Abstract
Atomic force microscopy (AFM) has been used to image, at single molecule resolution, transcription events by Escherichia coli RNA polymerase (RNAP) on a linear DNA template with two convergently aligned λpr promoters. For the first time experimentally, the outcome of collision events during convergent transcription by two identical RNAP has been studied. Measurement of the positions of the RNAP on the DNA, allows distinction of open promoter complexes (OPCs) and elongating complexes (EC) and collided complexes (CC). This discontinuous time-course enables subsequent analysis of collision events where both RNAP remain bound on the DNA. After collision, the elongating RNAP has caused the other (usually stalled) RNAP to back-track along the template. The final positions of the two RNAP indicate that these are collisions between an EC and a stalled EC (SEC) or OPC (previously referred to as sitting-ducks). Interestingly, the distances between the two RNAP show that they are not always at closest approach after ‘collision’ has caused their arrest.
INTRODUCTION
Convergent transcription results from the presence of two convergent promoters located on a DNA template. Such convergently aligned genetic structures, whilst not common, have been observed in a wide variety of organisms ranging from prokaryotes such as Escherichia coli (1–3), to eukaryotes as diverse as Saccharomyces cerevisiae (4,5), Drosophila and humans (6,7). Transcriptomic studies continue to discover further cases. Such arrangements were initially thought to be a consequence implicit in the highly compressed genetic organisation of an organism's genome. However, further study has prompted the hypothesis that such arrangements are evolutionarily selected for their ability to control gene expression at a fundamental level (1). In higher order eukaryotes, the presence of larger introns allows complete genes to be nested within (and in the opposite orientation to) the intron of a larger gene. This raises important questions in respect of the co-regulation of host and nested genes, including the possibility that transcription of one gene might preclude simultaneous transcription of the other (6).
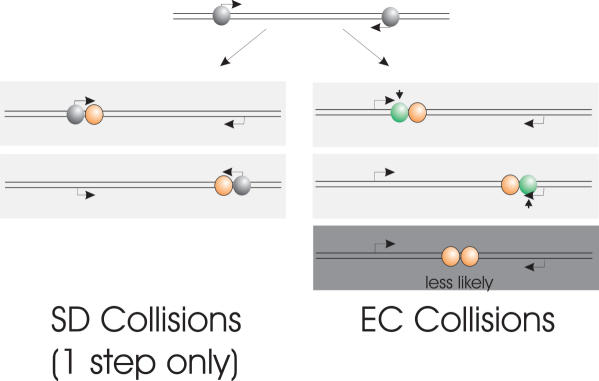
Transcriptional Interference (TI) between two convergently aligned promoters can be considered to occur through three mechanisms (1,8); (i) promoter occlusion, where the binding of an RNAP to a promoter is prevented by the presence of another RNAP initiated from a second convergently aligned promoter transcribing across the promoter region. (ii) A ‘sitting duck’ collision (SD), where an EC initiated from one promoter collides with an OPC which still remains at the other promoter. ‘Sitting ducks’ are defined as initiation complexes that are waiting to ‘fire’ at a promoter (1). However pre-open promoter complexes are unlikely to be sitting ducks as they are in rapid equilibrium with the promoter and their removal will not greatly inhibit subsequent transcription since a replacement RNAP is able to rapidly re-bind (9). (iii) EC collisions, where two ECs collide within the inter-promoter DNA region. Recent simulation (stochastic and mean field) shows that for strong promoters such as λPR , promoter occlusion is not significant (9). Stochastic simulation using the model of Sneppen et al. (9). shows that for the DNA template used in our study the effect of TI is likely to inhibit the production of full length RNA transcripts by a factor of over 10, illustrating the importance of TI in vivo.
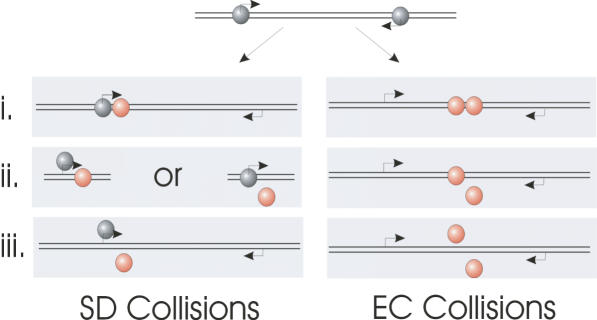
The research currently presented in the literature is unable to investigate the fate of the RNAP once they have undergone collision (either as SD or EC). The original in vivo work of Ward and Murray (3) argued that since the effects of TI were rapidly reversible, the collided RNAP (whether these were EC or SD collisions was not determined) must disengage from the template after collisions. When one of the convergent promoters was switched off they observed a rapid restoration of gene expression indicating that no stalled RNAP were present to act as an irreversible roadblock to further transcription. Alternatively a secondary process present in an in vivo system could rescue such stalled complexes to account for this reversibility. The Mutation frequency decline factor (Mfd) is able to clear stalled complexes from a DNA template (10), and Gene regulation factors GreA and GreB are able to rescue back-tracked complexes (11). After a collision of RNAPs the three possible outcomes are: (i) both RNAP dissociate from the template resulting in a complete loss of gene expression for both genes; (ii) one RNAP dissociates, i.e. is knocked off (possibly in a competitive manner) resulting in loss of expression of one gene; (iii) both the RNAP stall and remain bound to the DNA (6,8). None of these outcomes are mutually exclusive in the single molecule view and they are all summarised in Figure 1. The outcomes may differ depending on whether the RNAPs discussed above are SDs or ECs. It is also postulated that as a result of collision an EC could force the other EC to be back-tracked towards its promoter (9). Although less likely, at present it is conceivable that two convergent RNAPs may be able to pass each other on the same template by some as yet unknown mechanism (6).
Figure 1.
The possible outcomes of RNAP collisions resulting from convergent promoters, classified as sitting duck (SD) collisions and elongation complex (EC) collisions. SD complexes are shown in grey and EC in orange. Possible outcomes include (i) both RNAP remaining bound on the template, (ii) one RNAP being displaced, or (iii) both RNAP being displaced.
Any ensemble biochemical experiment can only report on molecular population sizes of at best nanomolar concentration (∼1014 molecules). Using these techniques (e.g. footprinting, transcription assays, reporter assays, transcriptional run-on, etc.) it is possible to say only whether each gene is being transcribed on average, but not if these transcripts are being transcribed simultaneously from the same template. Using a direct imaging technique such as atomic force microscopy (AFM) with single molecule resolution, we are addressing this crucial question. Using a λPR E.coli RNAP-σ70 holoenzyme system, AFM studies have previously been used to study the formation of OPCs (12) and stalled ECs, including their associated wrapping, and for the case of stalled ECs, the presence of extruded RNA transcripts (13). The action of DNA looping as a mechanism of transcriptional activation by a nitrogen regulatory protein (NtrC) has also been followed (14). Conversely the action of transcription inactivation has been visualised with the protein H-NS binding between the up and downstream sites on DNA causing the RNAP to become trapped (15).
The aim of this study was to use AFM to investigate simultaneous convergent transcription and hence TI at the level of individual molecular events. To achieve this we induce RNAP collisions and hence TI by two methods. The first method involved stalling ECs downstream of their convergently aligned promoters by nucleotide omission followed by re-initiation. By this method TI would occur only through EC collisions. Secondly, by omitting the stall site, we were able allow TI to occur through both EC and SD collisions. In both cases, we observed stalled complexes with two RNAP still bound to the DNA template and back-tracking of one RNAP as a result of collision with the other. This study represents the first experiments of convergent transcription by AFM, or indeed any single molecule technique and this resolution provides new insight into the molecular processes governing TI.
MATERIALS AND METHODS
DNA preparation and transcription complex formation
The 1150 bp DNA template used in this study was obtained by restriction digestion of plasmid pDSU (13) with HindIII endonuclease. The fragment containing the λPR promoter was purified in 1% agarose, eluted using MinElute gel extraction kit (Qiagen, Valencia, CA) and re-suspended in nuclease free water (Ambion, Austin, TX). The DNA concentration was determined by 1% agarose gel electrophoresis and confirmed by absorbance measurement of A260. The E.coli RNAP-σ70 subunit holoenzyme was purchased from Epicenter (Madison, WI).
Open promoter complexes (OPCs) were formed by incubating 200 fmol DNA and 400 fmol holoenzyme in 10 μl transcription buffer [20 mM Tris–HCl (pH 7.9), 50 mM KCl, 5 mM MgCl2, 1 mM DTT] containing 20 units of ribonuclease inhibitor, (RNaseIn, Promega, Madison, WI) for 15 min at 37oC. A 1:1 holoenzyme:promoter stoichometry was used to minimise non-specific binding at the DNA ends and non-promoter sequences. Collided complexes were formed in two ways; either by addition of all four NTPs from the OPC state or by addition of CTP after the formation of SECs. The formation of collided complexes with all NTPs is termed the one-step process, while intermediate SEC formation is termed the two-step process. In the one-step process ATP, CTP, GTP and UTP were added to a final concentration of 100 μM each and the reaction mixture incubated at room temperature for 20 min. In the two-step process, stalled elongation complexes (SECs) were formed by adding ATP, GTP and UTP (Ambion) to a final concentration of 133 μM each. Transcription up to the stall sites was carried out at room temperature for 20 min. On addition of the CTP the concentrations of other NTPs were adjusted to give a final total NTP concentration of 400 μM and the reaction mixture incubated at room temperature for a further 30 s, 10 min or 20 min. The reduction of each NTP concentration from 133 to 100 μM was shown not to affect transcriptional yield by in vitro transcription assay. All reactions were used immediately for AFM imaging.
Atomic force microscopy
Complexes prepared as above were diluted to a DNA concentration of 1–2 nM in 10 μL of deposition buffer [4 mM HEPES (pH 7.4), 10 mM NaCl, 2 mM MgCl2] and deposited onto freshly cleaved mica. The sample was incubated for 2 min and then rinsed with de-ionised water and dried with a weak flux of dry nitrogen. AFM images were collected in air with a Multimode Nanoscope IIIa™ AFM (Veeco, Santa Barbara, CA) operating in Tapping Mode™ using diving board cantilevers with integral etched probes (Olympus, Tokyo, Japan). Scans of 3 μm × 3 μm were collected at a scan line frequency of 2 Hz at 512 × 512 pixel resolution.
AFM images were analysed using the Nanoscope software. The centre of each RNAP was taken as the intersection of the two tangents to the incoming and outgoing DNA strands. The DNA contour length was measured manually following the helical backbone as a series of connected straight lines. DNA molecules containing either RNAP non-specifically bound at an end, or only 1 RNAP, or more than 2 RNAP were not considered. No further selection criteria were used to differentiate DNA molecules with 2 RNAP bound. Three measurements were taken from the complexes with 2 RNAP specifically bound: the contour length between the centres of the 2 RNAPs, termed the RNAP separation; and the two contour lengths from either RNAP centre to the closest end of the DNA fragment (the larger of which is termed the long arm and the smaller the short arm). The mean square end-to-end distance has been measured for a number DNA molecules with no RNAP bound and with an RNAP bound non-specifically at one end. These were comparable and hence showed that the RNAP was not affecting the binding and subsequent surface diffusion of the DNA (16).
In vitro transcription
DNA–RNAP complexes were formed by incubating 170 fmol of DNA and 600 fmol E Coli σ 70 holoenzyme (Epicentre) in transcription buffer in a total volume of 10 μl at 37oC for 15 min. After this, 5 μl of a NTP mix containing 200 μM of ATP and GTP, or ATP, GTP and CTP (run-on reactions), 20 μM UTP, 40 μCi [α-32 P] UTP and 200 μg/ml heparin was added. These reactions mixtures were incubated for 15 min at room temperature. In the case of re-initiation experiments, the omitted NTP was then added to a final concentration of 200 μM and the reaction incubated for a further 15 min at room temperature. Reactions were stopped by addition of 5 μl loading buffer (Roche Biosciences, UK, 80% formamide, 2 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue). Reactions were analysed with 6% denaturing PAGE.
RESULTS
AFM of transcriptional complexes
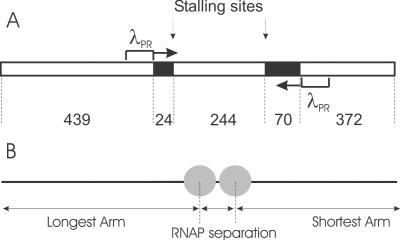
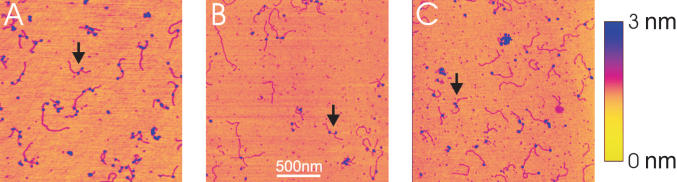
The DNA template contains two convergently aligned λPR promoters separated by 338 bp (Figure 2A). The position of the promoters on the DNA template is asymmetric allowing the ends to be distinguished when RNAP are bound at the promoters. When transcription is initiated the ability to distinguish the polarity of the template is lost because the DNA ends are not specifically labelled. Figure 2B illustrates the DNA contour length measurements from AFM images that were made in the statistical analysis. To image the complexes by AFM the deposition process onto mica and concentrations were optimised such that individual complexes were isolated and well resolved. A representative AFM image of the OPCs is shown in Figure 3A. DNA molecules with RNAP holoenzyme bound specifically at both promoters were identified as DNA molecules with two separate but equal sized globular features appropriately positioned towards the centre of the template. DNA molecules containing no holoenzyme, one holoenzyme or a number of holoenzymes at positions not corresponding to promoters were not included in subsequent analysis. The results indicated that single OPCs were present on ∼30% of DNA templates while 10% had two OPCs. The remaining DNA molecules had either no OPCs or a number of multiple (presumably non-specifically bound) holoenzymes.
Figure 2.
(A) Schematic representation of 1150 bp linear DNA template used in this study. The sequence deficient in C bases is shown in black. The position of each C-less cassette and the position and direction of promoters is shown in base pairs. (B) Schematic representation of the measurements of RNAP position along the template and RNAP-RNAP separation taken from the AFM images.
Figure 3.
Tapping mode atomic force microscopy images of convergent transcription complexes. The RNAP are visualised as globular features (blue) on the DNA template (magenta). (A) Open promoter complexes (OPCs) formed in the absence of NTPs. DNA molecules are observed with 0, 1 and 2 OPC present. RNAP in OPC are those where the RNAP are roughly one third from either end of the template. (B) Stalled elongation complexes (SECs) formed in the presence of ATP, GTP and UTP. RNAP in SEC are recognised as those that are now closer to the centre of the template. (C) Elongation complexes formed upon the addition of CTP to the SEC reaction mix. When a DNA molecule contains two SECs, these SECs can collide and sometimes stall against each other. Arrows denote DNA molecules harbouring two RNAP which were used in the statistical analysis of RNAP convergence and collision. Artificial height scale denotes yellow (low) to blue (high): Range = 3 nm.
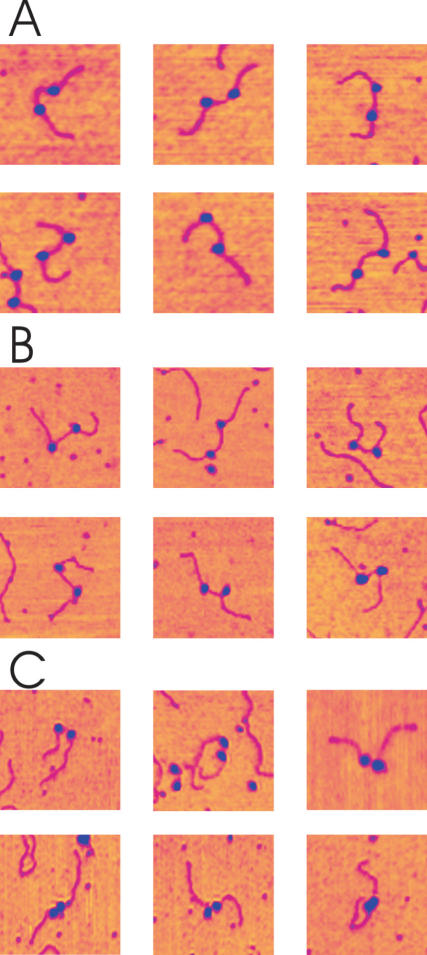
A montage of AFM images of DNA molecules with two OPCs is shown in Figure 4A. DNase I footprinting of an OPC on a single promoter template formed under similar solution conditions showed that these complexes have a footprint characteristic of an OPC with full protection in the melting region (approximately +20 to −10 nt) and partial protection in the recognition region (approximately −15 to −55 nt) in agreement with Metzger et al. (17,18) (data not shown). An enhanced periodic cleavage in the recognition domain implies DNA wrapping by the enzyme (19). This DNA wrapping can be observed as an apparent RNAP induced bending of the DNA in Figure 4A.
Figure 4.
Montage of AFM images of stalled elongation complexes (SEC) and collided complexes. (A) OPC formed in the absence of NTPs. (B) SEC formed in the presence of ATP, GTP and UTP. (C) Collided complexes formed by addition CTP to the reaction mix. Two populations of complexes exist with varying RNAP separations, ones with relatively large, non-contacting separations (top row), and others apparently in contact (bottom row). Image size: 250 × 250 nm.
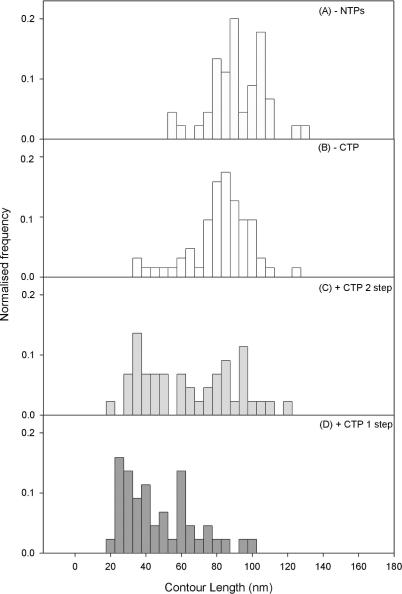
SECs were formed by the omission of CTP. The OPCs at each convergent promoter would then be expected to transcribe downstream (i.e. towards each other) until the first occurrence of a C base in the non-template strand. In our template, this C base occurred at positions +24 and +70 relative to the +1 sites of transcription initiation for each promoter respectively. The template was designed such that the sequence at each stall site was identical, hence the stalling and re-initiation behaviour of RNAP at either site should be identical. The appearance of the SECs by AFM was not greatly different to the OPC samples to the naked eye (cf. Figure 3A with 3B and 4A with 4B), but detailed DNA contour length analysis of the separation of the RNAPs along the template, reveals that the RNAP have converged consistent with the expected distance (see Figure 5). The number of DNA molecules on the mica surface with 1 or 2 SECs is reduced to about half of the OPC case, giving an overall yield of 15% of 1 SEC and 5% of 2 SECs with respect to the total number of DNA molecules observed.
Figure 5.
Inter RNAP–RNAP separation distributions along the DNA contour length. RNAP centre separation: (A) in the absence of all NTPs (n = 45), i.e. OPCs; (B) in the presence of ATP, GTP and UTP only (n = 131), i.e. SECs; (C) in the presence of ATP, GTP and CTP, followed by CTP after 20 min (n = 44); (D) formed by addition of all 4 NTPs together (n = 45) from the OPC state.
Collided complexes were first formed by the addition of CTP to the SEC reaction mix, in the second phase of this two-step process. This enables the stalled RNAPs to resume elongation and continue transcription until, either an obstacle to transcription, or the end of the template was encountered. This reaction mix was then deposited onto mica and imaged as previously described (Figures 3C and 4C). In cases where only one RNAP was initially bound to the template DNA, the RNAP is thought to run off the end of the DNA molecule, visualised as an increase in the proportion of bare DNA templates. In cases where two RNAP were bound as SECs, a collision would be expected after re-initiation, and a number of typical complexes resulting from such EC collisions are shown in Figure 4C. The number of DNA molecules with two ECs collided and stalled against each other is ∼50% of the number of DNA molecules that showed two SECs when CTP was omitted. This indicates that a significant proportion of collisions result in stalling. These data also highlighted that it is likely that a re-started EC would collide with an RNAP that remained an SEC. This would happen with significant frequency if the process of resuming elongation occurs on a time-scale comparable to that required for an RNAP to transcribe the inter-promoter region, or one of the two complexes became arrested, thus unable to resume transcription.
Collided complexes were also formed without first creating stalled intermediaries in a one-step process. In this case all four NTPs are added to the OPC reaction mix. The images of such complexes looked qualitatively the same as the previous case. This method will result in both EC–SD and EC collisions, where EC collisions can involve both EC–EC and EC–SEC collisions. In none of the analysed collided complexes (one or two-step processes) was the RNA transcript visible in the AFM images as the transcript is too short to be imaged reliably (13).
It was conceivable that the collided complexes observed do not result from transcription initiated from convergent promoters, but occur between an EC and a non-specifically bound RNAP holoenzyme, or indeed two non-specifically bound RNAP holoenzymes. To discount this possibility an additional template containing only 1 λPR promoter was used as a control. This 835 bp template had a 379 bp G-lessregion immediately downstream of the promoter. Using this template under identical experimental conditions no collided complexes were observed (data not shown).
Analysis of contour length between RNAPs
The DNA contour length between the apparent centres of the two RNAPs was measured for SECs and collided complexes formed by both methods (the two-step and one-step processes), see Figure 5. From these measurements we can infer how the position of each RNAP is changing relative to the other RNAP.
In the absence of all four NTPs, the RNAP separation distribution is centred at ∼92 ± 2 nm (292 ± 6 bp), assuming an experimental helical rise per base pair of 0.315 nm−1 (20). This value is less than the 338 bp expected from the DNA sequence (Figure 2A), because each RNAP bound as an OPC wraps the DNA by 30 nm (90 bp) (12) and therefore a contraction of this order of the apparent inter-promoter DNA is expected. In the two-step process, after addition of ATP, GTP and UTP, the RNAP separation distribution was monomodal, indicating that the majority of RNAPs on templates with two RNAP present have reached their stall sites and are SECs (Figure 5B). The distribution is now centred at ∼85.7 ± 1 nm (272 ± 3 bp), which is now slightly larger than the expected value of 244 bp. On transition from OPC to EC the wrapping of the DNA around the RNAP lessens from 30 to 20 nm, while the active site moves from 10 nm back in the wrap towards the front of the wrap (13). The consequence of this structural transition for the AFM measurements is that the length of DNA between the RNAPs (i.e. active site to active site) is underestimated for the OPC case and overestimated for the SEC case. Upon the addition of CTP in the two-step process the distribution of RNAP separation becomes bimodal, Figure 5C, where one population maintained the same RNAP separation, whilst in the other population the RNAP separation was greatly reduced. The population of constant RNAP separation is attributed to the case where both RNAP fail to resume elongation from their stall sites. Failures to resume elongation are expected and have been demonstrated by in vitro transcription assay on the DNA template shown in Figure 2 (data not shown). Where decreasing RNAP separation was seen, it is interpreted as the case where one or both RNAP resume elongation and collide at a given position on the template. In these cases, the RNAP remain bound on the DNA after collision and it is only this outcome of all collision events that can be unequivocally picked up in these ex situ discontinuous AFM imaging experiments. To study any outcomes of the RNAP encounters that involve displacement or passing will require a single molecule in situ approach, which is beyond current typical AFM scan rates. All subsequent analysis investigates this subset of collided complexes, in which two RNAP remain on the template.
When collided complexes were formed by simultaneous addition of all 4 NTPs (the one-step process) the distribution contains almost zero population corresponding to RNAPs at the stall sites (Figure 5D). This showed that the RNAP is able to pass the stall site when a full set of NTPs were present, indicating that stalling is a specific effect of CTP omission (21). Instead, RNAP were able to transcribe past this stall site and again a significant proportion of collided complexes were seen. Interestingly, the population of these collided complexes is somewhat different containing more complexes with much smaller RNAP separations. This population may contain collisions between two different classes of complexes; (i) an active EC and a SD and (ii) an active EC and another EC, either active or stalled, whereas the previously stalled complexes (Figure 5C) are only likely to contain class (i) (see discussion for more detail).
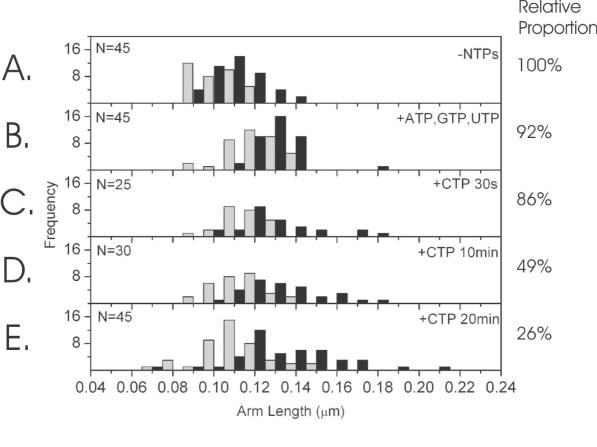
Analysis of DNA arm lengths
To investigate where this subset of collided and stalled RNAPs are on the template, the positions of the two RNAPs with respect to the DNA were measured for the two step process. The arm length is termed as the DNA contour length from the apparent centre of each RNAP to the closest DNA end. For each molecule, the smallest of the two measurements is designated the short arm and the largest, the long arm (see Figure 2). From this measurement we can infer how the position of each RNAP was changing relative to the DNA template. However, the DNA has not been specifically labelled to judge the polarity, so the distributions are not mutually exclusive, especially once the NTPs are added. Alongside each distribution is a percentage representing the proportion of DNA molecules within the distribution with 2 RNAP bound, compared to bare DNA and DNA with 1 RNAP bound. The values are normalised to the –NTPs case (panel A). As the RNAPs converge upon the addition of ATP, GTP and UTP and then finally CTP, the relative proportion decreases, probably due to a combination of RNAP disengaging from the template over the increased time course and the possibly less stable complexes being more susceptible to removal by deposition onto the mica surface. Since the relative proportion decreases through the experimental time course we can be confident that the complexes observed at long incubation times represent collisions of RNAP originating from each promoter, rather than some other class of artefactual complex that becomes more predominant over time. The distributions in the absence of NTPs (Figure 6A), showed arm lengths consistent with the RNAP holoenzymes positioned at their promoters. Upon addition of ATP, GTP and UTP the arm lengths increased as both RNAP moved downstream away from their respective ends to the stall sites (Figure 6B). The loss of DNA wrapping on the transition from OPC to SEC may also have contributed to the increase in arm length, although this is expected to be released downstream of each RNAP into the inter-RNAP distance rather than the DNA arm. Once the stall sites were reached, the asymmetry of the template is reduced, as seen by the reduced separation of the arm length distributions. Upon re-initiation following addition of CTP the shorter arm becomes progressively shorter over the 20 min time course studied, while in conjunction with this, the long arm length distribution broadens, with a significant proportion of the distribution moving to much longer lengths. The median value of the short arm contour length decreases by 23 nm with a 95% confidence interval of 17–30 nm and the median value of the long arm contour length increases by 13 nm with a 95 % confidence interval of 7–22 nm. The interpretation of these data is that one EC transcribes into the other EC, which is presumably still stalled and pushes it backwards along the DNA template until both EC stall. The addition of CTP causes one of the RNAP to continue transcribing with the associated arm length increasing, whereas the second RNAP appears to translocate in an upstream direction (i.e. backwards). In the previous section, we have shown that the addition of CTP causes the RNAP to collide and stall against each other. Using these two pieces of data we present a model in which the collision induces back-tracking in one of the RNAP (see Discussion).
Figure 6.
DNA arm contour length distributions: the grey distribution denotes the short arm and black, the long arm. The DNA contour lengths from the apparent centre of the RNAPs to the respective ends of the DNA molecule: (A) in the absence of NTPs (n = 45); (B) in the presence of ATP, GTP and UTP only (n = 45); and incubation with the final NTP, CTP for (C) 30 s, (D) 10 min and (E) 20 min (n = 25, 30, 45 respectively). The percentages on the right represent the proportion of DNAs within the distribution with 2 RNAP bound, compared to bare DNA and DNA with 1 RNAP bound. The values are normalised to the −NTPs case (panel A).
Clearly, for collisions between two RNAP to occur, one or both RNAP must resume elongation from its stall site. From the distributions in Figure 5B and C, it can be seen by cumulative counting of the histograms that in 50% of cases, both RNAPs fail to resume elongation and the DNA contour length between RNAPs remains constant. This explains why the long arm distribution in Figure 6 becomes progressively asymmetric during the time-course experiment and why the shift in the median value is not exactly comparable to the shift in the short arm distribution. For half of all complexes the RNAPs do not move at all and these data are superimposed on the data of the complexes where they have moved. In this sense, the distributions in Figure 6C–E are made up of at least two populations, the spatial resolution of this particular AFM technique is not sufficient to resolve these in the short arm distribution and although the long arm distribution has the increasing bimodal appearance at longer times, the two populations cannot be unequivocally separated. In essence, the remaining double SECs bias the long arm distribution median more significantly than the short arm one.
Since on 50% of the complexes neither RNAP resume elongation, the approximate probability of either one RNAP re-initiating, P, is obtained from solving (1−P)(1−P) = 0.5, resulting in P = 0.29. Thus the probability of both RNAPs re-initiating is P2 = 0.09 and the probability of only one re-initiating is . Thus in the population in which the DNA contour between RNAP decreases (i.e. a collision is observed) these collisions are mostly (∼80%) between a SEC and an active EC. This is consistent with the observation from the transcription assay, that a convergent promoter decreases re-initiation efficiency (data not shown).
DISCUSSION
Collision results in stalling
The presence of collided complexes in which two RNAPs appear stalled against each other on the DNA template, allow us to conclude that a significant proportion of converging RNAP do not pass each other or displace one another, but instead stall against each other. We know that these collided complexes are a result of convergent transcription for two reasons. Firstly, such collided complexes are only seen upon the addition of CTP in our two-step experiments. If such collision and stalling were as a result of collisions between RNAP searching for their promoters by a 1D diffusional search, (22) then they would be observed in the absence of NTPs or when just A, G, UTP were added. Secondly, to test if such collided complexes are a result of an EC ‘sweeping’ the downstream region until it collides with a non-specific RNAP holoenzyme, a template containing a single promoter was used in identical conditions (+ all NTPs). No such collided complexes were observed in this case so this hypothesis can be discounted.
The largest proportion of collisions are occurring at or very close to the stall site and hence providing a block to re-initiation after a certain time-scale. SEC that fail to resume elongation were termed arrested complexes by Komissarova and Kashlev (11). The reason for such arrests is unclear, but it is known that they occur more frequently at some stall sequences. These arrested complexes lose their active site due to a small amount of spontaneous back-tracking (∼6 nt) whilst the complex retains a broadly similar structure. The force generated by two opposing active ECs may well be larger than that generated by a single EC being opposed by a stationary EC (23), which may result in RNAP disengagement after ‘active–active’ collisions. As mentioned previously, such disengagement will not be visualised in this experiment, since a proportion of naked DNA is present at all times. Relating these results to possible outcomes that were suggested in Figure 1, we are able to present the outcomes actually observed using our experimental setup in Figure 7.
Figure 7.
Summary of the observed RNAP collisions in this study. EC collisions are only observed in the two-step process. EC and SD collisions are observed in the one-step process. EC collisions occur between SEC that have failed to resume elongation and an active EC as well as between two active ECs. It is shown that SEC–EC collisions are more likely. Whether EC–EC collisions are observed is unclear and the collision of two active ECs may result in disengagement. Grey: OPCs; Orange: active ECs; Green: SECs. Short vertical arrows denote the stall sites.
When collided complexes are formed in the absence of a stalled intermediary (the one-step process), a second class of collisions is expected to occur, i.e. those between an active EC and a SD. However these collisions will only be observed if both the SD and the EC RNAPs remain bound to the template after collision. Sneppen et al. (9) found that accurate modelling of the in vivo results of Callen et al. (1) required that SDs become disengaged from the template upon collision. However, they acknowledged that the disengagement of the collided SD may occur through an additional process present in vivo.
A transcribing RNAP is preceded by positive super-coiling and followed by negative super-coiling (24). Despite using a short linear template total dissipation of this super-coiling may not always be expected. This is because any bends present, either intrinsic or due to thermal fluctuations, will increase the viscous drag so as to prevent DNA rotation and therefore dissipation of supercoiling (25). The bending of the DNA by an RNAP may further prevent dissipation by a mechanism termed apical localisation (26,27). If transcription is faster than the dissipation of super-coiling then the build up of super-coiling may well stall the RNAPs irreversibly, such that they can be observed even after a 20 min incubation period, whether or not the super-coiling remains. The presence of domains of super-coiling that persist in our system could be sufficient to stall a proportion of RNAPs before a direct protein contact is made in a collision. In a significant proportion of the complexes, particularly in the experiment where all 4 NTPs are added to the OPC, the two RNAP remaining on the template are not in close contact but rather at separations between 20 and 60 nm (63–190 bp). Sometimes, the DNA between the RNAPs in these cases, appeared to be somewhat higher and have a broader width (not shown), which could indicate unusual DNA structure and/or possibly reflect the fact that this short portion of DNA cannot interact with the mica surface when suspended between the two RNAP. The latter idea is supported in the observation that this inter-RNAP DNA was often indistinct in images. In general, even at the distances of closest approach measured, ∼20 nm (see Figure 5), the RNAP are still not as close as one would expect from a hard sphere contact perspective. These observations lead us to think that the two RNAP may be able to sense each other's presence at a distance, either through long range forces directly between the RNAP (e.g. electrostatic) and/or through mediation of helical stress via the DNA linkage. There is a model proposed by Erie et al. in which ECs are shown to exist in two distinct classes, a slow class which is sensitive to stalls, and a fast class which is less sensitive to stalling (28,29). An EC's propensity to stall may affect the distance at which it stalls from another RNAP, particularly if it is encountering DNA regions with varying amounts of super-helical stress. These ideas are an interesting aspect relevant to convergent transcription systems, which will be explored in more detail in future work.
It is important to note that the presence of such collided complexes does not preclude any other possible outcomes. It is entirely possible that alternative scenarios exist in which the RNAPs disengage on collision, resulting in a DNA template with no bound RNAPs that would be lost in the ‘background’ signal of naked DNA. The histograms presented in Figures 5 and 6 represent a sub-population of the possible outcomes. If the potential energy barrier of disengagement is of comparable height to the energy of collision then whether or not a collision results in disengagement may be sensitively affected by factors such as DNA sequence at the point of collision. However, the crystal structure of an E coli OPC and the model structure of an EC (30–33), both reveal that the double stranded DNA is enveloped by the characteristic closed ‘claw’. For disengagement to occur a large structural rearrangement in the RNAP would be necessary and hence a relatively high potential energy barrier is expected. This barrier is further increased when an OPC makes the transition to an EC forming a complete RNA:DNA hybrid and RNA enters the exit channel (34).
The collision of single RNAP against stationary roadblocks such as a mutant restriction endonuclease deficient in DNA cleavage, leads to stalled complexes as determined by transcription assay and foot-printing (35). A restriction endonuclease roadblock is likely to present different perturbations in the downstream DNA topology in comparison with an RNAP. However this result shows that a restriction endonuclease is not an obstacle sufficient to displace the RNAP.
Collision results in back-tracking of one RNAP
By considering the analysis of DNA arm lengths, Figure 6, a model of back-tracking can be proposed. Back-tracking is a phenomenon that has been reported as a response to nucleotide mis-incorporation or transcriptional arrest for a single RNAP (11,28,36). In these cases, back-tracking occurs by the reverse threading of the RNA transcript with the loss of the 3′ RNA terminus from the enzyme's active site. This 3′ end emerges from a pore that is hypothesised to be used as a channel for incoming NTPs (30). Back-tracking has also been hypothesised as a response to RNAP collision (8), the extent of the back-tracking being limited by the length of the RNA transcript. Once the RNAP has returned to the promoter any further reverse motion will result in loss of the DNA–RNA hybrid crucial for complex stability (34). This loss of the DNA–RNA hybrid is likely to present a significant potential barrier that prevents further back-tracking past the promoter. Our results show a sample median back-tracking of 17 ± 6 nm (95% confidence interval) assuming an experimental helical rise per base pair of 0.315 nm−1 (37) and this equates to 54 ± 19 bp. It is likely that this helical rise could be affected by local distortions in the DNA structure around the collision region. The largest promoter-stall site distance is 70 bp implying back-tracking of this distance is not unfeasible, while the average distance from promoter to stall site for both promoters is 47 bp. These estimates of back-tracking distances along the DNA template could be affected by changes in the wrapping of DNA around the EC. If the collision were to cause unwrapping of the DNA from the EC being back-tracked (rather than moving the active site of the RNAP) this would lead to an overestimation of the back-tracking distance. An EC wraps ∼20 nm of DNA (13) so for unwrapping to account for all of the observed 17 ± 6 nm back-track, the EC would have to be almost totally unwrapped.
It is important to note that the distributions of Figure 6 also contain complexes in which SECs have failed to resume elongation. This resulted in a broadening of the distributions as the RNAPs were stalled and allowed to resume elongation, since a proportion remained at the stall sites. It is unlikely a RNAP will be able to re-bind to the promoter and undergo a second round of transcription as the rate of sigma factor recruitment is determined by 3D diffusion and this will be much slower than processes such as promoter recognition that benefit from facilitated diffusion mechanisms (i.e. sliding and hopping). This will effectively prevent an RNAP forming a holoenzyme after an initial round of transcription and undergoing multiple rounds of transcription. In support of this interpretation, we have never observed a DNA template at any incubation time that has more than two RNAP stalled together in close proximity.
The overall collision process was observed over a timescale of 20 min. Whilst an RNAP is capable of elongating at a fast rate, a number of transcriptional processes have been shown to occur on much slower time scales. The process of cooperative clearance of a restriction enzyme roadblock was shown to occur over a timescale of up to 10 min (38). Additionally the process of transcriptional arrest and back-tracking depended on the time the EC spent at a stall site. This time scale was shown to be up to 20 min (11).
An alternative explanation for the observed effect of back-tracking is that a large proportion of collisions result in disengagement. If this were the case the observed stalled complexes would become a relatively small sub-set of the population. If a subset of OPCs fail to clear the promoter and the size of this sub-set becomes comparatively large compared to the stalled complexes that remain, it will appear that the overall population's RNAP separation has shifted backwards. This interpretation requires that promoter clearance is inefficient and that a significant number of collisions result in disengagement. Although promoter clearance can be rate-limiting, the 20 min incubation period should be sufficient to allow promoter clearance. We cannot be certain of the promoter clearance efficiency by studying the transition from OPC to SEC through our AFM contour length measurements as the shortest promoter-stall site distance of 24 bp is comparable to the widths of our distributions. Instead we have evidence of this efficiency by studying the transition when collided complexes are formed directly from OPCs, where the distribution of RNAP separations shifts completely with no residual population remaining with an unchanged RNAP separation. This is in contrast to the re-initiation from stall sites, a process that is known to be inefficient (11) and which is reflected in the existence of a residual population in the RNAP separations. We are confident, therefore, that the observed back-tracking is a real effect and not a biasing of the distributions through population shifts.
The motion of an RNAP induced by a secondary RNAP has previously been observed in a model of cooperation between RNAP in which arrested complexes are rescued by a shunt from a following EC (20,38). In this situation, the arrested complex has already back-tracked and the forward shunting RNAP realigns the 3′ termini of the RNA transcript with the enzyme's active site. In this mechanism, the shunting RNAP does not have to overcome a force from the arrested RNAP's translocation machinery as this has disengaged as a consequence of back-tracking implicit in transcription arrest. As discussed previously the majority of collisions occur between a SEC and an EC. Hence the SEC will already have undergone a certain amount of back-tracking, possibly making it prone to further back-tracking.
In this study we are able to show directly that the presence of convergent promoters can cause RNAP to collide and form stable collision complexes in which the RNAPs stall against each other. Clearly this event prevents the formation of full length transcripts in vitro and hence the expression of the associated genes could be down regulated by TI. Convergently aligned promoters may be beneficial when the concurrent expression of two genes is detrimental to the cell, since each will cooperatively down regulate the others expression. In an alternative situation convergent promoters may be useful in cases where a ‘housekeeping gene’ which is expressed constitutively is convergently-aligned against a gene that requires specific tempero/spatial regulation to fulfil its biological function, see reference (6). In this scenario, the large number of RNAP on the house keeping gene will effectively inhibit transcription of the second, regulated gene. The backtracking that is observed in our experiments may well act as a safe-guard to ensure that RNAP that have been stalled are permanently deactivated even if the RNAP that is blocking transcription were to disengage. This work illustrates how gene expression cannot simply be considered in terms of static, isolated transcription units, instead the dynamic interplay between apparently unconnected transcription units must also taken into account. Only when this is considered will the often refractory mechanisms of gene expression be fully understood.
CONCLUSIONS
The processes of RNAP collision, stalling and back-tracking resulting from a convergent transcription model in vitro have been imaged at the single molecule level by AFM. A significant proportion of RNAP remain on the DNA template and are stalled in close proximity to each other, although other outcomes of the collision event cannot be ruled out and are not detected in this time-lapse approach.. We have visualised the process of back-tracking in one RNAP (either as a SEC or, less likely an OPC) induced by another RNAP as a convergently transcribing EC. This work shows the importance of convergently aligned promoters and the profound effect they can have on TI. The conclusion that a significant proportion of collided RNAP (either OPC or EC) remain bound to the DNA is unexpected. Such stalled complexes will provide a roadblock to any further transcription initiated from these promoters unless the roadblock can be cleared by an alternative mechanism in vivo. Whilst mechanisms exist to clear single stalled complexes by the factor mediated realignment of the 3′ RNA termini with the active site, how a stalled complex of two RNAPs could be cleared is unknown. Furthermore, a real-time single molecule experiment with sufficient time resolution will be required to ascertain whether the ‘colliding’ RNAPs, in some cases, can displace each other or pass.
Acknowledgments
The authors would like to thank the EPSRC and the University of Leeds for funding. Funding to pay the Open Access publication charges for this article was provided by a fund from the Leeds University Research Fellowship held by NHT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Callen B.P., Shearwin K.E., Egan J.B. Transcriptional interference between convergent promoters caused by elongation over the promoter. Molecular Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Horowitz H., Platt T. Regulation of transcription from tandem and convergent promoters. Nucl. Acids Res. 1982;10:5447–5465. doi: 10.1093/nar/10.18.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward D.F., Murray N.E. Convergent transcription in bacteriophage λ: interference with gene expression. J. Mol. Biol. 1979;133:249–266. [Google Scholar]
- 4.Prescott E.M., Proudfoot N.J. Transcriptional collision between convergent genes in budding yeast. Proc. Natl Acad. Sci. USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puig S., Perez-Ortin J.E., Matallana E. Transcriptional and structural study of a region of two convergent overlapping yeast genes. Curr. Microbiol. 1999;39:369–373. doi: 10.1007/s002849900474. [DOI] [PubMed] [Google Scholar]
- 6.Gibson C.W., Thomson N.H., Abrams W.R., Kirkham J. Nested genes: biological implications and use of AFM for analysis. Gene. 2005;350:15–23. doi: 10.1016/j.gene.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 7.Lehner B., Willams G., Campbell R.D., Sanderson C.M. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/s0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- 8.Shearwin K.E., Callen B.P., Egan J.B. Transcriptional interference : a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sneppen K., Dodd I.B., Shearwin K.E., Palmer A.C., Schubert R.A., Callen B.P., Egan J.B. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- 10.Roberts J., Park J.S. Mfd, the bacterial transcription repair coupling factor: translocation, repair and termination. Curr. Opin. Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Komissarova N., Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivetti C., Guthold M., Bustamante C. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivetti C., Codeluppi S., Dieci G., Bustamante C. Visualizing RNA extrusion and DNA wrapping in transcription elongation complexes of bacterial and eukaryotic RNA polymerases. J. Mol. Biol. 2003;326:1413–1426. doi: 10.1016/s0022-2836(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 14.Rippe K., Guthold M., von Hippel P.H., Bustamante C. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E.coli RNA polymerase center dot σ(54) holoenzyme by scanning force microscopy. J. Mol. Biol. 1997;270:125–138. doi: 10.1006/jmbi.1997.1079. [DOI] [PubMed] [Google Scholar]
- 15.Dame R.T., Wyman C., Wurm R., Wagner R., Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 2002;277:2146–2150. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 16.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 17.Craig M.L., Suh W.C., Record M.T. Ho-Center-Dot and Dnase I probing of E-σ (70) RNA polymerase-λ -PR Promoter Open Complexes - Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 18.Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA-polymerase translocation. EMBO J. 1989;8:2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox K.R. In: Drug–DNA interaction protocols. Fox K.R., editor. , Vol. 90. Totowa NJ: Humana press; 1997. pp. 1–22. [Google Scholar]
- 20.Epshtein V., Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 21.Kashlev M., Martin E., Polyakov A., Severinov K., Nikiforov V., Goldfarb A. Histidine-tagged RNA-polymerase: dissection of the transcription cycle using immobilized enzyme. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 22.Von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 23.Bar-Nahum G., Epshtein V., Ruckenstein A.E., Rafikov R., Mustaev A., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Bates A.D., Maxwell A. DNA Topology. 2nd ed. Oxford: Oxford University Press; 2005. [Google Scholar]
- 25.Nelson P. Transport of torsional stress in DNA. Proc. Natl Acad. Sci. USA. 1999;96:14342–14347. doi: 10.1073/pnas.96.25.14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L.F., Wang J.C. Supercoiling of the DNA-template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ten Heggeler-Bordier B., Wahli W., Adrian M., Stasiak A., Dubochet J. The apical localization of transcribing RNA polymerases on supercoiled DNA prevents their rotation around the template. EMBO J. 1992;11:667–672. doi: 10.1002/j.1460-2075.1992.tb05098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erie D.A., Yager T.D., Vonhippel P.H. The single-nucleotide addition cycle in transcription : a biophysical and biochemical perspective. Ann. Rev. Biophys. Biomol. Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- 29.Foster J.E., Holmes S.F., Erie D.A. Allosteric binding of nucleotide triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 30.Darst S.A. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 31.Darst S.A., Polyakov A., Richter C., Zhang G.Y. Insights into Escherichia coli RNA polymerase structure from a combination of x-ray and electron crystallography. J. Struct. Biol. 1998;124:115–122. doi: 10.1006/jsbi.1998.4057. [DOI] [PubMed] [Google Scholar]
- 32.Korzheva N., Mustaev A., Kozlov M., Malhotra A., Nikiforov V., Goldfarb A., Darst S.A. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 33.Polyakov A., Severinova E., Darst S.A. 3-Dimensional structure of Escherichia coli core RNA-polymerase : promoter-binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 34.Sidorenkov I., Komissarova N., Kashlev M. Crucial role of the RNA–DNA hybrid in the processivity of transcription. Molecular Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 35.Pavco P.A., Steege D.A. Elongation by Escherichia coli RNA-polymerase is blocked in vitro by a site-specific DNA-binding protein. J. Biol. Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- 36.Komissarova N., Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 37.Rivetti C., Codeluppi S. Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy. 2001;87:55–66. doi: 10.1016/s0304-3991(00)00064-4. [DOI] [PubMed] [Google Scholar]
- 38.Epshtein V., Toulme F., Rahmouni A.R., Borukhov S., Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]