavages by RNase III (lanes 3–7), and secondary processing to produce the mature termini of 16S rRNA (lane 1, 5′ end; lane 2, 3′ end) and 23S rRNA (lane 8, 5′ end; lane 9, 3′ end). Solid lines indicate mature RNAs; modified (36). (a) Lower panel, map of a plasmid used for mRNA stability test; derivative of a vector that contained a fragment of the rrnB operon including intergenic spacers. GFP was incorporated into the 23S rRNA sequence at AvaI cleavage sites into a position between nt 250 and 2773 (E.coli numbering, starting from 5′ end of 23S rRNA). Restriction sites and rrnB operon elements are indicated. (b) Total GFP synthesis from different plasmid–DNA constructs (semi-continuous system; reaction volume 1 ml). Squares, GFP-mRNA flanked by the 23S rRNA stability elements (stable); triangles, same as squares but with destroyed stability elements (unstable); closed diamonds, standard expression vector for GFP synthesis (GFP). For further explanations see text.
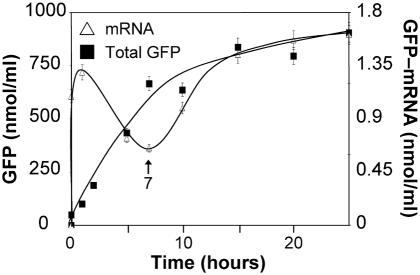
To test directly whether the mRNA stability in the coupled transcription–translation system is an issue, we determined the amount of GFP mRNA present during 25 h synthesis at 30°C via Northern blot hybridization and simultaneously the amount of synthesized GFP. The kinetics are shown in Figure 4. The rate of mRNA synthesis peaks after 1 h demonstrating the speed of T7 RNAP and then decreases, whereas the rate of GFP synthesis is maximal until the seventh hour of incubation. When the GFP synthesis ceases after the seventh hour, the amount of GFP-mRNA recovers again. These observations clearly indicate that NTPs are not limiting the reaction of transcription, which altogether confirm that neither synthesis of mRNA nor its half-life are limiting factors for protein synthesis in the bacterial cell-free system.
Figure 4.
Synthesis of GFP-mRNA and GFP in the RTS 500 system (1 ml reaction). Open triangles, amounts of GFP-mRNA synthesized and determined by Northern blotting; closed squares, total GFP synthesized.
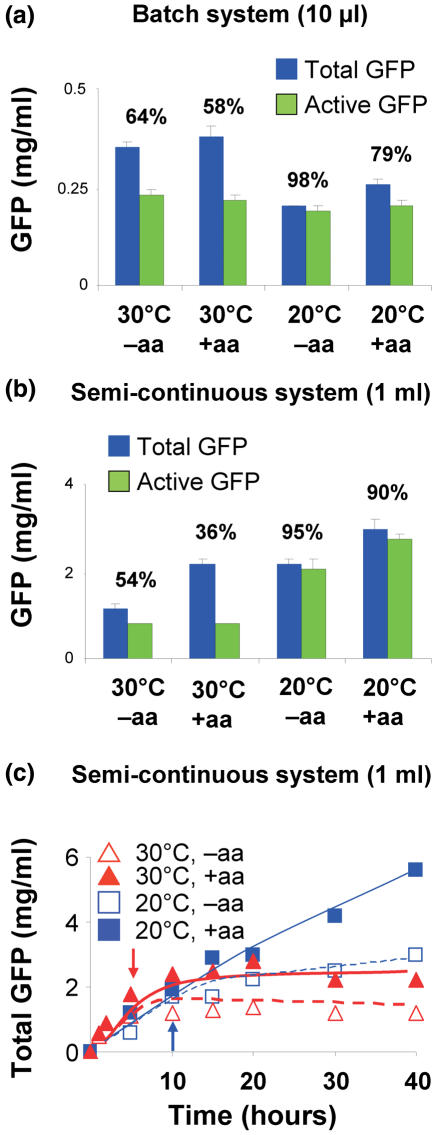
However, a shortage of amino acids could be the reason for a reduction of protein synthesis, since according to Jewett and Swartz (34) some amino acids are metabolized during the reaction of the cell-free system, e.g. cysteine, serine, threonine, glutamine and asparagine. Therefore, we added a mixture of all 20 amino acids in the middle of the incubation time. A little increase of total synthesis was observed in the batch system upon amino acid addition, whereas the amount of the active GFP remained unchanged (+aa; Figure 5a).
Figure 5.
GFP synthesis at 30 and 20°C with and without a second amino acid addition. (a) Batch system (10 μl). Blue bars, total GFP; green bars, active GFP; % values above bars. (b) Synthesis of GFP and active fraction after an incubation of 20 h. (c) Kinetics of GFP synthesis at 30°C (red triangles) and 20°C (blue squares, thick line). Closed symbols, amino acid additions (arrows) after 7 and 10 h at 30 and 20°C, respectively. Open symbols, no amino acid addition.
A strikingly different response to a second amino acid addition was seen in the semi-continuous system (1 ml reaction volume). At 30°C the total GFP was almost doubled without a concomitant increase of the active GFP thus reducing the active fraction to 36% (Figure 5b). However, the amounts synthesized at 20°C were a surprise, since they were larger than the corresponding 30°C values with and without a second addition of amino acids (incubation time 20 h), but with an active fraction of 90–95%. The best results were obtained with a second addition of amino acids after 10 h at 20°C incubation (Figure 5b and c). Kinetic analyses revealed that at 20°C the initial rate of GFP synthesis was slower, but the total amount even exceeded that at 30°C after 15 h incubation reaching values twice as large as the corresponding 30°C values (Figure 5c). A possible reason is that metabolization of amino acids runs faster at 30°C leading to a shortage of amino acids after 10–15 h in contrast to the situation at 20°C.
CONCLUSIONS
We show here that the efficiency of in vitro protein synthesis in a coupled transcription–translation system can be significantly increased to several mg per ml by incubating the reaction mix in a semi-continuous system at 20°C for ∼15 h and adding an amino acid mix after ∼10 h of the incubation. The protein synthesized is virtually 100% active, and therefore the low-cost bacterial system can be used under these conditions (i) for structural analysis such as crystallography or NMR after incorporation of, for example, 13C and 15N isotopes during the incubation and (ii) for folding and functional studies.
The optimization of the coupled system as it stands after these analyses can still be pushed forward. Let us compare the efficiency of the RTS (Roche) used here with that of an E.coli cell. The reaction mix before synthesis contains ∼40 mg/ml total protein, and after 10 h the amount of GFP synthesized approaches ∼10% of the total protein (4 mg/ml; Figure 1c). A continuation of this synthesis rate would lead to a doubling of the protein content in the reaction mixture (total proteins plus synthesized GFP) after 100 h. E.coli has a doubling time of 20 min under rich medium conditions, where it doubles the total protein synthesized and distributes it over the two daughter cells. It follows that the system used here is still 300-fold less efficient than protein synthesis in vivo in agreement with a recent report showing that the bulk protein synthesis rate in an optimized E.coli-based cell-free system is 200-fold slower than the in vivo rate (35).
Acknowledgments
We thank Dr Daniel N. Wilson for help and discussions. This work was supported by the BMBF, Project No. 031 2552 ‘Neue Anwendungspotentiale der in vitro-Proteinsynthese’, Teilprojekt AB, Gruppe Nierhaus. Proteomaster and RTS are trademarks of Roche. Funding to pay the Open Access publication charges for this article was provided by MPI für Molekulare Genetik.
Conflict of interest statement. None declared.
REFERENCES
- 1.Spirin A.S., Baranov V.I., Ryabova L.A., Ovodov S.Y., Alakhov Y.B. A continuous cell-free translation system capable of producing polypeptides in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 2.Kim D.M., Choi C.Y. A semi-continuous prokaryotic coupled transcription/translation system using a dialysis membrane. Biotechnol. Progr. 1996;12:645–649. doi: 10.1021/bp960052l. [DOI] [PubMed] [Google Scholar]
- 3.Jewett M.C., Voloshin A., Swartz J.R. Prokaryotic systems for in vitro expression. In: Weiner M.P., Lu Q., editors. Gene Cloning and Expression Technologies. Westborough, MA: BioTechniques Press; 2002. pp. 391–411. [Google Scholar]
- 4.Guillerez J., Lopez P.J., Proux F., Launay H., Dreyfus M. A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc. Natl Acad. Sci. USA. 2005;102:5958–5963. doi: 10.1073/pnas.0407141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He B., Rong M., Lyakhov D., Gartenstein H., Diaz G., Castagna R., McAllister W.T., Durbin R.K. Rapid mutagenesis and purification of phage RNA polymerases. Protein Express. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 6.Kim D.M., Swartz J.R. Prolonging cell-free protein synthesis by selective reagent additions. Biotechnol. Prog. 2000;16:385–390. doi: 10.1021/bp000031y. [DOI] [PubMed] [Google Scholar]
- 7.Crameri A., Whitehorn E.A., Tate E., Stemmer W.P. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli U.K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 9.Maniatis R.B., Fritsch E.F., Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 1982. [Google Scholar]
- 10.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 11.Yang F., Moss L.G., Phillips G.N., Jr The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 12.Ormo M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., Remington S.J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 13.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 14.Ward W.W., Bokman S.H. Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 15.Makino Y., Amada K., Taguchi H., Yoshida M. Chaperonin-mediated folding of green fluorescent protein. J. Biol. Chem. 1997;272:12468–12474. doi: 10.1074/jbc.272.19.12468. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa H., Inouye S., Tsuji F.I., Yasuda K., Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc. Natl Acad. Sci. USA. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerner M.J., Naylor D.J., Ishihama Y., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Chang H.C., Kaiser C.M., Hartl F.U., Barral J.M. De novo folding of GFP fusion proteins: high efficiency in eukaryotes but not in bacteria. J. Mol. Biol. 2005;353:397–409. doi: 10.1016/j.jmb.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 19.Sacchetti A., Cappetti V., Marra P., Dell'Arciprete R., El Sewedy T., Crescenzi C., Alberti S. Green fluorescent protein variants fold differentially in prokaryotic and eukaryotic cells. J. Cell Biochem. 2001;81:117–128. doi: 10.1002/jcb.1091. [DOI] [PubMed] [Google Scholar]
- 20.Dinos G., Wilson D.N., Teraoka Y., Szaflarski W., Fucini P., Kalpaxis D., Nierhaus K.H. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site tRNA binding. Mol. Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 21.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F.C., Curtiss R. III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikow W.S., Riley M., Schaechter M., Umbarger H.E., editors. Escherichia coli and Salmonella, 2nd edn. Vol. 2. Washington DC: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 22.Gowrishankar J., Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol. Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 23.Iost I., Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme. J. Biol. Chem. 1973;248:2235–2244. [PubMed] [Google Scholar]
- 25.Lewicki B.T.U., Margus T., Remme J., Nierhaus K.H. Coupling of rRNA transcription and ribosomal assembly in vivo—formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage-T7 RNA polymerase. J. Mol. Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 26.Bonner G., Lafer E.M., Sousa R. Characterization of a set of T7 RNA polymerase active site mutants. J. Biol. Chem. 1994;269:25120–25128. [PubMed] [Google Scholar]
- 27.Makarova O.V., Makarov E.M., Sousa R., Dreyfus M. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl Acad. Sci. USA. 1995;92:12250–12254. doi: 10.1073/pnas.92.26.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlieker C., Bukau B., Mogk A. Prevention and reversion of protein aggregation by molecular chaperones in the E.coli cytosol: implications for their applicability in biotechnology. J. Biotechnol. 2002;96:13–21. doi: 10.1016/s0168-1656(02)00033-0. [DOI] [PubMed] [Google Scholar]
- 29.Mohanty B.K., Kushner S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 30.Selinger D.W., Saxena R.M., Cheung K.J., Church G.M., Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liiv A., Tenson T., Remme J. Analysis of the ribosome large subunit assembly and 23S rRNA stability in vivo. J. Mol. Biol. 1996;263:396–410. doi: 10.1006/jmbi.1996.0584. [DOI] [PubMed] [Google Scholar]
- 32.Carpousis A.J., Dreyfus M. In: Protein Synthesis and Ribosome Structure. Nierhaus K.H., Wilson D.N., editors. Weinheim: Wiley-VCH Verlag GmbH & Co.; 2004. pp. 185–206. [Google Scholar]
- 33.Liiv A., Remme J. Importance of transient structures during post-transcriptional refolding of the pre-23S rRNA and ribosomal large subunit assembly. J. Mol. Biol. 2004;342:725–741. doi: 10.1016/j.jmb.2004.07.082. [DOI] [PubMed] [Google Scholar]
- 34.Jewett M.C., Swartz J.R. Substrate replenishment extends protein synthesis with an in vitro translation system designed to mimic the cytoplasm. Biotechnol. Bioeng. 2004;87:465–472. doi: 10.1002/bit.20139. [DOI] [PubMed] [Google Scholar]
- 35.Underwood K.A., Swartz J.R., Puglisi J.D. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol. Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 36.Srivastava A.K., Schlessinger D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 1990;44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]