Abstract
We synthesized C5-modified analogs of 2′-deoxyuridine triphosphate and 2′-deoxycytidine triphosphate and investigated them as substrates for PCRs using Taq, Tth, Vent(exo-), KOD Dash and KOD(exo-) polymerases and pUC 18 plasmid DNA as a template. These assays were performed on two different amplifying regions of pUC18 with different T/C contents that are expected to have relatively high barriers for incorporation of either modified dU or dC. On the basis of 260 different assays (26 modified triphosphates × 5 DNA polymerases × 2 amplifying regions), it appears that generation of the full-length PCR product depends not only on the chemical structures of the substitution and the nature of the polymerase but also on whether the substitution is on dU or dC. Furthermore, the template sequence greatly affected generation of the PCR product, depending on the combination of the DNA polymerase and modified triphosphate. By examining primer extension reactions using primers and templates containing C5-modified dUs, we found that a modified dU at the 3′ end of the elongation strand greatly affects the catalytic efficiency of DNA polymerases, whereas a modified dU opposite the elongation site on the template strand has less of an influence on the catalytic efficiency.
INTRODUCTION
Functional DNA molecules, such as DNA aptamers and DNAzymes with functions similar to antibodies and enzymes (1–6) could be useful as research tools for molecular biology and as indicators of specific substances for the analysis of clinical and food samples. Recent progress has been made at introducing additional functionalities into these DNA molecules to diversify their function and improve their activity (7–18). Such functionally modified DNAs can be produced by in vitro selection or SELEX (systematic evolution of ligands by exponential enrichment) techniques involving screening of the sequences with desired activity from large pools of random sequences and amplification of the selected pools by PCR (19–23).
Modified DNAs can be efficiently prepared from modified 2′-deoxynucleoside triphosphates in the absence of the corresponding natural substrates by symmetric PCR, which is capable of direct exponential amplification of DNA (24–36). Although, the production of modified DNAs is limited by the substrate specificity of the DNA polymerases, there are many examples of the enzymatic preparation of modified DNAs by primer extension or one-primer PCR from modified 2′-deoxynucleoside triphosphates or by symmetric PCR under coexistence of the corresponding natural substrates (37–44). Symmetric PCR is the most convenient method for preparing modified DNAs that can be subjected to in vitro selection. Thus, in the current study, we synthesized a variety of 2′-deoxynucleoside triphosphates to systematically analyze how the chemical structures of modified bases affect the synthesis of modified DNAs by symmetric PCR. We also examined the influence of the type of DNA polymerase on the production of modified DNAs. Finally, to identify the critical step in the synthesis of modified DNA, we measured single nucleotide insertion under single turnover conditions (45–52) by using primers containing modified dUs at their 3′ ends and templates containing modified dUs adjacent to the elongation site.
MATERIALS AND METHODS
General
A TC-312 thermal cycler (Techne) and an ALB-300 thermo-regulated bath (Iwaki) were used for PCR assays and kinetic experiments, respectively. PCR products were resolved by denaturing PAGE using a vertical electrophoresis unit(Atto) at 4°C. Reaction mixtures from kinetic studies were resolved by denaturing PAGE using a vertical electrophoresis unit (Nihon eido) at 36°C in an M-260F incubator (Taitech). Bands were imaged with a Molecular Imager® FX (Bio-Rad) equipped with an external laser module and quantified with Quantity One® software (Bio-Rad).
Materials
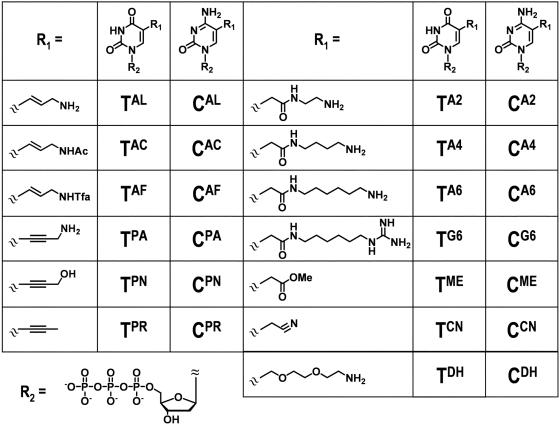
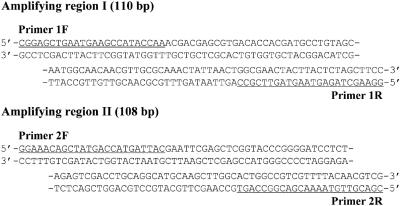
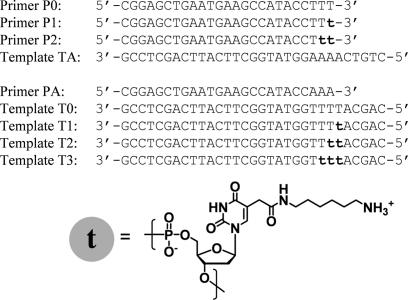
The following commercial available thermostable DNA polymerases were purchased: Taq (Takara Bio), Tth (Toyobo), Vent(exo-) (New England Biolabs) and KOD Dash (Toyobo). KOD(exo-) DNA polymerase was generously donated by Toyobo. Natural 2′-deoxynucleoside-5′-triphosphates(dATP, dGTP, dCTP and TTP) were obtained from Roche Diagnostics. All modified nucleoside triphosphates used for PCR assays are listed in Table 1. TAL, TPR, CAL and CPR were purchased from TriLink BioTechnologies; TPA was from Geneact; and CPA was from JENA Bioscience. The other modified nucleoside triphosphates were synthesized (see Supplementary Schemes 1 and 2). Primers 1F, 1R, 2F and 2R were purchased from JBioS, and pUC18 template DNA was obtained from TaKaRa Biomedicals. Primer sequences and the sequences of the two amplifying regions on pUC18 used for PCR assays are shown in Figure 1. Primers P0, P1, P2, PA and templates TA, T0, T1, T2 and T3 used for kinetic experiments were purchased from Geneact. The sequences of these oligonucleotides are shown in Figure 2. Primers P1 and P2 and templates T1, T2 and T3 contained more than one modified dU [5-(2-(6-aminohexylamino)-2-oxoethyl-2′-deoxyuridine]. These oligodeoxynucleotides were obtained by chemical oligomerization using the C5-modified dU amidite prepared according to our published procedure (53). To detect and quantify extension products, the 5′ ends of 1F, 2F, P0, P1, P2 and PA were labeled with 6-carboxyfluorescein (6-FAM). Plasmid DNAs for sequence analyses were prepared using pT7Blue T-vector and DNA ligation kit ver. 1 from Takara Bio, a competent cell kit (Competent high DH5) from Toyobo and a plasmid DNA purification kit (LaboPass™ Mini) from Cosmo Genetech.
Table 1.
Chemical structures and abbreviations of modified nucleoside triphosphates
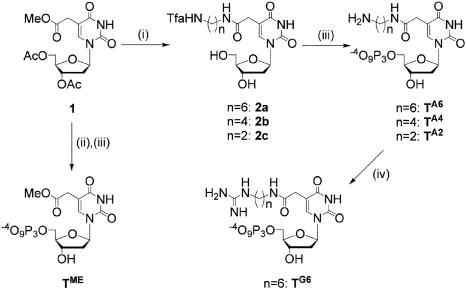
Scheme 1.
Synthesis of C5-modified dUTP analogs: (i) H2N-(CH2)n-NH2 (n = 2, 4, 6), methanol, 50°C, 2 h overnight, followed by ethyl trifluoroacetate, triethylamine, room temperature (rt), 1.5–3 h; (ii) sodium methoxide, methanol, rt, 1.5 h; (iii) POCl3, N,N,N′,N′-tetramethyl-1, 8-naphthalendiamine, trimethyl phosphate, 0°C, 45 min, followed by n-tributylamine pyrophosphate, dimethylformamide, rt, 1 h; (iv) S-ethylthiourea, triethylamine, dimethylformamide, rt, 4 h.
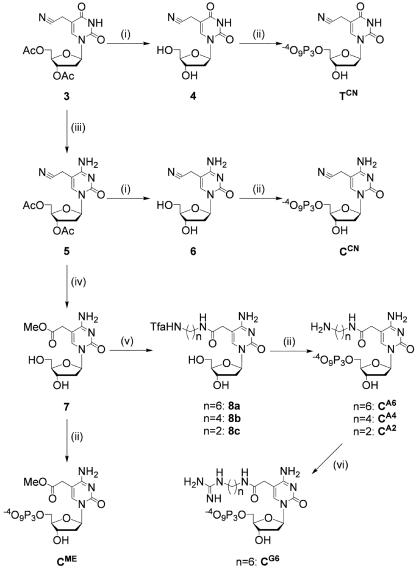
Scheme 2.
Synthesis of C5-modified analogs of dUTP and dCTP: (i) 7 M aqueous ammonia, rt, 1 h; (ii) POCl3, N,N,N′,N′-tetramethyl-1, 8-naphthalendiamine, trimethyl phosphate, 0°C, 45 min, followed by n-tributylamine pyrophosphate, dimethylformamide, rt, 1 h; (iii) POCl3, dry pyridine, rt, 4 h, then concentrated aqueous ammonia, 50°C, 2 h; (iv) 1 N aqueous KOH, 86°C, 1 h; chlorotrimethylsilane, dry methanol, 31°C, 1.5 h; (v) H2N-(CH2)n-NH2 (n = 2, 4, 6), methanol, 50°C, 2 h overnight, followed by ethyl trifluoroacetate, triethylamine, rt, 1.5–3 h; (vi) S-ethylthiourea, triethylamine, dimethylformamide, rt, 4 h.
Figure 1.
Sequences of amplifying regions of the pUC18 template DNA. Primer sequences are underlined.
Figure 2.
Sequences of primers and templates used for standing-start experiments. ‘t’ indicates 5-(2-(6-aminohexylamino)-2-oxoethyl)-dU.
Modified 2′-deoxynucleoside-5′-triphosphates
We used 26 different modified triphosphates for PCR assays, of which we synthesized the following 20 analogs: TAC, TAF, TPN, TA2, TA4, TA6, TG6, TME, TCN, TDH, CAC, CAF, CPN, CA2, CA4, CA6, CG6, CME, CCN and CDH. The synthetic routes for TA2, TA4, TA6, TG6, TME, TCN, CA2, CA4, CA6, CG6, CME and CCN are shown in Schemes 1 and 2. Analogs TA2, TA4, TA6 and TG6 were synthesized from 2′-deoxyuridine analog 1 bearing a 2-methoxy-2-oxoethyl group at the C5 position (53). Analogs TCN, CA2, CA4, CA6, CG6, CME and CCN were synthesized from 2′-deoxyuridine analog 3 bearing a cyanomethyl group at C5 position (54). We previously reported synthesis of starting material 1 and modified triphosphates TA2, TA6, TG6, TME, TDH and CDH (27,29,35). Detailed synthetic procedures and spectroscopic data including intermediates are provided in the Supplementary Data.
PCR assays
PCR experiments were performed in a 20 μl reaction volume containing 10 ng of pUC18 template DNA, 0.4 μM of each primer (1F and 1R or 2F and 2R), an appropriate concentration of thermostable DNA polymerase, reaction buffer supplied with enzyme (at 1× concentration), and nucleoside triphosphates at 200 μM each. A PCR with natural nucleoside triphosphates (dATP, dGTP, dCTP and TTP) was used as a positive control. The PCR assays were performed with one of the dUTP analogs (TAL, TAC, TAF, TPA, TPN, TPR, TA2, TA4, TA6, TG6, TME, TCN or TDH) in place of TTP; a PCR with dATP, dGTP, dCTP, and water in place of TTP was used as a negative control. Also, the PCR assays were performed with one of the dCTP analogs (CAL, CAC, CAF, CPA, CPN, CPR, CA2, CA4, CA6, CG6, CME, CCN or CDH) instead of dCTP; a PCR with dATP, dGTP, TTP, and water in place of dCTP was used as a negative control. The final concentration of a thermostable DNA polymerase in each reaction mixture was 0.025 U/μl for Taq, 0.025 U/μl for Tth, 0.050 U/μl for Vent(exo-), 0.025 U/μl for KOD Dash and 0.025 U/μl for KOD(exo-). At an enzyme concentration of 0.025 U/μl, the high polymerase activity of KOD Dash and KOD(exo-) polymerases caused an overreaction and decreased the yield of the PCR product for the positive controls; therefore, we reduced their concentrations to 0.0050 U/μl to provide an appropriate yield of the PCR product. All amplifications were performed with a hot start (1 min at 94°C), followed by amplification and final incubation for 5 min at 74°C. The amplification was carried out for 20 cycles of denaturation for 0.5 min at 94°C, annealing for 0.5 min at 52°C, and extension for 1 min at 74°C. The PCR products were resolved by denaturing PAGE, and gel images were recorded with excitation of the 5′-labeled fluorophore at 488 nm. To visualize molecular markers, ethidium bromide-stained gel images excited at 532 nm were also recorded (Figure 3). The relative yield of the full-length PCR product was calculated from the intensity of each band on gel images visualized by detection of the 5′-labeled fluorophore. The amount of full-length PCR product formed during the corresponding positive control was set at 1, and the calculated relative yields were the averages of three to five independent experiments. To achieve proper comparisons of the yields in the positive controls with those in other reactions, positive control reactions were performed under the conditions that would not entirely consume the 6-FAM primer; about 10–35% of the primer remained after the PCR as shown in 6-FAM images of the PCR product (see Supplementary Figure 1). In the positive controls, amounts of PCR products were estimated to be about 5.2–7.2 pmol per 20 μl of a reaction solution, and the amplified products were about 900- to 1200-fold from the template.
Figure 3.
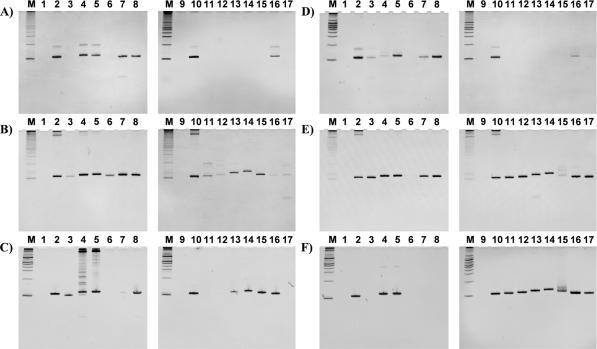
Representative ultraviolet images from ethidium bromide-stained PAGE gels of the 110 nt PCR product derived from pUC18 (amplifying region I) and the C5-modified nucleoside triphosphates. (A–C) The reaction mixtures containing dATP, dGTP, dCTP and TAL (lane 3), TAC (lane 4), TAF (lane 5), TPA (lane 6), TPN (lane 7), TPR (lane 8), TA2 (lane 11), TA4 (lane 12), TA6 (lane 13), TG6 (lane 14), TME (lane 15), TCN (lane 16), or TDH (lane 17) were separated by electrophoresis on denaturing PAGE. Except for the positive control, the reaction mixtures do not contain natural TTP. The PCR product was generated by the positive control (reaction mixture containing dATP, dGTP, dCTP and TTP; lanes 2 and 10) but not by the negative control (reaction mixture containing dATP, dGTP and dCTP; lanes 1 and 9). (D–F) Reaction mixtures containing dATP, dGTP, TTP and CAL (lane 3), CAC (lane 4), CAF (lane 5), CPA (lane 6), CPN (lane 7), CPR (lane 8), CA2 (lane 11), CA4 (lane 12), CA6 (lane 13), CG6 (lane 14), CME (lane 15), CCN (lane 16) or CDH (lane 17) were separated by electrophoresis on denaturing PAGE. Except for the positive control, the reaction mixtures did not contain natural dCTP. The PCR product was generated by the positive control (reaction mixture containing dATP, dGTP, dCTP and TTP; lanes 2 and 10) but not by the negative control (reaction mixture containing dATP, dGTP and TTP; lanes 1 and 9). The thermostable DNA polymerases used were Taq (A, D), Vent(exo-) (B and E), and KOD Dash (C and F).
Sequence analyses of PCR products
To analyze sequences of PCR products, the modified DNAs were converted to natural-type DNAs and incorporated into plasmid DNA as we have described previously (36). The modified DNA was purified by gel electrophoresis and eluted from the gel into TBE buffer (45 mM Tris–borate and 1 mM EDTA). The eluted sample was converted into natural DNA by a second PCR using the same polymerase as used for the PCR assay. After purification by gel electrophoresis and elution, the double-stranded DNA product was re-amplified by a third PCR using Taq DNA polymerase and subjected to the TA Cloning® method, which takes advantage of the nontemplate-dependent addition of a single 2′-deoxyadenosine at the 3′ end of the PCR product by Taq. The third PCR was omitted in the case of modified DNAs generated by the first PCR (PCR assay) using Taq. PCR products with a single dA at the 3′ end were inserted into p7Blue T-vector using a DNA ligation kit, after which cloning was performed using the competent cell kit according to the manufacturer's instructions. For sequencing, three samples were randomly chosen from the single colonies derived from modified DNA. Plasmids were extracted from the chosen single colonies and purified with a plasmid purification kit. Purified plasmids were sequenced by the Takara Bio gene analysis service.
Kinetic experiments
To study how a modified dU on the primer elongation terminus (3′ end) affects the DNA polymerase reaction, we performed standing-start experiments by using template DNA TA and 5′-(6-FAM)-labeled primers (P0, P1 or P2). Vent(exo-) DNA polymerase, which lacks 3′,5′ exonuclease activity, was used as the enzyme. The reaction was performed at 40°C because the reactions proceeded too fast to be monitored at the optimal temperature of the enzyme (∼75°C). These three primers (P0–2) have the same sequence, but P1 contains one modified dU (‘t’) and P2 has two consecutive modified dUs (‘tt’) at each 3′ end of the primers (Figure 2). Reaction mixtures (18 μl) containing one of the primers (P0, P1 or P2) at 0.5 μM, template DNA TA at 0.625 μM, an appropriate concentration (5–200 μM) of thymidine or thymidine analog triphosphate (TTP or TA6), and reaction buffer supplied with enzyme (at 1× concentration) were denatured at 95°C for 1 min with the thermal cycler and then annealed at room temperature for 1 h. Subsequently, enzyme solution (0.015–0.25 U) was added to the mixture, and the reaction tube was quickly placed in the thermo-regulated bath and incubated at 40°C during the primer extension reaction. After the reactions were started, reaction tubes were removed from the bath chronologically and immediately quenched by freezing in liquid nitrogen. The frozen reaction mixtures were then mixed with 4 μl of 40 mM EDTA containing 0.1% bromophenol blue and 24 μl of 7 M urea containing 3 mM EDTA and then melted into a homogeneous solution by vortexing. The sample solutions were resolved by denaturing PAGE, and gel images were recorded on the imager. The amount of reactant and product was measured from the intensity of each band with excitation at 488 nm to visualize the 5′-labeled fluorophore.
Similarly, to study the effect of modified dU on the template strand adjacent to the elongation terminus on the polymerase reaction, we performed standing-start experiments by using template DNAs (T0, T1, T2 or T3) and 5′-(6-FAM)-labeled primer PA. These four template DNAs (T0–3) have the same sequence, but T1 contains one modified dU (‘t’), T2 has two consecutive modified dUs (‘tt’), and T3 has three consecutive modified dUs (‘ttt’) near the opposite site of the primer's 3′-terminus (Figure 2). Reaction mixtures (18 μl) were prepared containing primer PA at 0.5 μM, one of the template DNAs (T0, T1, T2 or T3) at 0.625 μM, an appropriate concentration (10–800 μM) of dATP, and reaction buffer supplied with enzyme (at 1× concentration) in a 500 μl reaction tube. The sample was denatured at 95°C for 1 min with the thermal cycler and then annealed at room temperature for 1 h. Subsequently, solution containing Vent(exo-) DNA polymerase (0.015 U) was added, and the reaction tube was quickly placed in the thermo-regulated bath and incubated at 40°C to carry out the primer extension reaction. After the reactions started, the reaction tubes were removed from the bath chronologically and quenched immediately by freezing in liquid nitrogen. Sample preparation, PAGE and detection of bands were performed as described above. The amount of reactant and product was determined from the intensity of each band as visualized by detection of the 5′-labeled fluorophore. The initial rate and kinetic parameters of the polymerase reaction were determined from the time-dependent changes of these band intensities.
RESULTS
Comparison of substrate properties of various modified triphosphates in PCRs
We performed PCR assays to investigate the effects of template sequences, kinds of polymerases, and chemical structures of modified bases on the production of modified DNA by PCR. To examine the effect of template sequences, PCRs were made both on amplifying region I, flanked by primers 1F and 1R, and on amplifying region II, flanked by primers 2F and 2R (Figure 1). Excluding the primer sequences, amplifying region I of the double-stranded template (110 bp) contains 47 modified dU insertion sites (T content 27%) and 45 modified dC insertion sites (C content of 26%). The modified dU insertion sites include 10 instances of 2 consecutive T nucleotides (‘TT’) and 2 instances of 3 consecutive T nucleotides (‘TTT’). The modified dC insertion sites include 5 instances of 2 consecutive C nucleotides (‘CC’). Excluding the primer sequences, amplifying region II of the double-stranded template (108 bp) contains 40 modified dU insertion sites (T content of 24%) and 48 modified dC insertion sites (C content of 29%). The modified dU insertion sites include 4 instances of 2 consecutive T nucleotides (‘TT’), 1 of 3 consecutive T nucleotides ‘TTT’ and 1 of 4 consecutive T nucleotides (‘TTTT’). The modified dC insertion sites involve 7 instances of 2 consecutive C nucleotides (‘CC’), 1 of 3 consecutive C nucleotides (‘CCC’) and 1 of 4 consecutive nucleotides (‘CCCC’). Inferring from the of the T or C content and the number of sequences with consecutive T or C nucleotides on the template, region I has a relatively high barrier for incorporation of modified dU, and region II has a relatively high barrier for incorporation of modified dC.
To investigate the effect of kinds of polymerases, we examined five kinds of thermostable polymerases belonging to families A (Taq and Tth) and B (Vent(exo-), KOD Dash and KOD(exo-)). As examples, we have provided ultraviolet images from ethidium bromide-stained PAGE gels of the 110 nt PCR product derived from pUC18 (amplifying region I) and the C5-modified nucleoside triphosphates in Figure 3. Although the relative yields of full-length products were calculated from the PAGE gel images by 6-FAM detection, they almost corresponded to those from the ultraviolet images in main cases (see Supplementary Figure 1). Furthermore, in this experiment, a PCR that provides the product even in a relative yield of 10% can be estimated to achieve at least 90-fold amplification. Therefore, the amount of full-length product obtained from the forward primer labeled with 6-FAM would be as much as that from the reverse primer with no labeling. As longer strands, DNA fragments, in principle, are heavily stained by ethidium bromide and observed as a thicker band. For example, the bands with a slow mobility on lanes 4 and 5 in Figure 3C may be conspicuous, however, they are found to be minor products according to the corresponding PAGE gel images by 6-FAM detection. These long fragments could be byproducts from overreactions, because they were not produced if the enzyme concentration or number of reaction cycles was reduced (data not shown).
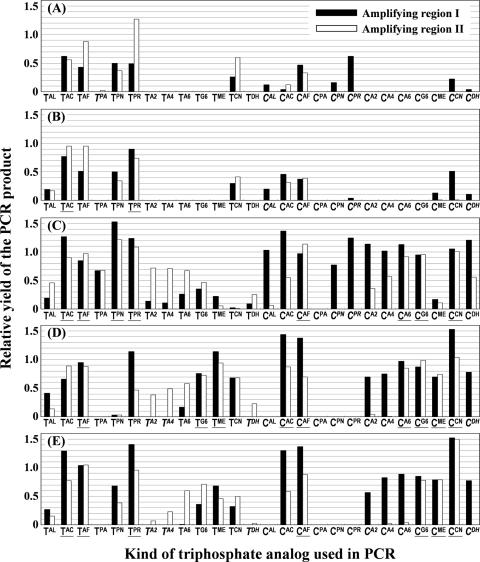
As shown in Figure 4, 20 combinations of enzyme and triphosphate (e.g. Taq/CPR and Vent(exo-)/CPN) were greatly affected by the template sequences. The PCRs using these combinations generated modified DNAs only from region I or II. Overall, it appeared that combinations resulting in lower relative yields likely depend on the template sequences. In contrast, of 130 combinations, the 26 combinations underlined in Figure 4 generated the products with higher relative yields (>70%) from both region I and II; PCR products were generated by only 2 (Tth/TAC and Tth/TPR) of the 52 possible combinations with family A polymerases but with 24 (Vent(exo-)/TAC, TAF, TPN, TPR, CAF, CA6, CG6, CCN; KOD Dash/TAF, TG6, TME, CAC, CAF, CA6, CG6, CME, CCN; KOD(exo-)/TAC, TAF, TPR, CAF, CG6, CME, CCN) of the 78 combinations with family B polymerases. These results suggest that the family B polymerases have broader substrate specificities than the family A polymerases. This is consistent with a previous report by Held et al. (28) describing the substrate properties of some C5-modified dUTPs with C≡C (triple bond) and C-C (single bond) linker arms for thermostable DNA polymerases. The present study clearly demonstrates a similar tendency in substrate specificities for the dCTP analogs.
Figure 4.
Relative yield of the modified DNAs generated by PCR using various triphosphate analogs together with (A) Taq DNA polymerase, (B) Tth DNA polymerase, (C) Vent(exo-) DNA polymerase, (D) KOD Dash DNA polymerase and (E) KOD(exo-) DNA polymerase. The x-axis indicates the kind of triphosphate analog used, and the y-axis indicates the relative yield of the PCR product. The black and white bars indicate the relative yield of the PCR product generated from amplifying regions I and II, respectively. The relative standard deviations were less than ±6% for all reactions.
Among family B polymerases, Vent(exo-) was found to have broader substrate specificities than KOD Dash and KOD(exo-) because the former accepted TPA, CAL, CPN and CPR as a PCR substrate but the latter did not. KOD(exo-) is an enzyme genetically engineered to eliminate the 3′,5′ exonuclease activity from KOD and KOD Dash is a mixture of KOD and KOD(exo-). In our previous study (27), Vent(exo-) was found to be more effective for the synthesis of modified DNA by PCR than Vent (which has 3′,5′ exonuclease activity). Therefore, we expected that KOD(exo-) would tolerate broader modifications at the C5 position of pyrimidine nucleotides than KOD Dash; however, we did not observe a significant difference in substrate specificity between the two polymerases.
We compared the relative yields of PCR products from the modified pyrimidine nucleotides to clarify the effect of the chemical structure of the substituent groups (Figure 4). Among six analogs with (E)-prop-1-enyl linkers, those with an amino protecting group, such as an N-trifluoroacetyl and N-acety group (TAC, TAF, CAC and CAF) were found to be better substrates than analogs with an amino group (TAL and CAL) for PCR by all of the DNA polymerases examined. Because the amino group in TAL and CAL is protonated under the PCR conditions, the results indicate that an electrostatic effect rather than steric hindrance between the enzyme and the substrate likely causes a dominant-negative effect on the production of modified DNA. A similar effect was also observed for dUTP analogs with prop-1-ynyl linkers. TPA, which has an amino group, generated a far lower yield of PCR products than TPN and TPR, which lack amino groups. Among the analogs with 2-oxoethyl linkers, those with longer linkers were found to be superior PCR substrates in main cases (TA2, TA4, TA6, CA2, CA4 and CA6; Figure 4C and E). Analogs with shorter linkers may be more negatively affected by electrostatic interactions, because the cation is located closer to the nucleobase. Furthermore, the relative yields of PCR products varied considerably depending on whether the substitution was on dU or dC. For example, in most cases, the dUTP analogs with a prop-1-ynyl linker arm (TPA, TPN and TPR) were better PCR substrates than the corresponding dCTP analogs (CPA, CPN and CPR). In contrast, the dCTP analogs with a 2-oxoethyl linker arm (CA2, CA4, CA6, CG6 and CME) were better substrates than the corresponding dUTP analogs (TA2, TA4, TA6, TG6 and TME).
Verification of accurate incorporation of nucleotides in the PCR products
To determine which nucleotides were accurately and effectively incorporated, we sequenced the modified DNAs that were generated by PCR using the 32 combinations of enzyme and triphosphates. Three colonies were randomly selected for each combination, and the results of the sequence analyses are shown in Table 2. The original sequence was preserved in all samples prepared from positive controls in PCR assays according to the above mentioned protocol. Therefore, the mutations found do not simply reflect the nature of polymerase used but rather interactions between modified groups and polymerases.
Table 2.
Verification of accurate incorporation of nucleotides in the PCR products
| DNA polymerase | Clone | Basea | Mutationb | Clone | Basea | Mutationb | Clone | Basea | Mutationa |
|---|---|---|---|---|---|---|---|---|---|
| Taq | |||||||||
| 1 | TAF | None | 7 | TPR | None | 13 | CPR | None | |
| 2 | TAF | None | 8 | TPR | None | 14 | CPR | None | |
| 3 | TAF | None | 9 | TPR | None | 15 | CPR | 61(A/T)→(G/C) | |
| 4 | TCN | None | 10 | CAF | None | 16 | CCN | None | |
| 5 | TCN | None | 11 | CAF | None | 17 | CCN | None | |
| 6 | TCN | None | 12 | CAF | None | 18 | CCN | None | |
| Tth | |||||||||
| 19 | TAF | None | 24 | TPR | 50(G/C)→(A/T) | 29 | CAF | None | |
| 20 | TAF | None | 25 | TCN | None | 30 | CAF | None | |
| 21 | TAF | 45(T/A)→(C/G) | 26 | TCN | None | 31 | CCN | None | |
| 22 | TPR | None | 27 | TCN | None | 32 | CCN | None | |
| 23 | TPR | None | 28 | CAF | None | 33 | CCN | 59 (A/T)→(G/C) | |
| Vent(exo-) | |||||||||
| 34 | TAF | 78 (G/C)→(T/A) | 47 | CPR | 39 (G/C)→(A/T) | 58 | CCN | 44 (G/C)→(A/T) | |
| 35 | TAF | 31 (T/A)→(A/T) | 48 | CPR | 40 (C/G)→(T/A) | 59 | CCN | 44 (G/C)→(A/T) | |
| 36 | TAF | 70 (G/C)→(T/A) | 57 (C/G)→(T/A) | 60 | CCN | 40 (C/G)→(T/A) | |||
| 78 (G/C)→(A/T) | 49 | CPN | 84 (C/G)→(A/T) | 73 (G/C)→(A/T) | |||||
| 37 | TPR | None | 50 | CPN | 63 (G/C)→(A/T) | ||||
| 38 | TPR | None | 85 (G/C)→(A/T) | ||||||
| 39 | TPR | 32 (A/T)→(G/C) | 51 | CPN | 33 (G/C)→(C/G) | ||||
| 40 | TG6 | None | 49 (T/A)→(A/T) | ||||||
| 41 | TG6 | None | 82 (G/C)→(A/T) | ||||||
| 42 | TG6 | None | 52 | CA6 | None | ||||
| 43 | CAF | None | 53 | CA6 | None | ||||
| 44 | CAF | None | 54 | CA6 | 64 (C/G)→(G/C) | ||||
| 45 | CAF | 47 (G/C)→(A/T) | 81 (C/G)→(G/C) | ||||||
| 67 (C/G)→(A/T) | 55 | CG6 | None | ||||||
| 81 (G/C)→(C/G) | 56 | CG6 | None | ||||||
| 46 | CPR | None | 57 | CG6 | None | ||||
| KOD Dash | |||||||||
| 61 | TAF | None | 70 | TCN | None | 78 | CA6 | 72 (T/A)→(A/T) | |
| 62 | TAF | None | 71 | TCN | None | 79 | CG6 | None | |
| 63 | TAF | None | 72 | TCN | None | 80 | CG6 | None | |
| 64 | TPR | None | 73 | CAF | None | 81 | CG6 | 56 (G/C)→(A/T) | |
| 65 | TPR | None | 74 | CAF | None | 82 | CCN | None | |
| 66 | TPR | 78 (G/C)→(A/T) | 75 | CAF | 52 (C/G)→(T/A) | 83 | CCN | 45 (T/A)→(C/G) | |
| 67 | TG6 | None | 71 (G/C)→(A/T) | 84 | CCN | 61 (A/T)→(G/C) | |||
| 68 | TG6 | None | 76 | CA6 | None | 85 (G/C)→(A/T) | |||
| 69 | TG6 | None | 77 | CA6 | None | ||||
| KOD(exo-) | |||||||||
| 85 | TPR | None | 89 | TG6 | None | 93 | CA6 | 84 (C/G)→(T/A) | |
| 86 | TPR | None | 90 | TG6 | None | 94 | CG6 | None | |
| 87 | TPR | None | 91 | CA6 | None | 95 | CG6 | None | |
| 88 | TG6 | None | 92 | CA6 | None | 96 | CG6 | None | |
aModified nucleotide used for the synthesis of modified DNA.
bSite and type of mutation. The site of mutation is numbered from the 5′ end of primer 1R on amplifying region I (110 bp). ‘None indicates no mutation observed. More than one site was mutated in clones 36, 45, 48, 50, 51, 54, 60, 75 and 84.
Interestingly, mutations were more frequent using dCTP than dUTP analogs. Although, Vent(exo-) DNA polymerase accepted a broader range of analogs as substrates and generated the corresponding modified DNAs by PCR, mutations were found when TAF, TPR, CAF, CPR, CPN, CA6 or CCN were used. In particular, one or more mutations were found in every sequence of modified DNAs produced using TAF, CPN and CCN. The sequence analysis indicated that mutation occurred not only at the site of the base pair involving the modified nucleotide but also at other unrelated sites, presumably because of the presence of modified groups near the incorporation site. Compared with Vent(exo-) DNA polymerase, both KOD Dash and KOD(exo-) were less tolerant of C5 modifications but showed a lower frequency of mutations.
Kinetic study on polymerase reactions using modified primers/templates/triphosphates
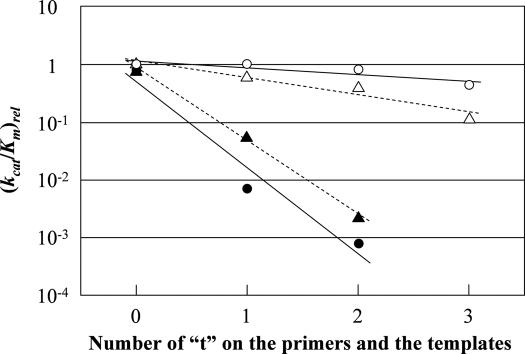
To determine the crucial step of the PCR, we carried out modified nucleotide standing-start experiments under conditions that achieve a single completed hit. Because the thermodynamic properties of oligonucleotides containing modified dU with (6-aminohexylamino)-2-oxoethyl group have been well-characterized (53), therefore we selected this type of a nucleotide for the experiments (Figure 2). The apparent values for Km and Vmax were obtained from the initial velocities of the standing-start primer extension reactions. The relative apparent reaction rate constant, (kcat)rel, is obtained by assuming that the Vmax value is proportional to total enzyme concentration and by setting the apparent relative reaction rate of a single incorporation of natural T to primer/template complex (P/T = P0/TA) at 1. The relative apparent catalytic efficiency, (kcat/Km)rel, is obtained by assuming that the apparent catalytic efficiency of a single natural T incorporation in P/T (P0/TA) is 1 (Table 3). Interestingly, the relative catalytic efficiency decreased at least 30-fold per modified dU residue when the modified dU was introduced in the growing terminus of the primer. On the other hand, it decreased only a few-fold per modified dU residue when the template contained modified dU (Table 3 and Figure 5). Of course, the apparent value for Km and Vmax obtained from this experiment can vary depending on the P/T sequence because the association–dissociation of the polymerase with P/T would influence the kinetic data; however, the apparent values of Km and Vmax for a single natural T incorporation in P/T (P0/TA) were almost equal to those of a single natural dA incorporation in P/T (PA/T0) under the same condition, indicating that the sequence difference in P/T would have little effect on the apparent value for Km and Vmax. Therefore, it appears that the decrease in relative apparent catalytic efficiency (kcat/Km)rel caused by the modified nucleotide on the primer and the template reflects the effects of the modified group on the true polymerization rate constant (kcat) and the rate constant for dissociation of enzyme from P/T (koff).
Table 3.
Kinetic parameters obtained from the standing-start experiments using Vent(exo-) DNA polymerase
| Primer/template | dNTP | Km (μM) | (kcat)rel | (kcat/Km)rel | Accuracy |
|---|---|---|---|---|---|
| P0/TA | TTP | 1.90 | 1a | 1c | 1c |
| TA6 | 2.10 | 0.85a | 0.77c | 0.77c | |
| P1/TA | TTP | 59.7 | 0.34a | 0.011c | 1e |
| TA6 | 91.2 | 0.34a | 0.0071c | 0.65e | |
| P2/TA | TTP | NA | NA | NA | |
| TA6 | 122 | 0.051a | 0.00080c | ||
| PA/T0 | dATP | 5.52 | 1b | 1d | |
| PA/T1 | dATP | 8.40 | 1.52b | 1.0d | |
| PA/T2 | dATP | 8.32 | 1.22b | 0.81d | |
| PA/T3 | dATP | 10.7 | 0.84b | 0.43d |
dNTP, deoxynucleotide triphosphate; NA, not available.
aRelative value of the apparent kcat obtained from the experiment using P0, TA and TTP.
bRelative value of the apparent kcat obtained from the experiment using PA, T0 and dATP.
cRelative value of the apparent kcat/Km obtained from the experiment using P0, TA and TTP.
dRelative value of the apparent kcat/Km obtained from the experiment using PA, T0 and dATP.
eRelative value of the apparent kcat/Km obtained from the experiment using P1, TA and TTP.
Figure 5.
Decrease in the apparent relative catalytic efficiencies (kcat/Km)rel according to the numbers of modified dUs (t) on the primers and templates. Values were obtained from standing-start experiments using the following: P0–2, TA, TA6 and Vent(exo-) DNA polymerase (filled circles); PA, T0–3, dATP and Vent(exo-) DNA polymerase (open circles); P0–2, TA, TA6 and KOD(exo-) DNA polymerase (filled triangles); or PA, T0–3, dATP, and KOD(exo-) DNA polymerase (open triangles).
The relative apparent catalytic efficiency (kcat/Km)rel for a single modified dU incorporation in P/T (P0/TA) had the same order of efficiency as that for a single natural T incorporation in P0/TA, and a large difference in catalytic efficiency was not seen. The ratio of relative apparent catalytic efficiencies (accuracy), defined as the ratio of modified to natural nucleotide insertion efficiencies [(kcat/Km)rel]modified/[(kcat/Km)rel]natural was 0.77, indicating that the modified group on the substrate nucleoside triphosphate has little effect on the relative apparent catalytic efficiency. Similar results were also observed for a single incorporation of the modified dU and natural T triphosphate in P1/TA, wherein the modified dU is present at the 3′ end of the primer. The ratio of relative apparent catalytic efficiencies [(kcat/Km)rel]modified/[(kcat/Km)rel]natural was 0.65 in the case of P1/TA. These results are consistent with the finding that the modified dUTP, TA6, is incorporated into DNA with high fidelity as found by sequence analysis of the PCR products (27). The present results clearly demonstrate that the modified group has a large effect on the polymerase reaction when it is at the C5 position on the primer than when it on the template and the substrate nucleoside triphosphate.
DISCUSSION
The efficiency of a PCR depends on a variety of parameters including temperature (denature, annealing and extension), reaction time and the number of reaction cycles. The PCR assays were performed under conditions optimized for the positive control using the natural 4 nt triphosphates. Therefore, some analogs that did not act as PCR substrates under these assay conditions could provide the corresponding PCR products if the parameters were separately optimized. The amount of PCR product depends on the substrate and enzyme concentration as well as the sequences and length of the amplifying region; however, comparison of the yields obtained by PCR using modified and natural analogs under the same conditions should supply useful information on the efficiency of the modified analog as a PCR substrate.
Our current finding that family B DNA polymerases accept a wider range of modified nucleoside triphosphates as PCR substrates than family A DNA polymerases agrees with previous reports (28,29). As shown in Figure 4A and B, family A DNA polymerases do not prefer substituents involving 2-oxoethyl linker arm, such as (2-aminoethylamino)-2-oxoethyl (A2), (4-aminobutylamino)-2-oxoethyl (A4), (6-aminohexylamino)-2-oxoethyl (A6), (6-guanidinohexylamino)-2-oxoethyl (G6) and 2-methoxy-2-oxoethyl (ME) as PCR substrates as well as analogs with the different type of C-C (single bond) linker arms (25,28,29). These results together with rigidity of the linker arm suggest that the difference in PCR efficiency between family A and B polymerases reflects the palm shape of the DNA polymerase complex bound to a DNA duplex (55–58); it is predicted that family A DNA polymerases cannot accommodate a flexible modified group close to the nucleobase in the P/T or the nucleoside triphosphate; however, a rigid linker arm providing the proper orientation of a modified group, even if it is long and bulky (24), would not prevent association of the polymerase with the P/T or formation of the transition state in the enzyme reaction. This prediction, however, does not agree with the fact that the relative yields of the PCR products varied depending on whether the substitution was on dU or dC. This difference may instead be due to other effects, such as changes in the pKa at the N4 position of the cytosine base or differences in the contribution of the C5 substituent to base-stacking interactions (59,60).
The presence of a charge near the nucleobase decreased the yield of the PCR products. Our previous study showed that the analog TME, which has no charge on the modified group, acts as a good substrate for PCR catalyzed by KOD Dash, whereas 5-(2-hydroxy-2-oxoethyl)-dUTP, which has a negative charge on the modified group, does not (30). Therefore, the amount of PCR products is decreased by either a negative or positive charge on the modified group. Basic amino acid residues are found in the P/T binding region of the polymerases, and, thus, the mode of interaction between the enzyme and P/T may be important for the extension reaction. The values of kcat and koff should be experimentally determined by running start experiments using two kinds of P/Ts containing modified groups with positive or negative charges, and the kinetic data should be compared to know which step of the reaction is more negatively affected by the presence of the modified group on P/T. Differences in the chemical structure of the modified nucleotide will alter the kcat and koff value in different ways.
Because the P/T forms a helix and the polymerase recognizes the base pair duplexes, how modified groups located on the upstream portion of the primer and on the downstream portion of the template affect the polymerase reaction should also be closely examined (61). The present standing-start experiments clearly revealed, however, that the modified group has a greater affect on the polymerase reaction when it is adjacent to the elongation terminus than when it is on the template. These tendencies were also observed in similar experiments using KOD(exo-) DNA polymerase (Figure 5 and see Supplementary Table 1).
Recent studies have focused on the use of enzymatic methods for fluorescently labeling or functionalizing DNA molecules because they are more convenient than chemical methods. Useful modifications of DNA molecules include conferring membrane permeability by incorporation of TG6 (62) in the polymerase reaction or by postsynthetic modification of modified DNAs prepared by PCR using TME (30). Furthermore, TAF and CAF, which were much better as substrates in the PCR than the conventional substrates TAL and CAL, could be useful for preparing modified DNAs with a high density of functional groups if the trifluoroacetyl protecting group is removed and the DNA is then subjected to postsynthetic modification. The kinetic study indicated that the existence of the modified group after incorporation into DNA greatly affected the enzymatic reaction rate. Therefore, it is not surprising that generation of PCR products was suppressed depending on the amplifying sequences. For screening of functional molecules by in vitro selection of modified DNA libraries, any bias except for the intended selection pressures is unfavorable. Therefore, determining which combination of enzyme and substrate provides modified DNA in high yields and with low sequence dependency will be necessary for constructing a selection system that reduces unsuitable biases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (B) and (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by PRESTO from the Japan Science and Technology Agency. The authors would like to extend their thanks to Toyobo Ltd for generously providing KOD(exo-) DNA polymerase. Funding to pay the Open Access publication charges for this article was provided by PRESTO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Breaker R.R. DNA enzymes. Nat. Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 2.Breaker R.R. DNA aptamers and DNA enzymes. Curr. Opin. Chem. Biol. 1997;1:26–31. doi: 10.1016/s1367-5931(97)80105-6. [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Breaker R.R. Deoxyribozymes: new players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol. 1999;9:315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 4.Famulok M., Mayer G., Blind M. Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc. Chem. Res. 2000;33:591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 5.Emilsson G.M., Breaker R.R. Deoxyribozymes: new activities and new applications. Cell. Mol. Life Sci. 2002;59:596–607. doi: 10.1007/s00018-002-8452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Famulok M. Allosteric aptamers and aptazymes as probes for screening approaches. Curr. Opin. Mol. Ther. 2005;7:137–143. [PubMed] [Google Scholar]
- 7.Latham J.A., Johnson R., Toole J.J. The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine. Nucleic Acids Res. 1994;22:2817–2822. doi: 10.1093/nar/22.14.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battersby T.R., Ang D.N., Burgstaller P., Jurczyk S.C., Bowser M.T., Buchanan D.D., Kennedy R.T., Benner S.A. Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog. J. Am. Chem. Soc. 1999;121:9781–9789. doi: 10.1021/ja9816436. [DOI] [PubMed] [Google Scholar]
- 9.Santoro S.W., Joyce G.F., Sakthivel K., Gramatikova S., Barbas C.F., III RNA cleavage by a DNA enzyme with extended chemical functionality. J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 10.Kusser W. Chemically modified nucleic acid aptamers for in vitro selections: evolving evolution. Rev. Mol. Biotechnol. 2000;74:27–38. doi: 10.1016/s1389-0352(99)00002-1. [DOI] [PubMed] [Google Scholar]
- 11.Perrin D.M., Garestier T., Hélène C. Bridging the gap between proteins and nucleic acids: a metal-independent RNAse A mimic with two protein-like functionalities. J. Am. Chem. Soc. 2001;123:1556–1563. doi: 10.1021/ja003290s. [DOI] [PubMed] [Google Scholar]
- 12.Lermer L., Roupioz Y., Ting R., Perrin D.M. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- 13.Verma S., Jäger S., Thum O., Famulok M. Functional tuning of nucleic acids by chemical modifications: tailored oligonucleotides as drugs, devices, and diagnostics. Chem. Rec. 2003;3:51–60. doi: 10.1002/tcr.10047. [DOI] [PubMed] [Google Scholar]
- 14.Ting R., Thomas J.M., Lermer L., Perrin D.M. Substrate specificity and kinetic framework of a DNAzyme with an expanded chemical repertoire: a putative RNaseA mimic that catalyzes RNA hydrolysis independent of a divalent metal cation. Nucleic Acids Res. 2004;32:6660–6672. doi: 10.1093/nar/gkh1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ting R., Lermer L., Perrin D.M. Triggering DNAzymes with light: a photoactive C8 thioether-linked adenosine. J. Am. Chem. Soc. 2004;126:12720–12721. doi: 10.1021/ja046964y. [DOI] [PubMed] [Google Scholar]
- 16.May J.P., Ting R., Lermer L., Thomas J.M., Roupioz Y., Perrin D.M. Covalent schiff base catalysis and turnover by a DNAzyme: a M2-independent AP-endonuclease mimic. J. Am. Chem. Soc. 2004;126:4145–4156. doi: 10.1021/ja037625s. [DOI] [PubMed] [Google Scholar]
- 17.Sidorov A.V., Grasby J.A., Williams D.M. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 2004;32:1591–1601. doi: 10.1093/nar/gkh326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masud M.M., Kuwahara M., Ozaki H., Sawai H. Sialyllactose-binding modified DNA aptamer bearing additional functionality by SELEX. Bioorg. Med. Chem. 2004;12:1111–1120. doi: 10.1016/j.bmc.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 20.Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breaker R.R. In vitro selection of catalytic polynucleotides. Chem Rev. 1997;97:371–390. doi: 10.1021/cr960008k. [DOI] [PubMed] [Google Scholar]
- 22.Osborne S.E., Ellington A.D. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 23.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 24.Sakthivel K., Barbas C.F., III Expanding the potential of DNA for binding and catalysis: highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew. Chem. Int. Ed. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Lee S.E., Sidorov A., Gourlain H., Mignet N., Thorpe S.J., Brazier J.A., Dickman M.J., Hornby D.P., Grasby J.A., Williams D.M. Enhancing the catalytic repertoire of nucleic acids: a systematic study of linker length and rigidity. Nucleic Acids Res. 2001;29:1565–1573. doi: 10.1093/nar/29.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gourlain T., Sidorov A., Mignet N., Thorpe S.J., Lee S.E., Grasby J.A., Williams D.M. Enhancing the catalytic repertoire of nucleic acids. II. Simultaneous incorporation of amino and imidazolyl functionalities by two modified triphosphates during PCR. Nucleic Acids Res. 2001;29:1898–1905. doi: 10.1093/nar/29.9.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawai H., Ozaki A.N., Satoh F., Ohbayashi T., Masud M.M., Ozaki H. Expansion of structural and functional diversities of DNA using new 5-substituted deoxyuridine derivatives by PCR with superthermophilic KOD Dash DNA polymerase. Chem. Commun. 2001;24:2604–2605. [Google Scholar]
- 28.Held H.A., Benner S.A. Challenging artificial genetic systems: thymidine analogs with 5-position sulfur functionality. Nucleic Acids Res. 2002;30:3857–3869. doi: 10.1093/nar/gkf500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuwahara M., Takahata Y., Shoji A., Ozaki A.N., Ozaki H., Sawai H. Substrate properties of C5-substituted pyrimidine 2′-deoxynucleoside 5′-triphosphates for thermostable DNA polymerases during PCR. Bioorg. Med. Chem. Lett. 2003;13:3735–3738. doi: 10.1016/j.bmcl.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Masud M.M., Ozaki N.A., Kuwahara M., Ozaki H., Sawai H. Modified DNA bearing 5(methoxycarbonylmethyl)-2′-deoxyuridine: preparation by PCR with thermophilic DNA polymerase and postsynthetic derivatization. Chembiochem. 2003;4:584–588. doi: 10.1002/cbic.200200539. [DOI] [PubMed] [Google Scholar]
- 31.Roychowdhury A., Illangkoon H., Hendrickson C.L., Benner S.A. 2′-Deoxycytidines carrying amino and thiol functionality: synthesis and incorporation by Vent (exo-) polymerase. Org. Lett. 2004;6:489–492. doi: 10.1021/ol0360290. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto A., Tanaka K., Nishiza K., Saito I. Synthesis of an artificial hole-transporting nucleoside triphosphate, dMDATP, and its enzymatic incorporation into DNA. Bioorg. Med. Chem. 2004;12:5875–5880. doi: 10.1016/j.bmc.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 33.Jäger S., Famulok M. Generation and enzymatic amplification of high-density functionalized DNA double strands. Angew. Chem. Int. Ed. 2004;43:3337–3340. doi: 10.1002/anie.200453926. [DOI] [PubMed] [Google Scholar]
- 34.Jäger S., Rasched G., Kornreich-Leshem H., Engeser M., Thum O., Famulok M. A versatile toolbox for variable DNA functionalization at high density. J. Am. Chem. Soc. 2005;127:15071–15082. doi: 10.1021/ja051725b. [DOI] [PubMed] [Google Scholar]
- 35.Obayashi T., Kuwahara M., Hasegawa M., Kasamatsu T., Tamura T., Sawai H. Expansion of repertorie of modified DNAs prepared by PCR using KOD Dash DNA polymerase. Org. Biomol. Chem. 2005;3:2463–2468. doi: 10.1039/b504330a. [DOI] [PubMed] [Google Scholar]
- 36.Kuwahara M., Hanawa K., Ohsawa K., Kitagata R., Ozaki H., Sawai H. Direct PCR amplification of various modified DNAs having amino acids found in proteins: convenient preparation of DNA libraries with high-potential activities for in vitro selection. Bioorg. Med. Chem. 2006;14:2518–2526. doi: 10.1016/j.bmc.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 37.Ortiz A., Ritter E. A rapid method for detecting specific amplified PCR fragments in microtiter plates. Nucleic Acids Res. 1996;24:3280–3281. doi: 10.1093/nar/24.16.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godovikova T.S., Kolpashchikov D.M., Orlova T.N., Richter V.A., Ivanova T.M., Grochovsky S.L., Nasedkina T.V., Victorova L.S., Poletaev A.I. 5-[3-(E)-(4-azido-2,3,5,6-tetrafluorobenzamido)propenyl-1]- 2′-deoxyuridine-5′-triphosphate substitutes for thymidine-5′-triphosphate in the polymerase chain reaction. Bioconjugate Chem. 1999;10:529–537. doi: 10.1021/bc980144r. [DOI] [PubMed] [Google Scholar]
- 39.Perrin D.M., Garestier T., Hélène C. Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities. Nucleosides Nucleotides. 1999;18:377–391. doi: 10.1080/15257779908043083. [DOI] [PubMed] [Google Scholar]
- 40.Augustin M.A., Ankenbauer W., Angerer B. Progress towards single-molecule sequencing: enzymatic synthesis of nucleotide-specifically labeled DNA. J Biotechnol. 2001;86:289–301. doi: 10.1016/s0168-1656(00)00420-x. [DOI] [PubMed] [Google Scholar]
- 41.Thum O., Jäger S., Famulok M. Functionalized DNA: a new replicable biopolymer. Angew. Chem. Int. Ed. 2001;40:3990–3993. doi: 10.1002/1521-3773(20011105)40:21<3990::AID-ANIE3990>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 42.Ohbayashi T., Masud M.M., Ozaki A.N., Ozaki H., Kuwahara M., Sawai H. Enzymatic synthesis of labeled DNA by PCR using new fluorescent thymidine nucleotide analogue and superthermophilic KOD dash DNA polymerase. Bioorg. Med. Chem. Lett. 2002;12:1167–1170. doi: 10.1016/s0960-894x(02)00111-7. [DOI] [PubMed] [Google Scholar]
- 43.Giller G., Tasara T., Angerer B., Muhlegger K., Amacker M., Winter H. Incorporation of reporter molecule-labeled nucleotides by DNA polymerases. I. Chemical synthesis of various reporter group-labeled 2′-deoxyribonucleoside-5′-triphosphates. Nucleic Acids Res. 2003;31:2630–2635. doi: 10.1093/nar/gkg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasara T., Angerer B., Damond M., Winter H., Dorhofer S., Hubscher U., Amacker M. Incorporation of reporter molecule-labeled nucleotides by DNA polymerases. II. High-density labeling of natural DNA. Nucleic Acids Res. 2003;31:2636–2646. doi: 10.1093/nar/gkg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodman M.F., Creighton S., Bloom L.B., Petruska J. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 46.Creighton S., Bloom L.B., Goodman M.F. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Meth. Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 47.Lutz S., Burgstaller P., Benner S.A. An in vitro screening technique for DNA polymerases that can incorporate modified nucleotides. Pseudo-thymidine as a substrate for thermostable polymerases. Nucleic Acids Res. 1999;27:2792–2798. doi: 10.1093/nar/27.13.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyce C.M., Benkovic S.J. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda S., Romesberg F.E. Optimization of interstrand hydrophobic packing interactions within unnatural DNA base pairs. J. Am. Chem. Soc. 2004;126:14419–14427. doi: 10.1021/ja047291m. [DOI] [PubMed] [Google Scholar]
- 50.Hwang G.T., Romesberg F.E. Substituent effects on the pairing and polymerase recognition of simple unnatural base pairs. Nucleic Acids Res. 2006;34:2037–2045. doi: 10.1093/nar/gkl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sintim H.O., Kool E.T. Remarkable sensitivity to DNA base shape in the DNA polymerase active site. Angew. Chem. Int. Ed. 2006;45:1974–1979. doi: 10.1002/anie.200504296. [DOI] [PubMed] [Google Scholar]
- 52.Potapova O., Chan C., DeLucia A.M., Helquist S.A., Kool E.T., Grindley N.D., Joyce C.M. DNA polymerase catalysis in the absence of Watson–Crick hydrogen bonds: analysis by single-turnover kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozaki H., Nakamura A., Arai M., Endo M., Sawai H. Novel C5-substituted 2′-deoxyuridine derivatives bearing amino-linker arms: synthesis, incorporation into oligodeoxyribonucleotides, and their hybridization properties. Bull. Chem. Soc. Jpn. 1995;68:1981–1987. [Google Scholar]
- 54.No Z., Shin D.S., Song B.J., Ahn M., Ha D. A facile one-pot synthesis of 2,3′-anhydro-2′-deoxyuridines via 3′-O-imidazolylsulfonates. Synth. Commun. 2000;30:3873–3882. [Google Scholar]
- 55.Eom S.H., Wang J., Steitz T.A. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto H., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Inoue T., Kai Y. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 2001;306:469–477. doi: 10.1006/jmbi.2000.4403. [DOI] [PubMed] [Google Scholar]
- 57.Kim T.W., Delaney J.C., Essigmann J.M., Kool E.T. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc. Natl Acad. Sci. USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim T.W., Brieba L.G., Ellenberger T., Kool E.T. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 59.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okumoto Y., Tanabe Y., Sugimoto N. Factors that contribute to efficient catalytic activity of a small Ca2-dependent deoxyribozyme in relation to its RNA cleavage function. Biochemistry. 2003;42:2158–2165. doi: 10.1021/bi020364z. [DOI] [PubMed] [Google Scholar]
- 61.Hendrickson C.L., Devine K.G., Benner S.A. Probing minor groove recognition contacts by DNA polymerases and reverse transcriptases using 3-deaza-2′-deoxyadenosine. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohmichi T., Kuwahara M., Sasaki N., Hasegawa M., Nishikata T., Sawai H., Sugimoto N. Nucleic acid with guanidinum modification exhibits efficient cellular uptake. Angew. Chem. Int. Ed. 2005;44:6682–6685. doi: 10.1002/anie.200500904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.