Abstract
G-quadruplexes are higher-order DNA and RNA structures formed from G-rich sequences that are built around tetrads of hydrogen-bonded guanine bases. Potential quadruplex sequences have been identified in G-rich eukaryotic telomeres, and more recently in non-telomeric genomic DNA, e.g. in nuclease-hypersensitive promoter regions. The natural role and biological validation of these structures is starting to be explored, and there is particular interest in them as targets for therapeutic intervention. This survey focuses on the folding and structural features on quadruplexes formed from telomeric and non-telomeric DNA sequences, and examines fundamental aspects of topology and the emerging relationships with sequence. Emphasis is placed on information from the high-resolution methods of X-ray crystallography and NMR, and their scope and current limitations are discussed. Such information, together with biological insights, will be important for the discovery of drugs targeting quadruplexes from particular genes.
INTRODUCTION
The knowledge that guanine-rich nucleic acids can self-associate has a long history, pre-dating the double helix itself by almost 50 years. For much of that time, the gels formed by such sequences were more of nuisance value than scientific worth. The molecular basis for the association was subsequently determined by fibre diffraction (1–3) and biophysical (4) studies using the concept (5,6) that the Hoogsteen hydrogen-bonded guanine (G)-tetrad (also termed a G-quartet) is the basic structural motif (Figure 1a). The synthetic polynucleotides poly(dG) and poly(G) were determined in these studies to form four-stranded helical structures (Figure 1b) with the G-tetrads stacked on one another, analogous to Watson–Crick base pairs in duplex DNA. These structures remained largely laboratory curiosities until it was found that short G-rich sequences at the ends of telomeric DNA in eukaryotic chromosomes can associate together in physiological ionic conditions to form discrete four-stranded structures (variously termed quadruplexes, tetraplexes or G4 structures) that incorporate the fundamental structural feature of having at least two contiguous G-tetrads stacked one on another (7,8).
Figure 1.
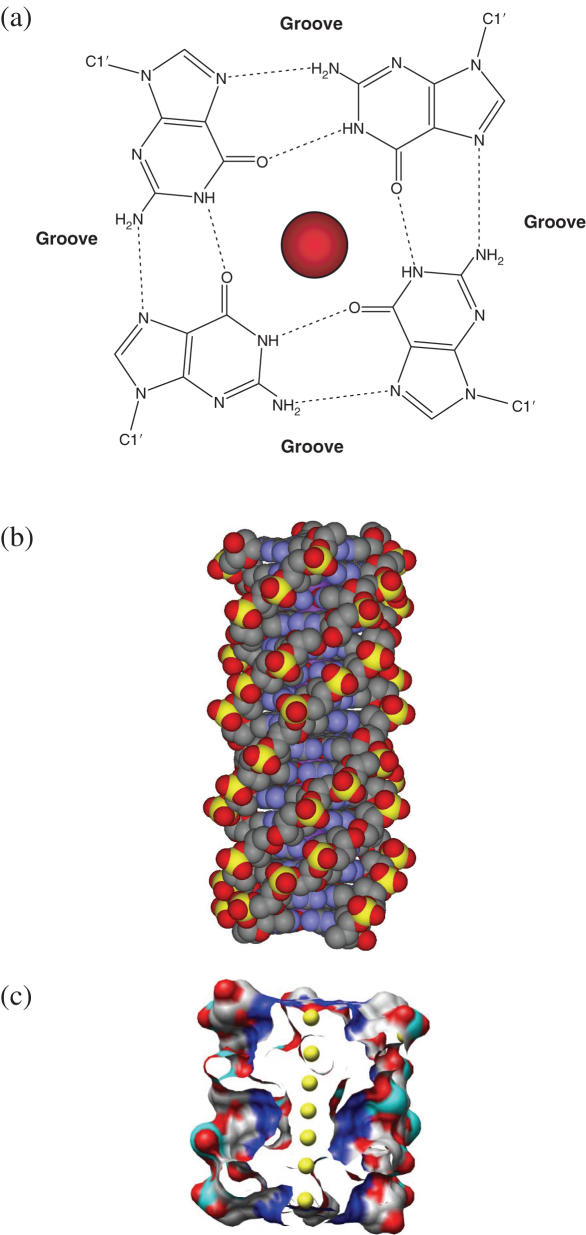
(a) The arrangement of guanine bases in the G-quartet, shown together with a centrally placed metal ion. Hydrogen bonds are shown as dotted lines, and the positions of the grooves are indicated. (b) The poly(dG) four-fold, right-handed helix. (c) Surface view representation of a quadruplex structure comprising eight G-quartets, with the central channel exposed to show an array of metal ions (coloured yellow).
Formation of these quadruplex structures at telomere ends is possible since the terminal nucleotides at the 3′ ends of all telomeric DNAs are single-stranded (9,10), albeit in association with single-strand-binding proteins, such as hPOT1 in Homo sapiens (11,12), where the single-strand overhang is ca. 100–200 nt long. Telomeric DNA sequences (13) comprise G-rich tandem repeats (Table 1), i.e. are not pure G sequences, and have short non-G tracts regularly interspersing the G ones. A few prokaryotic species, such as Streptomyces also have linear chromosomes, with repetitive DNA at the ends, but with distinct sequences that can form inverted repeat structures (14). A second category of quadruplexes involve oligonucleotide aptamers comprising quadruplex-forming sequences, which have the ability to selectively act as inhibitors of signal transduction or transcription via binding to particular targets, such as Stat3 (15) or nucleolin (16) in cancer cells. Few 3D structures of quadruplexes formed from aptamer sequences have been fully characterized; that of the thrombin-binding sequence d(GGTTGGTGTGGTTGG) is a notable exception (17). The third category comprises potential quadruplexes that may be formed from appropriate G-rich sequences that are present within a wide range of genes (and very extensively in non-coding regions of many genomes). Now that extensive sequence data are available on a large number of eukaryotic and prokaryotic genomes, it is apparent that such sequences are highly prevalent (18–21), and an increasing number of quadruplexes arising from them have been reported. This survey will focus on some of the underlying principles and emerging issues concerning (i) sequence (primary structure), (ii) the diverse patterns of folding, i.e. quadruplex topology (secondary structure) and (iii) more detailed structural information (tertiary structure) on both telomeric and non-telomeric quadruplexes, especially those from the high-resolution methods of crystallography, molecular simulation and NMR.
Table 1.
Some known telomeric DNA sequences
| Group | Organism | Telomeric repeat |
|---|---|---|
| Vertebrates | Human, mouse, Xenopus | TTAGGG |
| Filamentous fungii | Neurospora crassa | TTAGGG |
| Slime moulds | Physarum, Didymium, Dictyostelium | TTAGGG AG(1–8) |
| Kinetoplastid protozoa | Trypanosoma, Crithidia | TTAGGG |
| Ciliate protozoa | Tetrahymena, Glaucoma | TTGGGG |
| Paramecium Oxytricha | TTGGG(T/G) | |
| Stylonychia, Euplotes | TTTTGGGG | |
| Apicomplexan protozoa | Plasmodium | TTAGGG(T/C) |
| Higher plants | Arabidopsis thaliana | TTTAGGG |
| Green algae | Chlamydomonas | TTTTAGGG |
| Insects | Bombyx mori | TTAGG |
| Roundworms | Ascaris lumbricoides | TTAGGC |
| Fission yeasts | Schizosaccharomyces pombe | TTAC(A)(C)G(1–8) |
| Budding yeasts | Saccharomyces cerevisiae | TGTGGGTGTGGTG (from RNA template) or G(2,3)(TG)(1–6)T (consensus) |
| Candida glabrata | GGGGTCTGGGTGCTG | |
| Candida albicans | GGTGTACGGATGTCTAACTTCTT | |
| Candida tropicalis | GGTGTA[C/A]GGATGTCACGATCATT | |
| Candida maltosa | GGTGTACGGATGCAGACTCGCTT | |
| Candida guillermondii | GGTGTAC | |
| Candida pseudotropicalis | GGTGTACGGATTTGATTAGTTATGT | |
| Kluyveromyces lactis | GGTGTACGGATTTGATTAGGTATGT |
GENERAL FEATURES OF QUADRUPLEX TOPOLOGY AND STRUCTURE
Quadruplexes can be formed from one, two or four separate strands of DNA (or RNA) and can display a wide variety of topologies, which are in part a consequence of various possible combinations of strand direction, as well as variations in loop size and sequence. They can be defined in general terms as structures formed by a core of at least two stacked G-tetrads, which are held together by loops arising from the intervening mixed-sequence nucleotides that are not usually involved in the tetrads themselves. The combination of the number of stacked G-tetrads, the polarity of the strands and the location and length of the loops would be expected to lead to a plurality of G-quadruplex structures, as indeed is found experimentally.
Potential unimolecular (i.e. intramolecular) G-quadruplex-forming sequences can be described as follows:
where m is the number of G residues in each short G-tract, which are usually directly involved in G-tetrad interactions. Xn, Xo and Xp can be any combination of residues, including G, forming the loops. This notation also implies that the G-tracts can be of unequal length, and if one of the short G tracts is longer than the others, some of the G residues will be located in the loop regions. The assumption that all G tracts within a quadruplex sequence are identical is true for vertebrate telomeric sequences, but is not always the case for non-telomeric genomic sequences, or even for all telomeric sequences in some lower eukaryotics (see Table 1). In principle bimolecular (dimeric) and tetramolecular (tetrameric) quadruplexes can each be formed from the association of non-equal sequences, although very few quadruplexes with such features have yet been studied in detail. Thus, almost all bimolecular quadruplexes reported to date are formed by the association of two identical sequences Xn Gm Xo Gm Xp, where n and p may or may not be zero. Tetramolecular quadruplexes may be formed by four Xn Gm Xo or GmXnGm strands associating together.
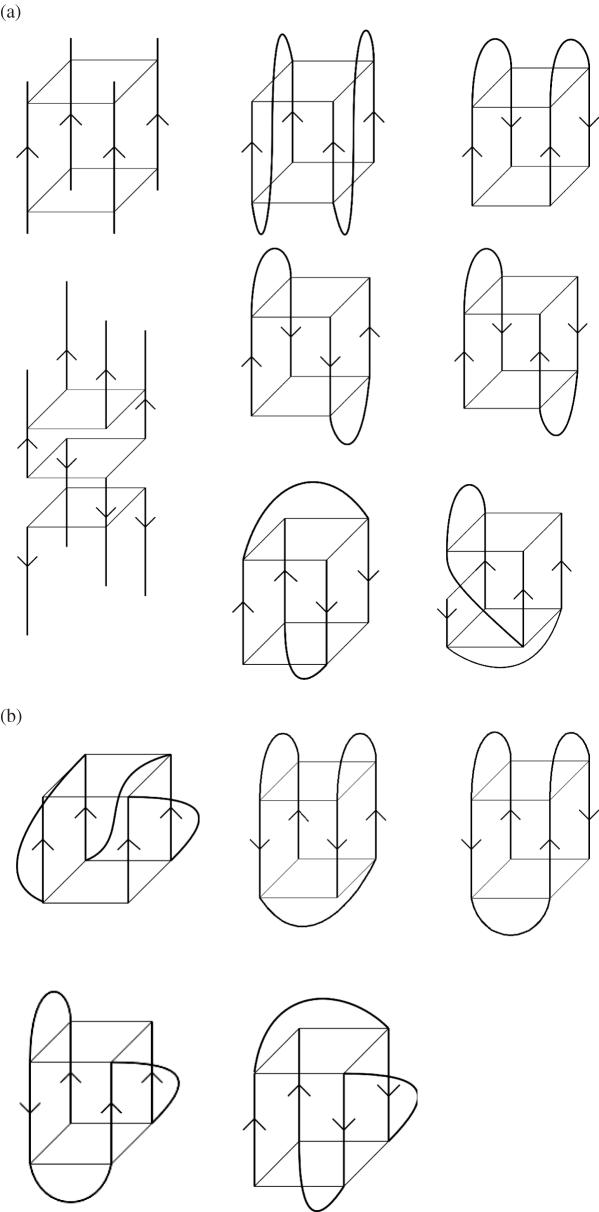
Quadruplex structures may be classified according to their strand polarities and the location of the loops that link the guanine strand(s) for quadruplexes formed either from a single-strand or from two strands. Adjacent linked parallel strands require a connecting loop to link the bottom G-tetrad with the top G-tetrad, leading to propeller type loops (these are sometimes termed strand-reversal loops but we prefer the simpler term since this describes the appearance of this loop and does not introduce any potential confusion about strand direction). This feature has been found both in crystal structures (22) and in solution (23) for quadruplexes formed from human telomeric DNA sequences (see below), and more recently in a number of non-telomeric quadruplexes. Quadruplexes are designated as anti-parallel when at least one of the four strands is anti-parallel to the others. This type of topology is found in the majority of bimolecular and in many unimolecular quadruplex structures determined to date. Two further types of loops have been observed in these structures, in addition to parallel loops. Lateral (sometimes termed edge-wise) loops join adjacent G-strands, as observed in the structures of both two asymmetric quadruplexes observed in solution by NMR for the d(TG4T2G4T) sequence (24) and in the bimolecular quadruplex structure formed by the sequence d(GGGCT4GGGC) (25). Two of these loops can be located either on the same or opposite faces of a quadruplex, corresponding to head-to-head or head-to-tail, respectively when in bimolecular quadruplexes (Figures 2a and 4). Strand polarities can vary, as in the example of the two distinct bimolecular quadruplexes formed by d(G4T3G4), with one being a head-to-tail lateral loop dimer in which all adjacent strands are anti-parallel, and the other is a head-to-head hairpin quadruplex with one adjacent strand parallel and the other is anti-parallel (26). The second type of anti-parallel loop, the diagonal loop joins opposite G-strands, as observed in the structure formed by the Oxytricha nova telomeric sequence d(G4T4G4) (27–31) In this instance the directionalities of adjacent strands must alternate between parallel and anti-parallel, and are arranged around a core of four stacked G-tetrads.
Figure 2.
(a) Some possible topologies for simple tetramolecular (on the left-hand side) and bimolecular quadruplexes. Strand polarities are shown by arrows. (b) Some possible topologies for simple unimolecular quadruplexes.
All parallel quadruplexes have all guanine glycosidic angles in an anti conformation. Anti-parallel quadruplexes have both syn and anti guanines, arranged in a way that is particular for a given topology and set of strand orientations, since different topologies have the four strands in differing positions relative to each other. All quadruplex structures have four grooves, defined as the cavities bounded by the phosphodiester backbones. Groove dimensions are variable, and depend on overall topology and the nature of the loops. Grooves in quadruplexes with only lateral or diagonal loops are structurally simple, and the walls of these grooves are bounded by monotonic sugar phosphodiester groups. In contrast, grooves that incorporate propeller loops have more complex structural features that reflect the insertion of the variable-sequence loops into the grooves (see Figure 5).
The formation and stability of G-quadruplexes is monovalent cation-dependent. This has been ascribed to the strong negative electrostatic potential created by the guanine O6 oxygen atoms, which form a central channel of the G-tetrad stack (4,32–34), with the cations located within this channel (Figure 1c). The precise location of the cations between the tetrads is dependent on the nature of the ion, with Na+ ions within the channel being observed in a range of geometries; in some structures, a Na+ ion is in plane with a G-tetrad whereas in others it is between two successive G-tetrads. K+ ions are always equidistant between each tetrad plane, and form the eight oxygen atoms in a symmetric tetragonal bipyramidal configuration. Other ions can substitute for these two. Thallium (1+), with an ionic radius close to that of the K+ ion, can substitute for it. The NMR structure (27) of the Tl+-containing bimolecular quadruplex formed from the O.nova sequence d(G4T4G4), shows identical quadruplex topology to that in the K+ ion form found in the crystalline state (28), which is itself identical with the NMR structures of the well-characterized Na+ form (29–31). On the other hand, there are a number of well-established examples where the change from Na+ to K+ induces profound structural alteration, implying high conformational flexibility for these particular quadruplexes. It is equally clear that some quadruplexes, such as the bimolecular d(G4T4G4) quadruplex (28–31) and the parallel-stranded structure formed by four d(TGGGGT) molecules (35,36), have very stable and unique topologies. A series of very long time-scale molecular dynamics simulations (0.5–1 µ s) have shown that these structures retain their integrity not only in simulated solution but also in the gas phase, provided the cations are present (37).
Methods for quadruplex topology and structure determination
A number of quadruplex studies have employed the methods of biophysical chemistry, notably circular dichroism (CD), to assign topology. The main attraction of CD spectroscopy is its potential to discriminate between quadruplex topologies having differences in parallel and anti-parallel strand orientation, arising from different arrangements of anti/syn glycosidic angles. CD therefore can be a useful and rapid method for establishing an overall fold. It requires very little sample (μM concentrations are sufficient) and is suited to examining a wide range of solution conditions and their influence on quadruplex formation. The method is, however, more sensitive than ultraviolet (UV) melting experiments to the buffer composition. Phosphate, acetate, sulphate and carbonate buffers should be avoided due to their strong absorbance at wavelengths commonly used for CD experiments. Many quadruplex-forming sequences have been studied using this technique and the majority of spectra conform to one of two characteristic spectral forms. Classic parallel and anti-parallel quadruplexes show similarly shaped traces but with maxima at distinct wavelengths. For quadruplexes assigned to be parallel-stranded, a maximum is present at ∼260 nm and a minimum at ∼240 nm; the maximum and minimum for an anti-parallel quadruplex are typically at around 290 and 260 nm, respectively. These assignments have predominantly been used to examine telomeric and telomere-like sequences (i.e. sequences with regular repeating loop regions). As more complex quadruplex-forming sequences are examined, the reliability of assigning topology based on the comparison of a spectrum with the CD signature of known parallel or anti-parallel telomeric quadruplexes cannot always be assumed since (i) topologies may not conform to those observed with telomeric quadruplexes, (ii) multiple species cannot readily be identified in CD spectra and (iii) non-telomeric loop sequences may perturb the CD spectra in unforeseen ways.
X-ray crystallography and high-field NMR spectroscopy offer in principle the possibility of both topological assignment and more detailed atomic-level structure determination. However sometimes even with these methods, caveats are required. Successful structure determination by NMR methods relies on the sequence forming a kinetically stable species in solution; the presence of multiple species limits the structural information obtained. This may be overcome by the use of mutated or modified sequences based on the original G-rich sequence, but which only form a single species in solution environments. It is common practice to screen up to several tens of mutated sequences and other variants from wild-type until one is found that produces a well-resolved NMR spectrum showing a single species amenable to analysis. Favoured mutations are of thymine by uracil or 5-bromo-uracil. Variations in both 5′ and 3′ flanking sequence are also commonly explored. Crystallography similarly uses site mutations and/or sequence scanning, to find sequences that will crystallize. It is also necessary to use bases with heavy-atom substitutions (as in 5-bromo-thymine) for phasing purposes when confronted with structures that cannot be solved by molecular replacement. The various structures formed in solution by variants of the human telomeric two-repeat sequence (23) show that such mutations and changes cannot always be relied upon to preserve a particular topology and will inevitably alter the equilibrium between different ones, sometimes by forming additional stabilizing interactions. Thus generalizations from any one NMR or crystal structure need to be made with care, and need to take due regard of the role played by the modified/additional nucleotides, especially in the absence of independent data or more than one corroborating structure.
Tetramolecular and bimolecular quadruplex structures
Tetramolecular G-quadruplexes comprises the simplest category of quadruplex nucleic acid (Figure 2a). Thus the crystallographic and NMR structures of d(TG4T)4 (35,36) and its RNA equivalent (UG4U)4 (38) show all the strands parallel to one another and the guanine glycosidic torsion angles are all in the anti conformation. However, even tetramolecular G-quadruplexes can form more complex structures, as shown by d(GGGT)8, in which eight strands form an interlocked bimolecular quadruplex (39) with two symmetric parallel tetramolecular d(GGGT) quadruplexes being linked by an external G-tetrad formed by slipped-out guanines from each quadruplex. The family of sequences d(GCGGXGGY) form tetramolecular structures comprising two unusual bistranded quadruplex monomer units containing G:C:G:C tetrads (40).
Association of two strands to produce bimolecular quadruplexes introduces increased topological variation (Figure 2a). The classic bimolecular quadruplex structure (Figure 3) is that formed by two strands of the O.nova sequence d(G4T4G4), with a diagonal T4 loop at each end of the symmetric quadruplex (28–31). It is remarkable that even apparently conservative changes in this sequence have major topological consequences: thus d(G3T4G4), with one guanine at the 5′ end less than in the wild-type sequence forms a bimolecular quadruplex having both lateral and diagonal loops (41). This is one of the few cases where a bimolecular quadruplex has an unequal number of parallel (three) and anti-parallel (one) strands; subsequent studies (42) showed that this topology is not dependent on the presence of ions, but is retained in K+ or Na+ solution, as does a mixed di-cation form (43). The sequence isomer, now with one guanine less at the 3′ end [i.e. d(G4T4G3)], also forms an asymmetric bimolecular quadruplex, but with less dramatic differences compared to the Oxytricha parent structure. This structure has a core of three stacked G-tetrads, so the two guanines not included in this core are involved in one of the two diagonal loops (44). Reducing the number of guanines still further, to d(G3T4G3), results in a more conventional diagonal-looped quadruplex, but with asymmetry in guanine glycosidic angles (45,46). Decreasing the size of the thymine loops also results in topological change, as observed in the crystal structures (Figure 4) of the bimolecular quadruplexes formed by d(G4T3G4), with lateral loops being consistently favoured (26). The implication of this, that loops with three or less nucleotides dis-favour diagonal in preference to lateral loops, is borne out by the exclusive presence of lateral loops in both interconverting bimolecular quadruplexes formed by the d(TG4T2G4T) Tetrahymena sequence (24). These are closely similar to the head-to-head and head-to-tail lateral loop bimolecular quadruplexes of d(G4T3G4) (26). Interestingly, the 5′ and 3′ flanking thymine residues in this pair of sequences have no effect on quadruplex topology.
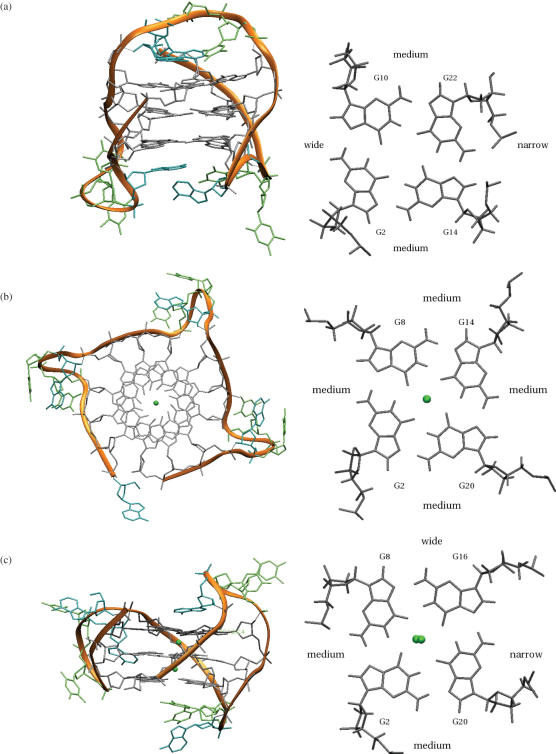
Figure 3.
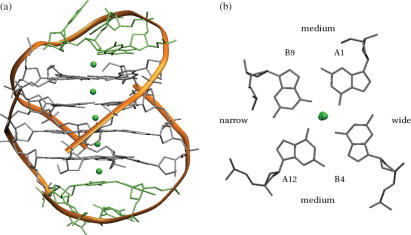
The crystal structure (28) of the bimolecular quadruplex formed by the O.nova telomeric sequence d(G4T4G4) (PDB entry 1JPQ). (a) Overall topology is indicated by the ribbon representation in orange. The details of the molecular structure are also shown. Potassium ions are shown as green spheres. (b) A projection down the central channel, indicating the relative widths of the four grooves
Figure 4.
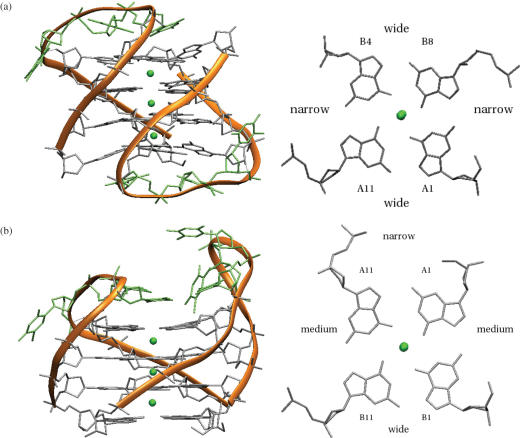
Crystal structure (26) of the two bimolecular quadruplexes found in the crystal structure of d(G4T3G4) (a) two views of the head-to-tail quadruplex (PDB entry 2AVH). (b) Two views of the head-to-head quadruplex (PDB entry 2AVJ).
It is not possible at present to define a comprehensive set of rules that specifies the folding of bimolecular G-quadruplexes, in the absence of much more structural and energetic information than is currently available, especially since in solution it is apparent that multiple structures sometimes exist in equilibrium. However, several significant contributing factors are apparent, notably loop length and sequence, and G-tract length (47,48). In general bimolecular quadruplex topology appears not to be markedly dependent on the nature of the cation, in striking contrast with unimolecular quadruplexes. Molecular dynamics simulations have been employed to model the stability of particular quadruplexes, such as that in the Oxytricha bimolecular topology (49–52). Simulations have suggested a set of preferences for thymine-containing loops (53), which are broadly in accord with the experimental observations from crystallographic and NMR studies, as outlined above, which show that T3 loops have a marked preference for lateral loop conformations. This is not consistently indicated by the free-energy calculations, which may be a consequence of the inadequacies of current force fields to fully account for the electrostatics of quadruplexes, and of the likely small energy differences between differing loop conformations. On the other hand, shorter T2 loops do restrict conformational flexibility of topological features. It also seems that differing numbers of guanines in the individual G-tracts results in quadruplexes with asymmetric topologies, which again, are not readily predictable at present.
Unimolecular quadruplexes
The same three loop types (propeller, lateral and diagonal) found in bimolecular quadruplexes also occur in unimolecular quadruplex structures (Figure 2b). For example the human telomeric sequence d[AG3(TTAGGG)3] forms an anti-parallel arrangement in Na+ solution (Figure 5a), with one diagonal and two lateral loops (54). In K+ solution, this sequence appears to be able to access a number of distinct folds, as described further below; the crystal structure of this sequence (22) shows all strands in parallel orientations and therefore with the three TTA tracts forming three propeller loops. This all-parallel topology has been observed for several other sequences in solution, e.g. for the aptamer sequence d(G4TG3AG2AG3T), which is a potent inhibitor of HIV-1 integrase. This aptamer forms an interlocked quadruplex dimer, each with three single-nucleotide propeller loops (55). Propeller loops are also found in conjunction with lateral or diagonal loops, as in the d(T2G4T2G4T2G4T2G4) and d(G2T4G2CAG2GT4G2T) NMR structures (56,57). The size of the loop can affect unimolecular quadruplex stability (48); the Oxytricha-like unimolecular sequence, d(G4T2G4TGTG4T2G4) has a more unfavourable ΔG0 value than its bimolecular counterpart, although the former's melting temperature is considerably higher due to lower entropic contributions (58).
Figure 5.
Structures of the human unimolecular telomeric quadruplex formed from the sequence d[AGGG(TTAGGG)3]. In each case two views are shown (a) one of the deposited structures of the Na+ form, determined by NMR (PDB entry 143D) (54), with a diagonal and two lateral loops. (b) K+ form A, determined by crystallography (PDB entry 1KF1) (23), with three strand-reversal loops (c) K+ form B, showing the topology determined by NMR (75,96), with one strand-reversal and two lateral loops. Nucleotide loop conformations for the detailed atomic structure shown here have been obtained from a molecular dynamics simulation performed by Sarah Burge that has used this topology as a starting-point. The NMR-derived structure of one of the sequences determined experimentally (96), is also available as PDB entry id 2KGU.
Few systematic studies have been reported of the effects of differing loop lengths and sequence on various unimolecular quadruplex folds and loop types. An analysis, restricted to loops with differing numbers of thymine residues, used molecular dynamics in conjunction with biophysical measurements (59), and has concluded that quadruplexes with three T1 loops are constrained to only form parallel topologies, whereas quadruplexes with three T2 loops can form both parallel and anti-parallel topologies (in this instance parallel structures are likely to be favoured). In addition, a single T1 loop in a quadruplex is compatible with both parallel and anti-parallel arrangements, but the parallel type is more energetically favoured. Quadruplexes with a single T2 to T6 loop are stable with either parallel or anti-parallel topologies; however, anti-parallel ones are likely to be slightly preferred. The conclusions regarding single-nucleotide loops are likely to be generally applicable to all four nucleotides A, T, G and C since loop size is the determinant of steric constraints on topology and energetics. However, the relative stabilities of loops with >1 nt are also dependent on relative nucleotide stacking energies within loops, as has been shown by a thermodynamic profiling study (60). Another factor has been highlighted by a study on the effects of ribonucleotide substitution for deoxynucleotide (61). Systematic substitutions in both loops and G-tracts have suggested that the greater tendency for ribonucleotides to be in an anti glycosidic conformation, resulting in a preference for parallel topologies in RNA quadruplexes.
Vertebrate telomeric quadruplexes
The large number of studies on the structure(s) adopted by repeats of the vertebrate telomeric sequence d(TTAGGG) have, in large part, focused on the topology adopted by the folding of the single-stranded repeats at the 3′ telomere end. The average length of this single-stranded overhang, of ca. 150 nt, corresponds to an assembly of 5–6 four-repeat unimolecular quadruplexes. Almost all considerations of the structural features of the ‘human quadruplex’ have focused on individual quadruplexes, especially the four-repeat unimolecular quadruplex(es) rather than the structure and dynamics of quadruplex assemblies, which are the more biologically relevant system (11,12). Apart from the 3′ single-stranded overhang, all telomeric DNA is double-stranded [and associated with a number of telomeric proteins in the ‘shelterin’ complex (62)]. In the absence of proteins or small molecules, the equilibrium for vertebrate telomeric DNA has been found (63) to favour duplex over dissociation into quadruplex and i-form motifs (the four-stranded arrangements formed by the complementary C-rich strand and organized around cytosine–cytosine base pairs).
There is good evidence from a range of biophysical techniques, that the four-repeat quadruplex formed by the sequence d(TTAGGG)4 (and variants on it, notably d[AGGG(TTAGGG)3]), adopt differing topologies in Na+ versus K+ solution (60,64–69). NMR analysis (54) of the species formed in Na+ conditions by the 22mer d[AGGG(TTAGGG)3] has shown that the structure has an anti-parallel fold with two lateral and one diagonal loops, each loop comprising the TTA triad sequence (Figure 5a). Subsequent crystallographic analyses of this sequence and the related 12mer (i.e. two-repeat) sequence d(TAGGGTTAGGGT), in K+ solution (22), showed that they form a unimolecular (Figure 5b) and a bimolecular quadruplex, respectively in the crystal lattice. Both have the same topology with parallel orientations for all four strands, and propeller loops formed by the TTA sequence [single occurrences of these type of loops had been observed previously (56,57)]. This all-parallel arrangement, which is radically different from the Na+ structure, was subsequently observed in solution by NMR (23) for the closely-related sequence d(TAGGGUTAGGGT), although the same study also showed that the dominant form for another modified sequence, d(UAGGGTBrUAGGGT), is that of an anti-parallel quadruplex with lateral loops. The propensity of telomeric quadruplexes for topological diversity is shown by the unusual asymmetric bimolecular quadruplex (69) formed by three telomeric repeats, with all three G-tracts of one strand associating with a single G-tract of another.
The unexpected nature of the quadruplex fold in the K+ crystal structures has led to a number of biophysical studies intent on identifying the nature of the species formed by d[AGGG(TTAGGG)3] in solution [see e.g. Refs. (64–68,70–72)]. It is unsurprising that some studies suggest the co-existence of several forms (59,60,67), especially in view of the ability of quadruplexes with 3 nt loops to readily adopt topologically distinct structures upon small changes in environment or sequence, suggesting that the various forms are energetically-similar. This is in accord with both experimental and simulation studies, which show that there is only a small free-energy difference between the human telomeric parallel and anti-parallel quadruplexes with TTA loops (59,67). Thus a particular set of conditions or sequence will favour a particular fold or mixture of folds, analogous to the process of crystallization, which selects one or a few particular low-energy form(s) from solution, that are best able to pack effectively to give a well-ordered crystal. One key feature of the crystal structure's parallel fold is that the open nature of the G-tetrad surfaces of individual quadruplexes, due to the absence of lateral or diagonal loops, facilitates their stacking together into a very compact and stereochemically acceptable arrangement. This feature would also enable the assembly of successive quadruplexes, as would occur in biological telomeric DNA, and the binding of appropriate small-molecule drugs.
A recent CD study (73) of the K+ form of the 22mer sequence has exploited the property of 8-bromo-guanosine to favour the syn glycosidic angle conformation, and has incorporated this modification at various positions in the sequence to determine topology from CD measurements. This is challenging since not all the CD spectra of individual modified sequences show behaviour consistent with the proposed structures. It was concluded that d[AGGG(TTAGGG)3] in solution is a mixture of two forms, one of which has a new topology for telomeric unimolecular quadruplexes, having anti-parallel/parallel strands with one propeller and two lateral loops. The unambiguous identification of all the species present in solution using CD alone may not be straightforward (74), so the topology of any other components have not yet been clarified. This fold (Figure 5c) has also been reported in two separate NMR analyses (75,96). Both have used sequences that have been slightly altered at the terminii from telomeric regularity: d[TTG3(TTAG3)3A] (96) and d[AAAG3(TTAG3)3AA] (75), since NMR finds that the native 22mer as used in the crystal structure determination, forms a mixture of species in K+ solution, which is not amenable to structure analysis. The structure of the former has been reported in detail (96), and shows that the extra flanking residues are involved in Watson–Crick and reverse Watson–Crick base pairs that are stacked one on each end of the core of G-tetrads, and help to stabilize this particular topology. This explains why the fold has not been observed to date with the 22mer, which cannot form such base pairs. Thus what remains still to be determined by fine structure methods is the precise nature of all the species present in the K+-solution of d[AGGG(TTAGGG)3].
UNIMOLECULAR NON-TELOMERIC QUADRUPLEXES
Sequence occurrences
The realization that potential quadruplex-forming sequences can occur in double-stranded non-telomeric regions of the human genome (and therefore in other eukaryotic and prokaryotic genomes), is not new, and they have been identified, e.g. in promoter and immunoglobulin switch regions and in recombination hot spots (76). There have been two recent systematic surveys of the complete human genome sequence, searching for potential unimolecular quadruplex-forming sequences (18,19). Both have used the same criteria for the definition of a potential quadruplex sequence and have agreed on the overall number of these sequences present, even though the statistical and analytical approaches used were quite different. These studies assumed that long-range and even medium length loops, although feasible are impractical to include because of the very large number of possibilities, which would be present. The criteria for a potential quadruplex sequences was therefore restricted to:
where NL1-3 are loops of unknown length, within the limits 1<NL1-3 <7 nt.
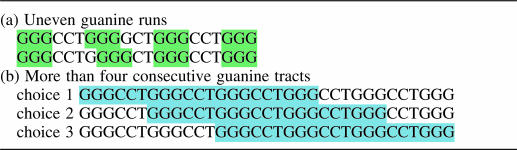
Potential quadruplex sequences are distributed throughout the human genome in exons, introns, in untranslated regions, in promoter sequences (sometimes though not invariably directly upstream of transcription start sites) and within gene desert regions. The majority of potential quadruplex sequences appear to be involved in more than one possible quadruplex, either as a result of being in a sequence with more than four consecutive runs of guanine (Table 2a) or because a lack of parity in the lengths of the loop sequence (Table 2b) means that some of the guanines in the G-runs have to be part of at least one of the loop sequences. Although it is possible that many of the potential topologies exist in dynamic equilibrium we cannot at present predict which are stable. This fold ambiguity will require much more extensive experimental data before generalized theoretical approaches to predicting folding can be reliable.
Table 2.
Examples of ambiguity in potential quadruplex sequences showing (a) uneven guanine runs creating a choice of loop sequence and (b) where more than four consecutive guanine tracts gives rise to more than one possible quadruplex fold for a sequence
When every possible combination is considered, a survey of the Ensemble database (V20 NCBI assembly 34c) yields 5 713 900 possible potential quadruplex sequences in the human genome. However, this corresponds to a maximum of 375 157 distinct non-overlapping potential quadruplex sequences. Since a unimolecular quadruplex sequence can be broken down into four equal sized G-tracts and three distinct loop regions we can characterize different quadruplex sequences by the contents of their loop regions. Over a range of loop lengths of 1 to 7 bases there are 21 844 possible sequences of which 20 492 are actually found at least once in the human genome. The large differences in the number of times that these loops occur indicates that some sequences are over-represented whereas others are highly under-represented within the entire population of potential quadruplex-forming sequences (Table 3).
Table 3.
The top 20 most frequently occurring loop sequences (18)
| Rank | Sequence | Population |
|---|---|---|
| 1 | A | 193 756 |
| 2 | T | 121 406 |
| 3 | C | 44 020 |
| 4 | AA | 40 026 |
| 5 | CT | 32 472 |
| 6 | CA | 32 070 |
| 7 | G | 29 623 |
| 8 | AT | 19 957 |
| 9 | AGA | 19 144 |
| 10 | TT | 17 089 |
| 11 | TA | 12 641 |
| 12 | CC | 10 955 |
| 13 | AGT | 9896 |
| 14 | AGGA | 9463 |
| 15 | AGGT | 9434 |
| 16 | TGA | 9237 |
| 17 | AAA | 7839 |
| 18 | CCT | 7151 |
| 19 | TGT | 6619 |
| 20 | CCA | 6269 |
Unsurprisingly there is a tendency for a high proportion of quadruplex sequences to occur within promoter sequences given that these are G-rich regions. This is reflected in the frequency of occurrence of each loop sequence. Small single-base loops are the most common and loops which are made up of guanines in the centre of a sequence are abundant e.g. AGGA and AGGT are very common loop sequences. In general, longer sequences occur less frequently within quadruplex loops. There are however notable exceptions. For example, CCTGTT and TAGCATT are highly over-represented among possible six and seven base pair loop sequences.
Unique sequence motifs have been identified in the human genome by examining how frequently a given loop sequence is found in the first, second or third loop position e.g. the sequence CCTGTT occurs in the human genome 1266 times as a first loop sequence, only 18 times as a second loop and just 9 times in the third loop position. There are several variants of this sequence with a similar pattern of loop distribution, with the common feature that they tend to have CCTGT within the sequence of the first loop. Although seemingly ubiquitous throughout the human genome, this sequence is also strongly represented in the Human Endogenous Retrovirus Database (77).
The findings that quadruplex sequences occur in the promoter regions of several cancer genes have stimulated a number of structural studies, which are outlined in a subsequent section. We list in Table 4 a selection of potential quadruplex sequences, mostly directly upstream of the transcription start site in a set of cancer-associated genes (from the Wellcome Trust Sanger Institute Cancer Genome Project web site (http://www.sanger.ac.uk/genetics/CGP).
Table 4.
Sequences in cancer-related genes that have been identified as forming quadruplex structures
The likely G-tracts that are involved in G-quartets are marked in red.
The concept that quadruplex formation may in particular provide a transcriptional regulatory signal has received support from the analysis of quadruplex occurrence in Escherichia coli and 17 other prokaryotic genomes (21), where G4 sequences are statistically significantly over-represented in promoter regions proximal to transcription start sites, and may be associated with global supercoiling-sensitive gene regulation. The occurrence of quadruplex sequences in alternatively spliced mammalian pre-mRNA sequences has been surveyed, and results are now available in the GRSDB database (78). There is yet rather little experimental data on RNA quadruplexes; since some RNA quadruplexes are likely to have high stability these would be more likely to be present in non-translated mRNA sequences. It has been suggested that the fragile X mental retardation protein (FMRP) binds with high affinity to G-rich mRNA targets in yet undefined genes, which can form quadruplex arrangements: one with a possible parallel topology has been identified (79). A survey of all 16 654 genes in the human gene database has found that there is a significant correlation between quadruplex sequence occurrence and classes of gene (80), with proto-oncogenes having a high quadruplex-forming potential compared with a low potential for tumour-suppressor genes. It is suggested (80) that this reflects genomic structure being selected based on gene function.
Potential quadruplex-forming sequences also occur within regions of chromosomal translocations. One such well-characterized example is the breakpoint region on human chromosome 14 associated with the lymphoma-associated bcl-2 gene translocating to chromosome 18 (81). The region just downstream of the breakpoint is G-rich with runs of short G-tracts characteristic of a quadruplex-type sequence. Analogous G-tracts have been mapped in the breakpoint region of the SHANK3 gene (82).
Much of the interest in non-telomeric quadruplex sequences and their possible structures are due to their occurrence in genes associated with proliferation, especially in c-myc and a number of oncogenes (outlined below). The biological implications of these sequences is as yet at an early stage of study, although the possibility is currently being actively explored that they may be involved in the regulation of gene expression, and that this might be amenable to exploitation by small-molecule therapy (83). Evidence from the small number of non-telomeric quadruplex structures available to date suggests that there is large diversity both in topology and molecular structure between topoisomers, which may be exploited in the future for therapeutic gain (84,85).
Topology and structure
Quadruplex formation has been examined in vitro in a number of non-telomeric sequences (Table 4). The NHE III1 G-rich sequence in the promoter region of the c-myc oncogene, which is responsible for 80–90% of its transcriptional activity, has been especially studied. The existence of a quadruplex within this promoter region was initially proposed (20) based on the data from chemical probe and gel mobility measurements, and from fluorescence resonance energy transfer spectroscopy (86). Subsequent studies established a relationship between quadruplex stabilization within this sequence and suppression of c-myc transcriptional activation (83), with the porphyrin TMPyP4 acting to stabilize the quadruplex structure. NMR in solution (87–89), as well as that of a porphyrin TMPyP4 complex (89) has determined topology and detailed structures of several c-myc quadruplex sequences. Non-telomeric G-rich regions often contain more than four consecutive G-tracts (see above) which, as in the case of c-myc, results in the formation of multiple quadruplex species in the native Pu27 region (this dynamic behaviour, which may involve shuffling between G-tracts, is distinct from the conformational rearrangements shown by the human telomeric 22mer, e.g.). Shorter sequences from within the Pu27 region have been successfully analysed by NMR methods. Myc-2345 and Myc-1245 each contain four G-tracts and form very stable parallel-stranded unimolecular quadruplexes in solution (Figure 6), with G-tracts joined through propeller loops, and with all guanines involved in the G-tetrads having an anti conformation (87). These structures therefore share features with the human telomeric 22mer K+ crystal structure (22). In both myc-1245 and myc-2345, structures the first and third loops are single-nucleotide propeller loops; the second (central) loop in myc-2345 is a GA propeller loop and that in the non-natural myc-1245 consists of a large six-base TTTTTA loop. This loop is destabilizing compared to that in the shorter myc-2345 sequence, as shown by the 16°C difference in their melting temperatures. Both of these c-myc quadruplexes have higher melting temperatures than the human telomeric quadruplexes in similar K+ conditions. One remarkable feature of the Pu24I NMR structure, which has five G-tracts, is the fold-back of the 3′ terminal G in the last G-tract, enabling participation in a G-tetrad and the establishment of a neighbouring G·A•G hydrogen-bonded triad that is positioned as a planar platform-like diagonal loop above this G-tetrad. The other face of the quadruplex has a stack of base pairs and bases arising from the 5′ end sequence. The NMR solution structure of a complex of the Pu241 quadruplex with the porphyrin ligand TMPyP4 (89) shows that it stacks on the other terminal G-tetrad, sandwiched against one of base pair platforms, with overall little perturbation from the ligand-free Pu241 quadruplex structure.
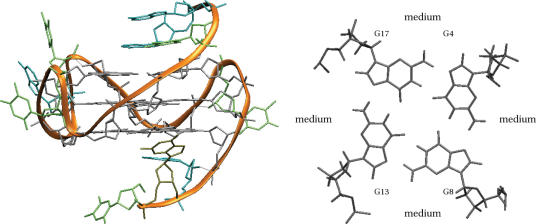
Figure 6.
NMR-derived topology and one of the deposited structures of the c-myc quadruplex (86) (PDB entry 1XAV).
NMR methods have been used to validate the existence of quadruplex species in two G-rich stretches of the promoter region of the c-kit kinase gene, an important therapeutic target in gastro-intestinal tumours (90,91). Unusually, NMR has observed only a single quadruplex species in K+ solution for one of these sequences (90), which is 87 bp upstream of the transcription start site, and therefore there will be no ambiguity as to the topology of the quadruplex species, once determined. This is probably due to both the four G-tracts constraining the number of quadruplexes, and the presence of topologically restraining single-nucleotide loops (Table 4). The second quadruplex sequence that has been identified in the promoter region of the c-kit gene (91) occupies a site required for core promoter activity. This sequence requires to be mutated in order to act as a single quadruplex species, probably with a parallel fold. Both these quadruplexes have high conservation across vertebrate species, suggestive of a functional role for them.
Chemical footprinting and CD methods have been used to characterize (92) a quadruplex species found in a nuclease-hypersensitive sequence within the vascular endothelial growth factor (VEGF) promoter region that is essential for basal promoter activity in human cancer cells. This quadruplex, which is induced to form from the duplex sequence by the ligands telomestatin or TMPyP4, has been assigned a parallel topology, which is consistent with the presence of two single-nucleotide loops. A parallel quadruplex has also been proposed (93) for a sequence in the hypoxia inducible factor 1α (HIF-1α) promoter region, based on footprinting and CD data; again the sequence (Table 4) has two likely single-nucleotide loops. The bcl-2 oncogene has a major transcriptional promoter sequence ca. 1400 bp upstream of the transcription start site, that has been characterized as showing quadruplex characteristics (94,95). A potential loop sequence in this quadruplex region has the sequence AGGA in common with one of the c-kit sequences (90); this sequence was predicted (18) to have a high frequency of occurrence. An NMR study of the bcl-2 quadruplex (94) shows that one of the topologies for this mixed parallel/anti-parallel unimolecular quadruplex has two lateral loops and one propeller loop, analogous to one of the 22mer vertebrate telomeric quadruplex topologies (75,96), but with a reversed order of loops. Putative quadruplexes have been assigned within the k-ras (97) and neuroblastoma oncogenes (98).
It is striking that a high proportion of these quadruplex topological assignments suggest folds with all strands parallel [for which the vertebrate telomeric 22mer crystal structure (22) is the paradigm]. This is the inevitable consequence of the presence of at least two single-nucleotide loops, which energetically disbar these sequences from forming lateral or diagonal-looped anti-parallel structures. Although we know little as yet about the structural basis for loop sequence preferences, it is also reassuring that the sequences with high occurrences in the human genome overall are among those that have been observed (in the admittedly rather small sample base accumulated to date). This may suggest that these are the loop sequences with greatest stability. The NMR structure of the G-quadruplex from the c-myc promoter sequence with five G-tracts (89) shows that the presence of the fifth (very short) tract can produce unexpected and significant changes in topology compared to analogous sequences with four tracts, so topology and structure prediction for new quadruplexes based on the very small number of known quadruplex structures is probably premature at present.
CONCLUDING REMARKS
The quadruplexes studied to date by crystallography and NMR has revealed a diversity of topology and structure not shown by any other type of DNA. The symmetric telomeric quadruplexes comprise a small group of structures, although the topology and fine structure of consecutive unimolecular quadruplexes formed on the vertebrate single-stranded telomeric overhang remain to be established. The potentially much larger group of genomic quadruplexes have inherent sequence diversity (and usually asymmetry), and this will undoubtedly be reflected in high structural diversity once further structures beyond the c-myc quadruplex, are established. A note of caution is needed. Even more than with telomeric quadruplexes, double helical (or mRNA for quadruplexes in transcribed sequences) sequence context will ultimately need to be taken into account when describing genomic quadruplex form and function. The question of the extent to which quadruplex structural complexity can rival that of RNA folding (and possibly even have catalytic activity), remains to be fully answered, but again, sequence context may play a role. The folding of RNA itself into unimolecular quadruplex structures is largely unexplored, and it would be unsurprising if these RNA quadruplexes were to have greater complexity, given the propensity of complex RNAs for example to exploit the 2′-hydroxy group in folding. Other backbone chemistries, such as in peptide nucleic acids (PNA) and conformationally locked nucleic acids (LNA), can also form tetramolecular and bimolecular quadruplexes (99–102) that do not always conform to the topological patterns shown by DNA ones, and again it is to be expected that unimolecular quadruplexes with such backbones may also have topological novelty.
This review has not discussed the possible genomic quadruplex-related arrangements that may be formed by the expansion of triplet repeats, such as CCG, CTG, CAG or GAA, which are found in a range of genetic disorders repeats. They share at least some features with the G-quadruplexes discussed here [see e.g. Refs. (103,104)], together with the added complexity of non-G base associations that can form motifs, such as base pentads and heptads.
Whether or not duplex genomic G-rich sequences may form quadruplex-based structures in vivo remains to be fully demonstrated, although supportive data are starting to emerge (105). Data from the c-myc sequences (84) when embedded in plasmids, which are then transfected in cells is strongly indicative of quadruplex formation in a more complex environment than the NMR tube; however, the difficulty of observing endogenous DNA quadruplex structures in live cells is a major challenge for the future. Resolution of the requirement for duplex unwinding in promoter sequences in order for the G-rich strand to transiently become single-stranded, may be achieved by strand scission; it has been recently reported that DNA topoisomerase II produces double-strand breaks in promoter sequences (106), which could be a suitable mechanism for achieving this. Even if the propensity for native sequences to form quadruplexes is low, it may be that their induction is achievable with ligands with selectivity not only for particular topologies but also for the detailed structural features of individual quadruplexes. The demonstration (107) that a c-myc quadruplex can be induced by the porphyrin TMPyP4 to form in preference to its duplex sequence, is an important step in this direction. Thus, a major focus for future quadruplex studies is as a target for selective therapeutic intervention. For example stabilization of a G-quadruplex structure in upstream regions important for maximum promoter activity, as in the case of the NHE III sequence in the c-myc gene (83), would result in down-regulation of gene expression. The large number of potential quadruplex sites in a genomes, implies that many target quadruplexes may have unique architectures, and therefore that selective stabilizing ligands can be devised. The number of known quadruplex structures is as yet very limited compared to the large number of quadruplex-forming sequences (18,19), strongly suggesting that we can look forward to the determination of a large number of diverse quadruplex topologies and structures.
Acknowledgments
The authors are grateful for support from Cancer Research UK, the Association for International Cancer Research and the School of Pharmacy, and to Keith Fox and the anonymous reviewers for their constructive comments. Funding to pay the Open Access publication charges for this article was provided by JISC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman S.B., Cohen G.H., Davies D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 3.Arnott S., Chandrasekaran R., Marttila C.M. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974;141:537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howard F.B., Frazier J., Miles H.T. Stable and metastable forms of poly(G) Biopolymers. 1977;16:791–809. doi: 10.1002/bip.1977.360160407. [DOI] [PubMed] [Google Scholar]
- 5.Williamson J.R. G-tetrad structures in telomeric DNA. Ann. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 6.Davies J.T. G-tetrads 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Intl. Edit. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 7.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 8.Sundquist W.I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 9.Wright W.E., Tesmer V.M., Huffman K.E., Levene S.D., Shay J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sfeir A.J., Chai W., Shay J.W., Wright W.E. Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 11.Lei M., Podell E.M., Cech T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nature Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 12.Zaug A.J., Podell E.R., Cech T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cech T.R. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 14.Huang C., Lin Y., Huang S., Chen C.W. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 1998;28:905–916. doi: 10.1046/j.1365-2958.1998.00856.x. [DOI] [PubMed] [Google Scholar]
- 15.Jing N., Zhu Q., Yuan P., Li Y., Mao L., Tweardy D.J. Targeting signal transducer and activator of transcription 3 with G-tetrad oligonucleotides: a potential novel therapy for head and neck cancer. Mol. Cancer Ther. 2006;5:279–286. doi: 10.1158/1535-7163.MCT-05-0302. [DOI] [PubMed] [Google Scholar]
- 16.McMicken H.W., Bates P.J., Chen Y. Antiproliferative activity of G-tetrad-containing oligonucleotides generated by a novel single-stranded DNA expression system. Cancer Gene Ther. 2003;10:867–869. doi: 10.1038/sj.cgt.7700652. [DOI] [PubMed] [Google Scholar]
- 17.Kelly J.A., Feigon J., Yeates T.O. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 18.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsson T., Pecinka P., Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawal P., Kummarasetti V.B., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S.K., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 23.Phan A.T., Patel D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties and folding/unfolding kinetics. J. Am. Chem. Soc. 2003;125:15021–15027. doi: 10.1021/ja037616j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan A.T., Modi Y.S., Patel D.J. Two-repeat Tetrahymena telomeric d(TGGGGTTGGGGT) sequence interconverts between asymmetric dimeric G-quadruplexes in solution. J. Mol. Biol. 2004;338:93–102. doi: 10.1016/j.jmb.2004.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettani A., Bouaziz S., Gorin A., Zhao H., Jones R.A., Patel D.J. Solution structure of a Na cation stabilized DNA quadruplex containing G.G.G.G and G.C.G.C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 26.Hazel P., Parkinson G.N., Neidle S. Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex. J. Amer. Chem. Soc. 2006;128:5480–5487. doi: 10.1021/ja058577+. [DOI] [PubMed] [Google Scholar]
- 27.Gill M.L., Strobel S.A., Loria J.P. 205Tl MNR methods for the characterisation of monovalent cation binding to nucleic acids. J. Amer. Chem. Soc. 2005;127:16723–16732. doi: 10.1021/ja055358f. [DOI] [PubMed] [Google Scholar]
- 28.Haider S., Parkinson G.N., Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 29.Smith F.W., Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 30.Schultze P., Hud N.V., Smith F.W., Feigon J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G4T4G4) Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultze P., Smith F.W., Feigon J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG) Structure. 1994;2:221–233. doi: 10.1016/s0969-2126(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 32.Clay E.H., Gould I.R. A combined QM and MM investigation into guanine quadruplexes. J. Mol. Graphics Modelling. 2005;24:138–146. doi: 10.1016/j.jmgm.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 33.van Mourik T., Dingley A.J. Characterization of the monovalent ion position and hydrogen-bond network in guanine tetrads by DFT calculations of NMR parameters. Chemistry. 2005;7:6064–6079. doi: 10.1002/chem.200500198. [DOI] [PubMed] [Google Scholar]
- 34.Dingley A.J., Peterson R.D., Grzesiek S., Feigon J. Characterization of the cation and temperature dependence of DNA quadruplex hydrogen bond properties using high-resolution NMR. J. Amer. Chem. Soc. 2005;127:14466–14472. doi: 10.1021/ja0540369. [DOI] [PubMed] [Google Scholar]
- 35.Aboul-ela F., Murchie A.I., Lilley D.M. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T) Nature. 1992;360:280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- 36.Phillips K., Dauter Z., Murchie A.I., Lilley D.M., Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 37.Rueda M., Luque F.J., Orozco M. G-quadruplexes can maintain their structure in the gas phase. J Amer. Chem. Soc. 2006;128:3608–3619. doi: 10.1021/ja055936s. [DOI] [PubMed] [Google Scholar]
- 38.Deng J., Xiong Y., Sundaralingam M. X-ray analysis of an RNA tetraplex (UGGGGU)4 with divalent Sr(2+) ions at subatomic resolution (0.61 Å) Proc. Natl Acad. Sci. USA. 2001;98:13665–13670. doi: 10.1073/pnas.241374798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnan-Ghosh Y., Liu D., Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Amer. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 40.Mergny J.-L., De Cian A., Amrane S., Webba de Silva M. Kinetics of double-chain reversals bridging contiguous tetrads in tetramolecular quadruplexes. Nucleic Acids Res. 2006;34:2386–2397. doi: 10.1093/nar/gkl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cmugelj M., Sket P., Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 42.Sket P., Cmugelj M., Plavec J. d(G3T4G4) forms unusual dimeric G-quadruplex structure with the same general fold in the presence of K+, Na+ or ions. Bioorg. Med. Chem. 2004;12:5735–5744. doi: 10.1016/j.bmc.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Sket P., Crnugelj M., Plavec J. Identification of mixed di-cation forms of G-quadruplex in solution. Nucleic Acids Res. 2005;33:3691–3697. doi: 10.1093/nar/gki690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cmugelj M., Hud N.V., Plavec J. The solution structure of d(G4T4G3)2: a bimolecular G-quadruplex with a novel fold. J. Mol. Biol. 2002;320:911–924. doi: 10.1016/s0022-2836(02)00569-7. [DOI] [PubMed] [Google Scholar]
- 45.Smith F.W., Lau F.W., Feigon J. d(G3T4G3) forms an asymmetric diagonally looped dimeric G-quadruplex with guanosine 5′-syn-syn-anti and 5′-syn-anti-anti N-glycosidic conformations. Proc. Natl Acad. Sci. USA. 1994;91:10546–10550. doi: 10.1073/pnas.91.22.10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahan G.D., Keniry M.A., Shafer R.H. NMR structure refinement and dynamics of the K+-[d(G3T4G3)]2 quadruplex via particle mesh Ewald molecular dynamics simulations. Biophys. J. 1998;75:968–981. doi: 10.1016/S0006-3495(98)77585-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cevec M., Plavec J. Role of loop residues and cations on the formation and stability of dimeric DNA G-quadruplexes. Biochemistry. 2005;44:15238–15246. doi: 10.1021/bi0514414. [DOI] [PubMed] [Google Scholar]
- 48.Risitano A., Fox K.R. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spackova N., Berger I., Sponer J. Nanosecond molecular dynamics simulations of parallel and antiparallel guanine quadruplex DNA molecules. J. Amer. Chem. Soc. 1999;121:5519–5534. [Google Scholar]
- 50.Spackova N., Berger I., Sponer J. Structural dynamics and cation interactions of DNA quadruplex molecules containing mixed guanine/cytosine tetrads revealed by large-scale MD simulations. J. Amer. Chem. Soc. 2001;123:3295–3307. doi: 10.1021/ja002656y. [DOI] [PubMed] [Google Scholar]
- 51.Stefl R., Cheatham T.E., III, Spackova N., Fadrna E., Berger I., Koca J., Sponer J. Formation pathways of a guanine-quadruplex DNA revealed by molecular dynamics and thermodynamic analysis of substates. Biophys. J. 2003;85:1787–1804. doi: 10.1016/S0006-3495(03)74608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fadrna E., Spackova N., Stefl R., Koca J., Cheatham T.E., III, Sponer J. Molecular dynamics simulations of guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hazel P., Parkinson G.N., Neidle S. Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures. Nucleic Acids Res. 2006;34:2117–2127. doi: 10.1093/nar/gkl182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 55.Phan A.T., Kuryavyi V., Ma J.B., Faure A., Andreola M.L., Patel D.J. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Patel D.J. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 57.Kuryavyi V., Majumdar A., Shallop A., Chernichenko N., Skripkin E., Jones R., Patel D.J. A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple. J. Mol. Biol. 2001;310:181–194. doi: 10.1006/jmbi.2001.4759. [DOI] [PubMed] [Google Scholar]
- 58.Petraccone L., Erra E., Esposito V., Randazzo A., Mayol L., Nasti L., Barone G., Giancola C. Stability and structure of telomeric DNA sequences forming quadruplexes containing four G-tetrads with different topological arrangements. Biochemistry. 2004;43:4877–4884. doi: 10.1021/bi0300985. [DOI] [PubMed] [Google Scholar]
- 59.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Amer. Chem. Soc. 2004;1286:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 60.Olsen C.M., Gmeiner W.H., Marky L.A. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-tetrad stacking. J. Phys. Chem. B. 2006;110:6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]
- 61.Tang C.F., Shafer R.H. Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in guanine quadruplexes. J. Amer. Chem. Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–1210. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 63.Phan A.T., Mergny J.L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marathias V.M., Bolton P.H. Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry. 1999;38:4355–4364. doi: 10.1021/bi982604+. [DOI] [PubMed] [Google Scholar]
- 65.Risitano A., Fox K.R. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 66.Rujan I.N., Meleney J.C., Bolton P.H. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ying L., Green J.J., Li H., Klenerman D., Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee J.Y., Okumus B., Kim D.S., Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18934–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhan N, Phan A.T., Patel D.J. (3+1) assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Amer. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balagurumoorthy P., Brahmachari S.K. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 71.Li J., Correia J.J., Wang L., Trent J.O., Chaires J.B. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang C.C., Chu J.F., Kao F.J., Chiu Y.C., Lou P.J., Chen H.C., Chang T.C. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 73.Xu Y., Noguchi Y., Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 74.Dapic V., Abdomericovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simonsson T. G-quadruplex DNA structures--variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 77.Paces J., Pavlicek A., Paces V. HERVd: database of human endogenous retroviruses. Nucleic Acids Res. 2002;30:205–206. doi: 10.1093/nar/30.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kostadinov R., Malhotra N., Viotti M., Shine R., D'Antonio L., Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D1124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zanotti K.J., Lackey P.E., Evans G.L., Mihaela-Rita M. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G tetrad forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 80.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raghavan S.C., Swanson P.C., Wu X., Hsieh C.-L., Lieber M.R. A non-B-DNA structure at the bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 82.Bonaglia M.C., Giorda R., Mani E., Aceti G., Anderlid B.M., Baroncini A., Pramparo T., Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J. Med. Genetics. 2005 doi: 10.1136/jmg.2005.038604. doi:10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu W., Sun D., Hurley L.H. Binding of G-quadruplex-interactive agents to distinct G-quadruplexes induces different biological effects in MiaPaCa cells. Nucleosides Nucleotides Nucleic Acids. 2005;24:1801–1815. doi: 10.1080/15257770500267238. [DOI] [PubMed] [Google Scholar]
- 85.Shafer R.H., Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 86.Simonsson T., Sjoback R. DNA tetraplex formation studied with fluorescence resonance energy transfer. J. Biol. Chem. 1999;274:17379–17383. doi: 10.1074/jbc.274.24.17379. [DOI] [PubMed] [Google Scholar]
- 87.Phan AT., Modi Y.S., Patel D.J. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Amer. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-myc promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 89.Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nature Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Amer. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fernando H., Reszka A.P., Huppert J., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun D., Guo K., Rusche J.J., Hurley L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Armond R., Wood S., Sun D., Hurley L.H., Ebbinghaus S.W. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 94.Dai J., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Amer. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dexheimer T.S., Sun D., Hurley L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Amer. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Amer. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cogio S., Xodo L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu Y., Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krishnan-Ghosh Y., Stephens E., Balasubramanian S. A PNA4 quadraplex. J. Amer. Chem. Soc. 2004;126:5944–5945. doi: 10.1021/ja031508f. [DOI] [PubMed] [Google Scholar]
- 100.Datta B., Bier M.E., Roy S., Armitage B.A. Quadruplex formation by a guanine-rich PNA oligomer. J. Amer. Chem. Soc. 2005;127:4199–4207. doi: 10.1021/ja0446202. [DOI] [PubMed] [Google Scholar]
- 101.Randazzo A., Esposito V., Ohlenschlager O., Ramachandran R, Mayola L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nielsen J.T., Arar K., Petersen M. NMR solution structures of LNA (locked nucleic acid) modified quadruplexes. Nucleic Acids Res. 2006;34:2006–2014. doi: 10.1093/nar/gkl144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsugami A., Okuizumi T., Uesugi S., Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 104.Khateb S., Weisman-Shomer P., Hershco L., Loeb L.A., Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Duquette M.L., Handa P., Vincent J.A., Taylor A.F., Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ju B.G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G. A topoisomerase IIß-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 107.Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]