Abstract
For a bacterium, Streptomyces coelicolor A3(2) contains a relatively large genome (8.7 Mb) with a complex and adaptive pattern of gene regulation. We discovered a correlation between the physical structure of the S.coelicolor genome and the transcriptional activity of the genes therein. Twelve genes were surveyed throughout 72 h of growth for both in vivo sensitivity to DNase I digestion and levels of transcription. DNase I-sensitivity correlated positively with transcript levels, implying that it was predictive of gene expression, and indicating increased accessibility of transcribed DNA. The genome was fractionated based on the sensitivity to DNase I digestion, with the low molecular weight (frequently cut) fraction highly enriched for actively transcribed sequences when compared to the infrequently cut fraction, which was representative of the entire genome. This approach will allow comparison of nucleoid proteins, and any modifications thereof, associated with transcriptionally active and inactive regions of the bacterial genome.
INTRODUCTION
The bacterial chromosome exists within the cell as a complex amalgam of predominantly nucleic acids and proteins that is visible by light microscopy and called the nucleoid. Much has been learnt about the function of other cellular organelles by studying their structure, and this can apply equally well to the nucleoid; understanding how different genes are packaged in vivo may provide new insights into mechanisms of bacterial gene regulation. A ready demonstration of the value of such an approach exists in the study of chromatin structure in eukaryotes. This has been a fertile field ever since the discovery that in vivo accessibility to DNase I was predictive of the level of expression of the underlying gene (1). Our goal was to determine whether similar approaches could be used to study prokaryotic gene regulation. Specifically, we applied our analysis to S. coelicolor A3(2), the most genetically characterized actinomycete. Streptomycetes are important sources of many clinically used antibiotics and other pharmaceutically important molecules (2). Biosynthesis of these compounds is tightly regulated at the transcriptional level by pathway-specific regulatory genes that serve to integrate both developmental and environmentally-sensitive regulatory networks in a growth phase-dependent manner (3). Thus streptomycetes provide an excellent model system for analysing how a prokaryotic genome regulates complex genetic and developmental processes.
S.coelicolor contains two variants of nucleoid-associated HU-like proteins. The latter are similar to eukaryotic histone proteins; they are small (12–16 kDa), abundant proteins that bind DNA cooperatively (forming HU dimers or heterodimers with other proteins) with little sequence specificity to constrain negative supercoils. In Escherichia coli, two variant HU-like proteins are expressed differentially, and the composition of the resultant dimers reflects the relative abundance of the two variants; heterodimers are formed with other binding partners, including transcription factors that are commonly induced as part of a stress response (4). There is genetic evidence that these proteins coordinate global transcriptional programmes but the mechanism of action remains controversial. We set out to assess whether a eukaryotic-like chromatin structure exists in S.coelicolor. Our studies reveal evidence for a dynamic chromatin-like structure that correlates well with levels of gene expression.
MATERIALS AND METHODS
Culture conditions
Spores of S.coelicolor strain M145 were germinated by heat treatment as described (5) and grown in R5 medium with shaking at 30°C for for up to 72 h. Measurement of antibiotic production was performed as described previously (5).
DNase I-digestion of the bacterial chromosome
Mycelium was digested with increasing concentrations of DNase I in a permeabilizing buffer, using standard methods (1), with some adaptations. Typically 50 ml of each culture were pretreated with 20 μl of RNase A (20 mg/ml) and 10 μl DNase I (80 U/ml) after supplementation with 5 mM CaCl2, and incubation at 30°C continued to remove contaminating nucleic acids. The cells were collected by centrifugation and rapidly washed three times in TES buffer [10 mM Tris–HCl, 1 mM EDTA and 20 mM sucrose (pH 8)] supplemented to 7.5 mM EDTA to inactivate endonucleases. The pellet was resuspended in 1/10th original volume of TES supplemented with 0.5% NP-40 and 0.5% Triton X-100 and the cells incubated at 30°C for a further 15 min. The cells were washed in the same volume of DNase I Digestion Buffer [DDB: 10 mM Tris–HCl, 2.6% sucrose, 10 mM MgCl2, 0.25 mM CaCl2, 0.1 mM DTT, 0.5% NP-40 and 0.5% Triton X-100), and split into five aliquots and digested with different amounts of DNase I (0 to 50 U of Roche's molecular biology grade enzyme) at 37°C for 3 min. The reactions were stopped by addition of an equal volume of a stop buffer consisting of 50 mM Tris–HCl, 5% SDS, 10 mM EDTA (pH 8) supplemented with 10 mg/ml Proteinase K. Genomic DNA was recovered following an overnight digestion at 55°C.
Fractionation of the genome by in vivo DNase I digestion
Mycellial cells were pretreatred with RNase and DNase I, washed and resuspended in DDB as described above. The recovered pellets of cells were simultaneously treated with lysozyme (20 μg/ml), 0.75% formaldehyde and a range of DNase I concentrations (0 to 50 U/ml) for 5 min at 37°C. The reactions were stopped by addition of an equal volume of TBE running buffer and subjected to 0.7% agarose/TBE gel electrophoresis. The nucleoprotein complexes that entered the gels were excised and the material recovered by electroelution in the same buffer. DNA was recovered by ethanol precipitation following thermal denaturation of the formaldehyde crosslinks, Proteinase K treatment and two rounds of phenol–chloroform extraction.
Southern hybridization DNase I-sensitivity assay
DNase I sensitivity was measured by Southern hybridization using either DIG-labeled (Roche) DNA or PCR products as probes. The restriction sites used are listed in the figures and the sequences of the primers in Table 1.
Table 1.
Primers used in this study
| Primers used for qrt-PCR | |||||
| actII-orf4 | 5085 | qa24f | CTTAAATCCTCGAAGGCGACCCAG | qa24r | TCCTCGAGCCGGTTCTCCTCG |
| afsK | 4423 | qafsKf | GATCAAGACGGTGCGCACGGAAC | qafsKr | GAGGGCTTCAGGTCGCGGTG |
| afsQ2 | 4906 | qaQ2f | TCCGGGCTGCGCTTCACCAG | qaQ2r | GTGCGCAGCGACTCGGGCA |
| cdaR | 3217 | qcdaRf | CCCGACCGCCGGTCTCGAATT | qcdaRr | CGGAGCGAGTCGAGTGAGTGG |
| hrdB | 5820 | qhrdBf | GCGCTCATTGAGCGGGGAAAGG | qhrdBr | GCGGGCGCCTCTTCCTCGA |
| fba | 3649 | qfbaf | TTCGCCTACCCCGCCATCAACGT | qfbar | GCCGGCCTTGACGCGCTCCT |
| rbnH | 5812 | qrbnHf | AAGATCATTGCCGGTGTCGACGAGG | qrbnHr | CTGCTGGTCACCGGCTTCGC |
| redD | 5877 | qredDf | AAAGATCGGGCGGCACCCCGT | qredDr | TTGAGGACGTTGTCGGGCGGG |
| rlpA | 4649 | qrlpAf | CTCCACGAGCAAGTTCGACGGC | qrlpAr | ACGGTGCCGGTCTTGGGGTTC |
| rpmG3 | 4632 | qrpmf | GTGGCTGCCACCGACGTCCG | qrpmr | CTATCGCGTTTCGCGGTGCGCG |
| pgk | 1946 | qpgkf | ACCTGAACGTCCCGCTGGACG | qpgkr | TAGACGTCCGCGAGGCCGG |
| sti1 | 0762 | qstif | GCAGCGACTCTGGGCCTGAC | qstir | CAGACGCCGTCGACGGTGAC |
| Primers used for Southerns | |||||
| actII-orf4 | a24f | AGAACGGCCACCAGGTCCTGG | a24r | CATCTCGACCCCACGGGTTTCG | |
| afsK | afsKf | CTCCGACGTCTTCCCCGCC | afsKr | GCGGAACAGCTGGTCCTCCG | |
| afsQ2 | aQ2f | GGCCAGATGATCGTCTCCGGC | aQ2r | GTCACCGGTGGCGAGGGC | |
| cdaR | cdaRf | GCTGGTCGGCGTCACCGCG | cdaRr | CCTGCCTGCAGTCGGAATAATACG | |
| hrdB | hrdBf | GGCCAGATGATCGTCTCCGGC | hrdBr | GTCACCGGTGGCGAGGGC | |
| fba | fbaf | TTCCGGGGCTCCAGCAGGAC | fbar | GCTCGTCGTCCGTGCGCC | |
| rbnH | rbnHf | CAGGGGGCCGTTCCCGCTT | rbnHr | GCGAAACCGAAGTCTGCATGGTC | |
| redD | redDf | GACGCCTCCGGTGGGGTC | redDr | GGCATGATGGTGGCTGCGGC | |
| rlpA | rlpAf | CTCCGACGTCTTCCCCGCC | rlpAr | GCGGAACAGCTGGTCCTCCG | |
| rpmG3 | rpmf | GACAGGCTGCCGGCCGTG | rpmr | AGCGGTGACCGGACTTGAACCG | |
| pgk | pgkf | CCCTGCGTCGCGTCCGC | pgkr | TGCGGGAGCCGCTTCGGC | |
| sti1 | stif | GGTGCATGATCGACAGCACACC | stir | CATCTCGACCCCACGGGTTTCG | |
Quantifying DNase I sensitivity by real time PCR
The extent of digestion by DNase I was measured by quantitative real time PCR (qrt-PCR). A full description of the method and a discussion of the analysis can be found in (6). The protocol was modified by the inclusion of 7% dimethyl sulfoxide (DMSO) in the PCR cocktail and was performed using the DNase I-digested DNA as template and the primer pairs listed in Table 1. All primer pairs were designed to start at the 5′ end of the gene and to amplify a 150 bp fragment. The percentage of copies of target remaining intact, relative to an undigested standard, was calculated by reference to the standard curve, and plotted versus the number of units of DNase I used to treat the cells.
Gene expression analysis
Absolute levels of expression were measured by multiplex qrt-PCR. RNA was prepared as described (7).
Slot blot analysis
Labeling was performed by a modification of the DeRisi method (8); 10 ng of genomic DNA or cDNA (generated by random priming as described above) was used in the first round of synthesis to generate template for PCR amplification using a dNTP mix seeded with DIG-11-dUTP. Southern hybridizations and colorimetric detection were performed according to the manufacturer's (Roche) instructions.
RESULTS
DNase I-sensitivity in the S.coelicolor chromosome
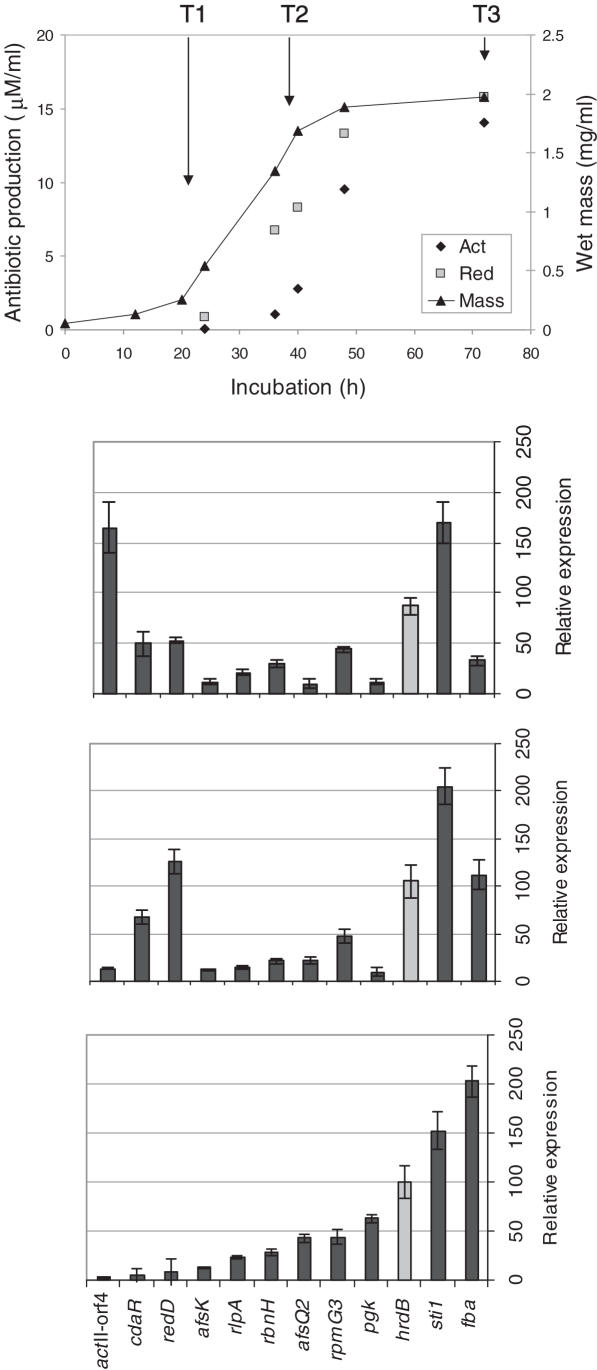
We performed a comparative study of in vivo DNase I-sensitivity and transcriptional activity with a panel of 12 genes. Expression data were collected at three time points during growth of S.coelicolor in liquid culture (Figure 1) corresponding to early rapid growth (T1), transition phase (T2) and stationary phase (T3) (identified by determining the wet mass of the culture per ml). The physiology of S.coelicolor changes in each of these phases, with the onset of secondary metabolism generally occurring as growth slows. Three pathway-specific regulatory genes were included in the panel that control the production of three distinct antibiotics and whose expression was known to increase as the culture reaches stationary phase: actII-orf4, the pathway-specific regulatory gene for the blue-pigmented polyketide actinorhodin (Act); redD, encoding the activator for production of the red-pigmented undecylprodiginine complex (Red); and cdaR, which controls production of the calcium-dependent antibiotic [CDA; see (3) for references]. The remaining genes were chosen for their different patterns of expression as determined in (9) using the same medium as in this study (R5). afsK, which also influences secondary metabolism, is expressed throughout growth. Genes expressed predominantly early in growth were pgk (phosphoglycerate kinase) and fba (fructose 1,6-bisphosphate aldolase), both involved in primary metabolism, while the major vegetative sigma factor gene, hrdB, commonly used as a reference for transcriptional analysis, and the highly expressed sti1, encoding a protease inhibitor, represented genes expressed at high levels throughout growth. Finally, rlpA and rpmG3, both encoding ribosomal proteins, the ribonuclease gene rbnH, and the histidine kinase gene afsQ2 were chosen as genes expressed at relatively low levels.
Figure 1.
Gene expression during growth of S.coelicolor strain M145. Spores were inoculated into R5 liquid medium and grown at 30°C for 3 days. The culture was monitored throughout the experiment for growth and production of the two pigmented antibiotics Act and Red (5; upper). Mycelium was harvested at the three time points indicated [early growth phase (T1), transition phase (T2) and stationary phase (T3)] to generate RNA samples for expression analysis by qrt-PCR (lower) and for use in subsequent DNase I-sensitivity experiments.
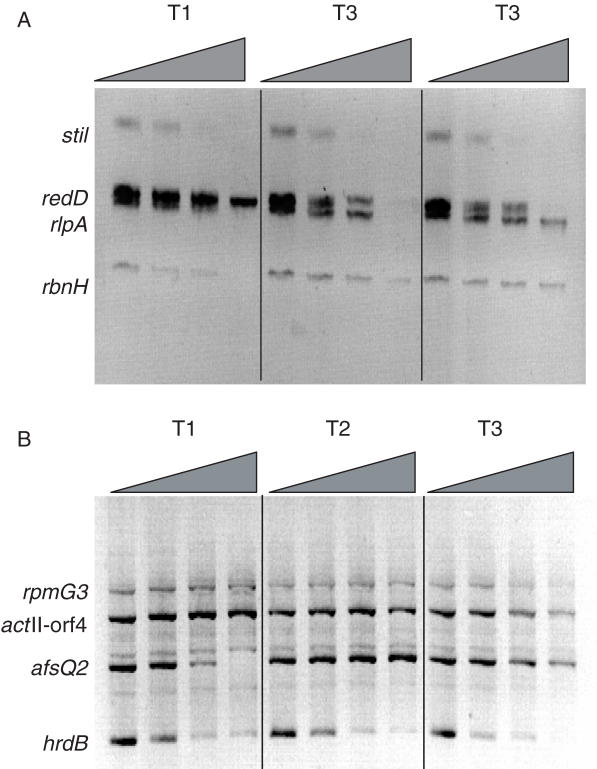
In vivo DNase I-sensitivity of each gene was initially examined by Southern hybridization. By analyzing four genes per blot, it was possible to assess differential changes in sensitivity with time (Figure 2). For each time point, increasing concentrations of DNase I were used to digest the bacterial chromosome (indicated by the wedge in Figure 2). Following in vivo digestion with DNase I, genomic DNA was recovered and digested to completion with a restriction enzyme to produce genomic bands of known size. Gene-specific bands were then detected by Southern hybridization with a mixture of DIG-labeled probes. Figure 2 shows the DNase I-sensitivity of regions of the genome containing a panel of eight genes at the three time points. Substantial changes in DNase I-sensitivity occurred with redD, which became more susceptible at timepoints T2 and T3 (Figure 2A), and afsQ2, which was most susceptible at T1 (Figure 2B). qrt-PCR confirmed that afsQ2 was highly expressed during early growth, but progressively reduced in expression, whereas redD had a complementary profile, being highly expressed at T2 and T3 (Figure 1B). Of the remaining probes sti1, rbnH, rpmG3 (encoding ribosomal protein L33) and hrdB were expressed at much the same level throughout growth and showed little change in DNase I sensitivity. However, whereas rbnH, expressed at low levels, remained relatively DNase I insensitive, the highly expressed sti1 was highly sensitive at each time point. Increased expression of the actII-orf4 gene at T3 coincided with increased sensitivity to DNase I digestion. Thus, genes with changing levels of expression during growth exhibited different levels of DNase I-sensitivity, with maximum sensitivity corresponding to increased levels of transcription. These results suggest that transcriptional induction and repression coincide with increases and decreases in DNase I sensitivity, respectively.
Figure 2.
In vivo DNase I sensitivity determined by Southern hybridization analysis. DNA harvested from mycelium taken from each time point was treated with increasing amounts of DNase I, digested to completion with restriction enzymes and used in Southern hybridization assays. Blots were hybridized with mixtures of probes for (A) rbnH, redD, rlpA and sti1 and (B) actII-orf4, afsQ2, hrdB and rpmG3. The positions of the genomic bands for each gene are indicated, with the amount of DNase I used increasing from left to right.
Conditions of DNase I digestion were varied extensively in an attempt to detect DNase I hypersensitive sites (seen on Southern blots as smaller, stable bands of a specific size that transiently appear on digestion of the parental bandand that denote a site of localized DNase I digestion), but none were detected. This implies that DNase I sensitivity is a property of the gene as a whole and that promoters are not marked by DNase I hypersensitive sites, as occurs in eukaryotes. Southern analysis using micrococcal nuclease instead of DNase I failed to produce the iterative pattern (the ‘micrococcal nuclease ladder’) characteristic of the nucleosomal structure of eukaryotic chromatin, implying that no similar structure exists in S.coelicolor.
DNase I-sensitivity is a dynamic property
Differences in the size of probes used and varying efficiencies with which they detect their target sequences compromise the ability to generate quantitative data for gene copy number by Southern analysis. Therefore, to test more accurately any correlation between DNase I sensitivity and level of gene expression, we adapted a previously described method for measuring DNase I-sensitivity (6) for use in Streptomyces. qrt-PCR can be used to calculate relative copy number loss at a specific site in the genome as a function of DNase I digestion. A key component of the assay is the identification of a DNase I-resistant gene to serve as a control for small differences for template used in each reaction and to standardize the extent of cutting. Southern analysis had already demonstrated that over the range of digestion conditions used, rbnH showed little change in DNase I-sensitivity using limiting amounts of DNase I (Figure 2B). As the qrt-PCR assay relies on template DNA generated in a DNase I-limited digest, it was evident that rbnH could be used as the non-digested control.
PCR primers were designed to generate ∼150 bp amplicons within the coding region of each of the 12 genes. The template DNA came from the DNase I digestion series at each of the three time points (the highest concentration digests were not analyzed). Typically, 500 pg of DNA from each digestion was analyzed in triplicate for a specific gene and the rbnH control. Amplification was performed on a Roche Lightcycler system, and the size and purity of the PCR product were assessed by agarose gel electrophoresis. For each gene, a three-point standard curve was derived using a serial dilution of genomic DNA (from 500 to 50pg). The number of copies of each gene present at each point in the DNase I-digestion series was then determined using the standard curves. To correct for small differences in the amount of template between reactions, the rbnH copy number was used as an internal standard. The final plot for each gene therefore shows relative copy number loss versus units of DNase I used in the digestion.
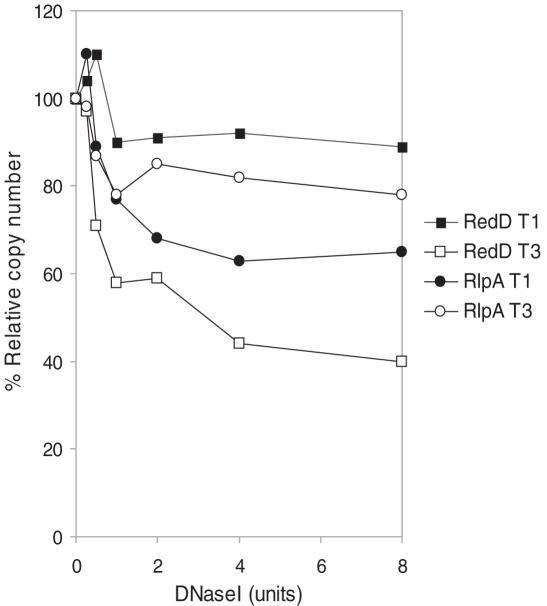
The digestion profiles show a relatively rapid loss in copy number before the curves level out (Figure 3). This is consistent with results obtained with eukaryotic chromatin, where the final value represents accessibility of the chromatin, and allows an accurate comparison of the DNase I-sensitivity of each gene. Figure 3 shows the DNase I sensitivity of two of the genes analyzed by Southern blots in Figure 2A, redD and rlpA, at time points T1 and T3. Under limiting conditions of digestion, redD shows 8 and 58% relative copy number loss at T1 and T3, consistent with the late expression of the gene. The earlier expressed rlpA shows the opposite pattern, with 36 and 18% relative copy number loss at T1 and T3, respectively. These data correspond well with the Southern blots shown in Figure 2A.
Figure 3.
Quantifying changes in DNase I-sensitivity with qrt-PCR. qrt-PCR was used to detected small changes in DNase I-sensitivity in two reciprocally expressed genes (redD and rlpA) at the earliest and latest time points. In vivo digestions were performed as described in Figure 1; qrt-PCR is described elsewhere (6). DNase I sensitivity was estimated by the percentage loss of copy number after the rate of digestion had reached a steady level (commonly after 4 U of enzyme had been used).
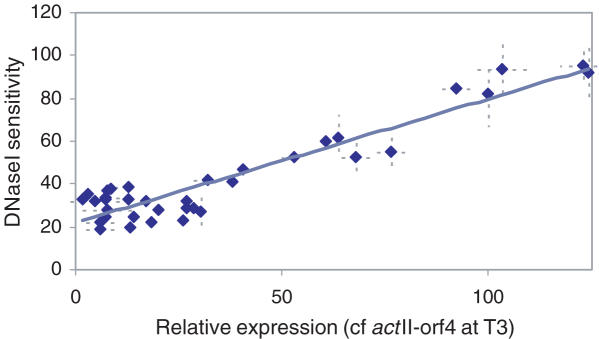
DNase I-sensitivity correlates with gene expression
All 12 genes were analyzed by this method with samples collected at each time point and the percentage loss at 4 U of DNase I digestion was taken as a measurement of accessibility. Figure 4 shows a plot of DNase I-sensitivity against level of gene expression relative to the mean level of expression of the actII-orf4 gene at the T3 timepoint. There is a strong positive correlation between the two properties (R2 = 0.882), confirming the hypothesis that in vivo DNase I-sensitivity of genomic sequences is predictive of transcriptional activity.
Figure 4.
Transcriptionally regulated genes in S.coelicolor show a positive correlation between DNase I-sensitivity and level of expression. qrt-PCR was used to measure both relative DNase I sensitivity and RNA abundance (Figure 1, lower) for each of the twelve genes used in this study at the three timepoints described. Expression data is relative to the value of actII-orf4 gene at T3 timepoint. Each point is the average of at least three independent measurements with standard deviations shown as dotted lines.
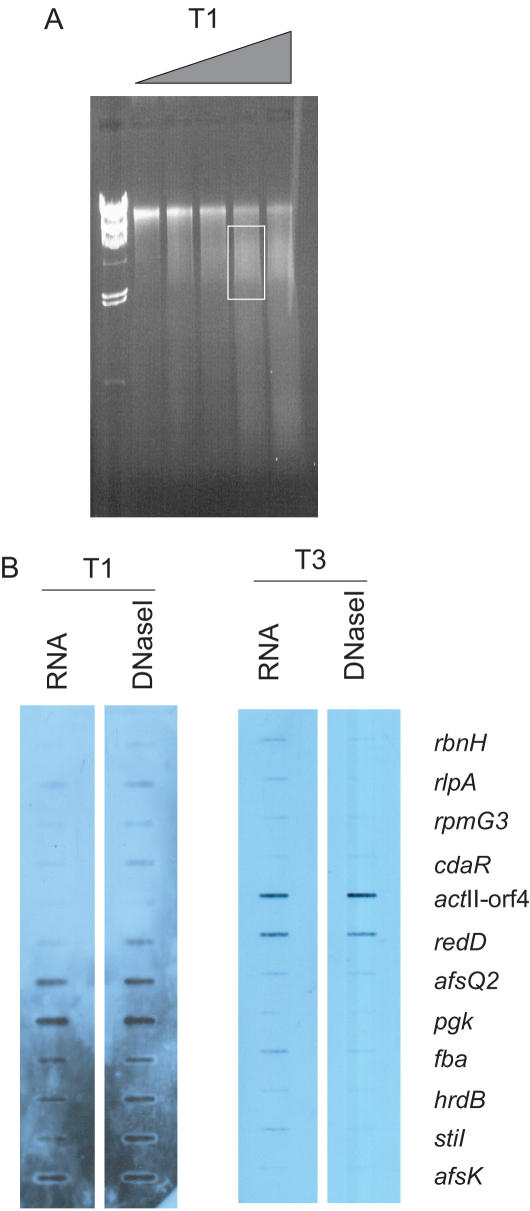
If this is so, it should be possible to use DNase I fractionation of the genome to divide the bacterial chromosome into transcriptionally active and inactive parts. A genomic region susceptible to DNase I digestion would accumulate more cuts and could be separated from the remainder of the genome by size selection. To test this hypothesis, we repeated the growth curve and withdrew samples at time points equivalent to T1 and T3. Half was used to generate RNA and subsequently cDNA samples for expression analysis and the remainder subjected to in vivo DNase I digestion. Following digestion, the mycelium was lysed gently at neutral pH, and the released material was collected by slow centrifugation. The supernatant fraction contained low molecular weight genomic DNA (0.5 to 2 kb) that had been preferentially solubilized by the action of DNase I. The genomic DNA remaining in the pelleted cells was largely of high molecular weight. DNA released into the supernatant by DNase I digestion, along with cDNA generated from the RNA samples, were labeled by random PCR priming and used as hybridization probes with a slot blot containing the 12 profiled genes (Figure 5). There was a good positive correlation between the genes detected in the DNase I-sensitive portion of the genome (Figure 5B, DNase I) and actively transcribed genes (Figure 5B, RNA) at all three time points. There was little correspondence between time points, consistent with the changing patterns of gene expression. However, the DNase I-insensitive fraction consistently detected all of the targets on the filter, demonstrating that this component was largely unfractionated genomic DNA (data not shown).
Figure 5.
In vivo DNase I digestion of the S.coelicolor genome results in fractionation according to transcriptional activity. (A) HCHO-crosslinked DNA–protein complexes from S.coelicolor collected at T1 (shown as an example with the triangle representing increasing amounts of DNase I) and T3 were digested in vivo with increasing amounts of DNase I, and the nucleoprotein complexes separated by 0.7% agarose gel electrophoresis. DNA was extracted from the soluble fraction, defined as that resolved by the gel and indicated by a box, and used as a probe in the subsequent slot-blot experiment. (B) DNA and RNA samples were labeled with DIG-dUTP and used as probes in slot-blot hybridizations with 12 ∼500 bp PCR products corresponding to the panel of genes used in the experiment. At each time point, the RNA and DNA probes detect the same targets, demonstrating that in vivo DNase I treatment had fractionated the genome based on transcriptional activity. Different targets were detected by samples from time points T1 and T3, consistent with the transcriptional programme changing as S.coelicolor enters stationary phase.
DISCUSSION
We have shown that, in vivo, some sequences within the S.coelicolor genome are more accessible than others to DNase I digestion, and that this reflects the transcriptional activity of the gene. Consequently, it is conceivable that the physical structure of the bacterial chromosome plays a role in regulating transcription. This observation parallels the seminal description of the correlation between in vivo DNase I-sensitivity and the expression of eukaryotic genes that established the hypothesis that chromatin structure is central to the control of eukaryotic transcriptional regulation [reviewed in (10)]. Our ongoing work shall investigate to what extent changes to the in vivo DNase I-sensitivty of S.coelicolor genes are causative of increased transcriptional activity.
Bacterial genomes are tightly packed to form visible structures in the cytoplasm, the nucleoids, which are the templates for numerous functions, including transcription, replication and recombination. Eukaryotic chromatin structure is determined by the physical necessity for the genome to be compacted and by the need of DNA-binding proteins to gain access to the underlying sequences. Similar forces may influence the structure of the bacterial nucleoid, but it will be an interesting point of comparison to see if the nucleoid is specially adapted to enable bacteria to thrive in environments in constant flux, where signal transduction mechanisms rapidly induce genetic pathways in response to environmental stress.
Analysis of the structure and function of eukaryotic chromatin (10) has provided fundamental insights into how genes are controlled in vivo. If a bacterial analogue exists, will similar studies reveal new insights into the mechanisms of prokaryotic genetic regulation? Our knowledge of the physical structure of bacterial chromatin lags behind that of eukaryotic chromatin. It is clear that bacterial genomes associate with proteins to form visible nucleoids. Enough is known at the gross level to conclude that the packaging of the genome is ordered, that the nucleoid has a distinct physical structure and that this structure changes in response to environmental stimuli (11). There is also evidence for a higher-order level of organization. For example, six macrodomains capable of independently maintaining superhelical density were described in the E.coli genome (12). These appear to be fixed in size and position but the interactions necessary to maintain the structure are unknown. The existence of smaller domains, ranging in size from 5 to 50 kb, has been inferred from Atomic Force Microscopy studies that have recorded ‘loops’ of DNA fibres emanating from detergent-treated nucleoid preparations (13). Similar structures were described in eukaryotic nuclei and gave rise to the idea of transcriptional units containing several genes (domains) physically tethered to a proteinacieous matrix, or scaffold, allowing close control of local superhelical density which in turn influences transcriptional activity. As in eukaryotes these loops may be tethered or alternatively reflect supercoiling boundaries determined by topoisomerases. Interestingly, antibiotic-induced DNA cleavage by bacterial type II topoisomerase results in the release of 50 to 100 kb sized fragments that may represent the ‘loops’ described by microscopists (14). There is also evidence from microarray data that prokaryotic genomes contain domains: there are periodic maxima and minima of expression along the length of the E.coli genome, with the peaks and troughs separated by ∼50 kb (15). These examples provide good evidence for the bacterial genome having higher-order structure, but the effect of this physical organization on the regulation of the genome is less clear. Perhaps one of its functions is to arrange the nucleoid so that highly expressed genes are close to the tips of the loops allowing transcription factors and gene products to diffuse more readily into and out of the cytoplasm; alternatively it may serve to bring together genes and distant regulatory elements at the base of the loops (16). Such interpretations would favour a model where the size and position of the loops remained relatively stable. An alternative model, proposed by Cook, predicts that the loops are mobile and are created by the association of highly transcribed sequences with an immobilized polymerase factory (17). What is clear is that at a gross level the nucleoid structure influences or reflects gene expression. By studying the local structure of bacterial chromatin, and by developing protocols for its biochemical fractionation, we aim to better understand not only nucleoid structure, but also the mechanisms responsible for changes in it and correlate that with changes in gene expression.
Other studies of genome accessibility have been performed. For example, Struhl and colleagues assessed the accessibility of the E.coli genome by using a ChIP-on-chip protocol to map in vivo LexA binding sites (18). All 27 predicted occurrences of the site were occupied, and 22 novel sites were also detected. The authors concluded that since all sites appeared to be occupied, there must be no issues of accessibility to different parts of the genome. However, by using an established method of investigating in vivo chromatin structure (DNase I digestion), we are employing a system with large precedent that is easy to quantify, applicable to a large number of genes, and uses a probe (DNase I) of variable sensitivity (in contrast, the sensitivity of the antibody-binding step is difficult to control in ChIP protocols).
Bacterial histone-like or nucleoid-associated proteins have been proposed to combine with genomic DNA to form an ordered structure at least in some ways comparable to eukaryotic chromatin (19–21). Our work supports this hypothesis and strengthens the analogy with eukaryotic chromatin by demonstrating that the in vivo accessibility of a gene is positively correlated with its level of expression, providing evidence that the structure of the bacterial nucleoid is dynamic and may be a determinant of transcriptional activity.
This paper describes an approach to fragmenting the genome according to the transcriptional activity of the genes, and provides an opportunity, which we are pursuing, to characterize and compare the (histone-like) proteins associated with the expressed and non-expressed portions of the nucleoid. In addition, our work on the regulation of antibiotic production has provided potential examples of epigenetic phenomena that we are eager to study with this method. For example, antibiotic production can differ between two genetically identical strains depending on the medium from which the spores were derived. Is this evidence of Streptomyces having some ‘memory’ of its history, and if so, are the histone-like proteins implicated in ‘remembering’? The protocol we have developed, together with a method to isolate nucleoprotein complexes on the basis of DNA sequence (M. McArthur and M. J. Bibb, manuscript in preparation), can be used to isolate histone-like proteins associated with antibiotic regulatory genes to assess whether the proteins are the same and whether there is any evidence of post-translational modification. This work would answer the question whether or not a system of epigenetic modification exists in prokaryotes, as it does in eukaryotes, where study of the ‘histone code’ has revolutionized the understanding of how genes are regulated in vivo.
Acknowledgments
The authors thank David Hopwood for comments on the manuscript. This work was funded by Plant Biosciences Limited and a grant to the John Innes Centre from the BBSRC. Funding to pay the Open Access publication charges for this article was provided by the John Innes Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1.Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 2.Berdy J. Bioactive microbial molecules. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M.J. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Pettijohn D.E. Histone-like proteins and bacterial chromosome structure. J. Biol. Chem. 1988;263:12793–12796. [PubMed] [Google Scholar]
- 5.Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. 2000. Practical Streptomyces Genetics. John Innes Foundation. [Google Scholar]
- 6.McArthur M., Gerum S., Stamatoyannopoulos G. Quantification of DNase I-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryding N.J., Anderson T.B., Champness W.C. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 2002;184:794–805. doi: 10.1128/JB.184.3.794-805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi J., Penland L., Brown P.O., Bittner M.L., Meltzer P.S., Ray M., Chen Y., Su Y.A., Trent J.M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Lih C.J., Pan K.H., Cohen S.N. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001;15:3183–3192. doi: 10.1101/gad.943401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 11.Thanbichler M., Viollier P.H., Shapiro L. The structure and function of the bacterial chromosome. Curr. Opin. Genet. Dev. 2005;15:153–162. doi: 10.1016/j.gde.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Valens M., Penaud S., Rossignol M., Cornet F., Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Yoshimura S.H., Hizume K., Ohniwa R.L., Ishihama A., Takeyasu K. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu Y.-H., Chung M.-W., Li T.-K. Distribution of gyrase and topoisomerase IV on bacterial nucleoid: implications for nucleoid organization. Nucleic Acids Res. 2006;34:3128–3138. doi: 10.1093/nar/gkl392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen T.E., Price N.D., Joyce A.R., Palsson B.O. Long-range periodic patterns in microbial genomes indicate significant multi-scale chromosomal organization. PLoS Computat. Biol. 2005;2:e2. doi: 10.1371/journal.pcbi.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kepes F. Periodic transcriptional organization of the E. coli genome. J. Mol. Biol. 2004;340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 17.Cook P.R. Predicting three-dimensional genome structure from transcriptional activity. Nature Genet. 2005;32:347–352. doi: 10.1038/ng1102-347. [DOI] [PubMed] [Google Scholar]
- 18.Wade J.T., Reppas N.B., Church G.M., Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dame R.T., Goosen N. HU: promoting or counteracting DNA compaction? FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 20.Dame R.T., Wyman C., Wurm R., Wagner R., Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 21.Dorman C.J., Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]