Abstract
Here we describe a novel endonuclease IV (Endo IV) based assay utilizing a substrate that mimics the abasic lesions that normally occur in double-stranded DNA. The three component substrate is characterized by single-stranded DNA target, an oligonucleotide probe, separated from a helper oligonucleotide by a one base gap. The oligonucleotide probe contains a non-fluorescent quencher at the 5′ end and fluorophore attached to the 3′ end through a special rigid linker. Fluorescence of the oligonucleotide probe is efficiently quenched by the interaction of terminal dye and quencher when not hybridized. Upon hybridization of the oligonucleotide probe and helper probe to their complementary target, the phosphodiester linkage between the rigid linker and the 3′ end of the probe is efficiently cleaved, generating a fluorescent signal. In this study, the use of the Endo IV assay as a post-PCR amplification detection system is demonstrated. High sensitivity and specificity are illustrated using single nucleotide polymorphism detection.
INTRODUCTION
Nucleic acid assays utilizing cleavage enzymes to generate a fluorescent signal from dual-dye-labeled oligonucleotide probes have been extensively used in diagnostics (1–3). In these assays, the ability of fluorescent dyes to transfer energy absorbed from light to nearby molecules forms the basis of homogeneous nucleic acid based assays (1,4,5). In the unhybridized state the fluorophore and quencher are within close proximity, which allows the quencher to absorb energy from the fluorophore to affect quenching. The use of fluorogenic probes in a 5′-nuclease polymerase assay, where the probe is enzymatically cleaved to release the fluorophore (2), is well known. The signal amplification reaction utilizes invasive cleavage with structure-specific 5′-nucleases (6). The method requires annealing of two oligonucleotides, called the upstream oligonucleotide and the probe, to a target sequence, which results in the formation of a unique substrate for the 5′-nuclease. In the DNAzyme-PCR strategy the primer contains a target-specific sequence and harbors the antisense sequence of a 10–23 DNAzyme. During amplification, amplicons are produced containing active sense copies of DNAzymes that cleave a reporter substrate included in the reaction mixture (7). A target nucleic acid sequence can be amplified exponentially in vitro under isothermal conditions by using three enzymatic activities essential to retroviral replication: reverse transcriptase, RNase H (RNA cleavage) and a DNA-dependent RNA polymerase (8). Cycling probe technology represents a simple method for the detection of DNA target sequences, utilizing a chimeric DNA–RNA–DNA probe which is cleaved by RNase H when hybridized with its complementary target (9).
Apurinic/apyrimidinic (AP) sites in DNA arise via spontaneous or mutagen-induced hydrolysis of the N-glycosylic bond, or through the repair activity of DNA glycosylases (10). If left unrepaired, AP sites are potentially lethal or mutagenic (11). To cope with the deleterious consequences of AP sites, organisms possess AP endonucleases that initiate the repair of these DNA lesions (10). Class II AP endonucleases initiate repair by catalyzing the hydrolysis of the 5′-phosphodiester of an abasic site to generate a 3′-OH group and a 5′-abasic phosphate residue (10). The substrate specificity of human apurinic endonuclease on synthetic substrate analogs has been reported previously (12).
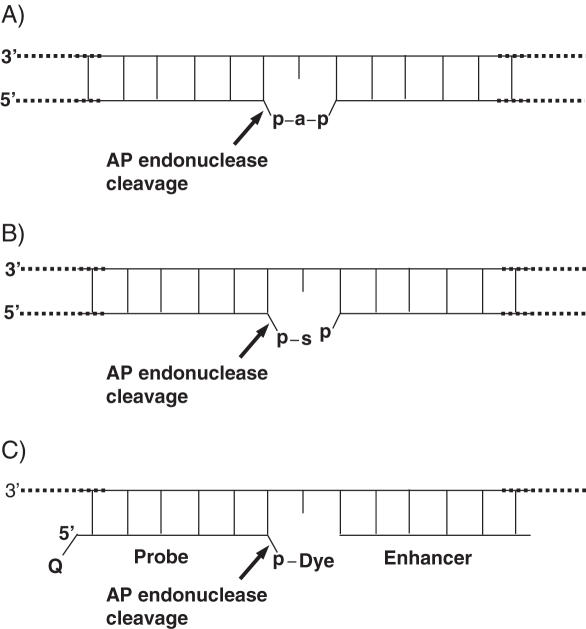
Here we describe a novel endonuclease IV (Endo IV) assay substrate (Figure 1C) that mimics the abasic lesions that normally occur in double-stranded DNA (Figure 1A and B). The first lesion (Figure 1A) is a typical abasic (apurinic or apyrimidinic site generated by spontaneous or enzymatic loss of a nucleic acid base. The second lesion (Figure 1B) is an atypical abasic site appearing as a result of inherent instability of the 3′-phosphodiester bond in abasic deoxyribose in lesion 1 or its cleavage by a Class I AP endonuclease. As shown by the arrow, AP endonucleases cleave the phosphodiester linkage at the abasic sites. In the novel Endo IV assay a short probe with a fluorophore linked through a phosphate at the 3′ end and a short enhancer oligonucleotide generate an artificial, lesion 2 type Endo IV substrate. This arrangement allows the specific and efficient cleavage of the phosphodiester bond of the probe by Endo IV and release of fluorescent dye (Figure 1C). The addition of a quencher at the 5′ end of the probe allows quenching of the fluorescence in the uncleaved probe. The specific cleavage of the phosphate bond and the generation of fluorescence is the basis of the new assay.
Figure 1.
(A) First double strand DNA lesion where ‘a’ represents an abasic 2′-deoxyribose residue and ‘p’ is a phosphate group. (B) Second DNA lesion, formed by spontaneous or enzymatic cleavage of the phosphodiester bond between the 3′-hydroxyl group of the 2′-deoxyribose (abasic site) and the nearest nucleotide of the DNA strand. ‘s’ represents several products of 2′-deoxyribose degradation. (C) Artificial abasic lesion generated by the novel probe and enhancer. Arrows indicated the specific cleavage by AP endonuclease.
In this study we used endonuclease IV from Escherichia coli (Endo IV). The enzyme functions as a Class II AP endonuclease and as a 3′ terminal phosphodiesterase (13,14). Endo IV is enzymatically similar to exonuclease III, the major AP endonuclease of E.coli, but it differs from the latter enzyme mainly in lacking the associated exonuclease activity (14). It was also found that E.coli Endo IV is fairly heat stable to ∼70°C (15,16).
MATERIALS AND METHODS
Reagents
Full characterization for all the compounds described below can be found in the Supplementary Data.
Nucleic acid templates
A total of 102 unrelated Centre Etude Polymorphism Humaine DNA samples were obtained from the Coriell Institute of Medical Research (http://locus.umdnj.edu/) after specifying that the DNA samples should be used for research purpose only. The list of templates used is available at http://snp500cancer.nci.nih.gov.
Synthesis of fluorophore-modified controlled pore glass supports
For the synthesis of the fluorophore-modified controlled pore glass(CPG) supports, please see the Supplementary Data.
Synthesis of oligonucleotide conjugates
Fluorogenic probes were synthesized on fluorophore-linker-based CPG supports (Scheme 1) using standard 3′-DNA-phosphoramidites. An Eclipse Dark Quencher was introduced at the 5′ end using the corresponding phosphoramidite (www.glenresearch.com). Probes were purified by reverse-phase high-performance liquid chromatography (HPLC), dried and re-dissolved in 1× TE buffer. A nearest-neighbor model was applied to calculate extinction coefficient (ɛ260) of oligonucleotides (17). A260 measurements were made in PBS (pH 7.2) at ambient temperature and assumed to be a random coil DNA structure in solution. For each Eclipse Quencher, fluorophore 1 (FL1), fluorophore 2 (FL2) or fluorophore 3 (FL3) substitution an ɛ260 correction of +6600, +18 500, or +25 100 or 28 600 M−1 cm−1 was used, respectively.
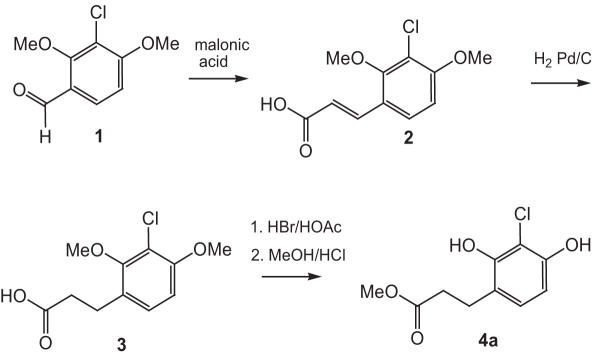
Scheme 1.
Synthesis of methyl 3-(3-chloro-2,4-dihydroxyphenyl)propanoate (4a) required for the preparation of PFP esters 11(a and c).
Preparation of labeled probes with aminohexanoyl-extended hydroxyprolinol linker (FI 4, Table 1)
3′-Hydroxyprolinol modified probe precursor was synthesized using a modified CPG support (18), and 3′-DNA-phosphoramidites and Eclipse Quencher phosphoramidite on a 0.2 μmol scale. The probe was purified by reverse-phase HPLC using a gradient of acetonitrile in 0.1 M triethylammonium bicarbonate buffer (pH 8–9) followed by drying in a SpeedVac evaporator. The probe precursor was re-dissolved in 50 μl of dry DMSO and treated with 10 μmol of pentafluorophenyl N-FMOC-6-aminohexanoate (19) for 10 h. The FMOC-aminohexanoyl modified probe was isolated by reverse-phase HPLC and deprotected by treatment with concentrated ammonium hydroxide for 1 h at room temperature. The deprotected oligonucleotide was further purified by reverse-phase HPLC using the triethylammonium bicarbonate buffer as described above, dried and re-dissolved in 50 μl of dry DMSO. To this solution, 10 μmol compound 8c (Scheme 2) and 1 μl triethylamine were added. After being kept at room temperature for 10 h the reaction was combined with 1 ml of 2% solution of NaClO4 in acetone. The precipitated crude conjugate was further purified by reverse-phase HPLC, dried and re-constituted in 1× TE buffer. An additional molar absorbance (ɛ260) of 28 500 M−1 cm−1 was used in the probe concentration calculation to correct for the presence of the dye.
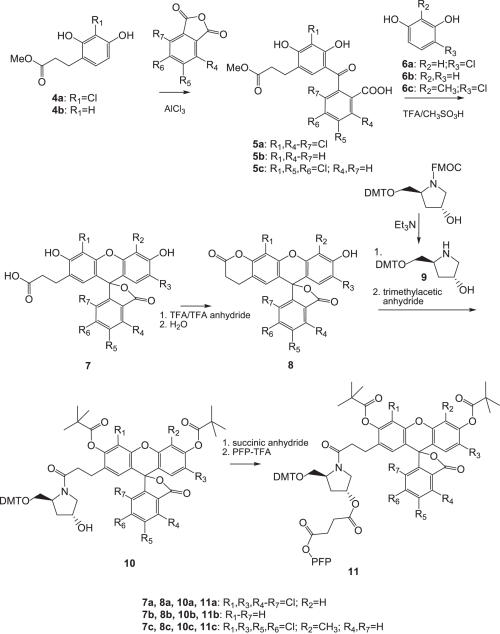
Scheme 2.
Synthesis of the pentafluorophenyl esters 11(a–c) required for the preparation of dye-modified CPG supports.
Synthesis of the dye-modified CPG supports
The synthesis of special CPG supports for the preparation of required oligonucleotide probes are shown in Schemes 1–3.
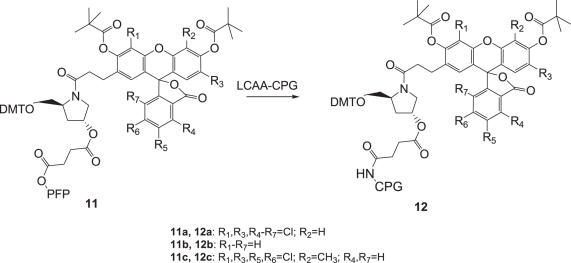
Scheme 3.
Synthesis of the dye-modified CPG supports 12(a–c).
Methyl 3-(3-chloro-2,4-dihydroxyphenyl) propanoate (4a) (Scheme 1) was synthesized starting from 3-chloro-2,4-dimethoxybenzaldehyde (1) (20), which was converted to substituted cinnamic acid 2 by Knoevenagel condensation. Intermediate 2 was hydrogenated in the presence of Pd/C to yield substituted phenyl propionic acid 3, which was first demethylated with HBr/HOAc and then converted to the methyl ester 4a.
Synthesis of the pentafluorophenyl ester dye intermediates 11 required to synthesize the dye-modified CPG supports 12 is shown in Scheme 2. Compounds 4a and b were reacted with substituted phthalic anhydrides in the presence of AlCl3 to give the benzophenones 5, which were reacted with the resorcinol analogs 6 (a–c) in trifluoroacetic acid/methanesulfonic acid to yield the carboxyethyl substituted dyes 7. Treatment of compounds 7 with trifluoroacetic anhydride generated lactones 8, which were first reacted with the DMT-protected hydroxyprolinol 9 (21) and then with trimethylacetic anhydride to afford the protected dyes 10. Reaction of intermediates 10 first with succinic anhydride and then with pentafluorophenyl trifluoroacetate, yielded the PFP esters 11.
The reaction of the PFP ester 11 with the long chain aminoalkyl CPG (LCAA-CPG) yielded the desired CPG supports 12 as shown in Scheme 1.
Optimization of Endo IV enzyme activity
Endo IV cleavage activity was measured in real-time as shown in Figure 2. The model system in Figure 3A was used to determine optimum E.coli Endo IV (Trevigen, Gaithersburg, MD) concentration, magnesium concentration and pH; the latter two were determined to be at 20 mM Tris buffer, pH 8.6. Results are shown in the Supplementary Data.
Figure 2.
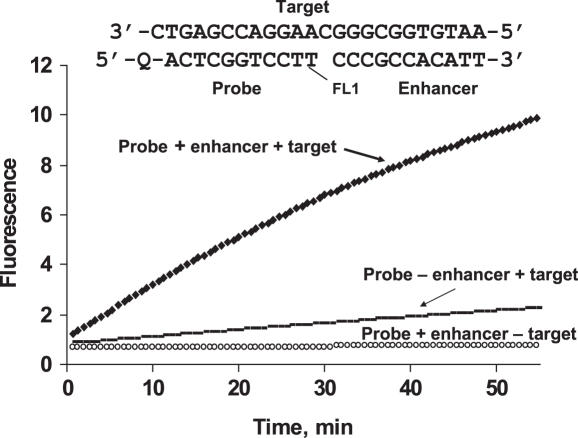
Monitoring of fluorescence by the Endo IV cleavage reaction was performed in 5 mM MgCl2, 20 mM Tris–HCl (pH 8.5). The three component substrate complex used in these experiments is shown. FL1 is a fluorescein derivative and Q is the Eclipse® Dark Quencher, structures of which are shown in Figure 3B and D, respectively. The reaction mixture contained Endo IV at 0.04 U/μl concentration, probe and enhancer at 150 nM, and the target at 5 nM. The experiment was performed on LightCycler.
Figure 3.
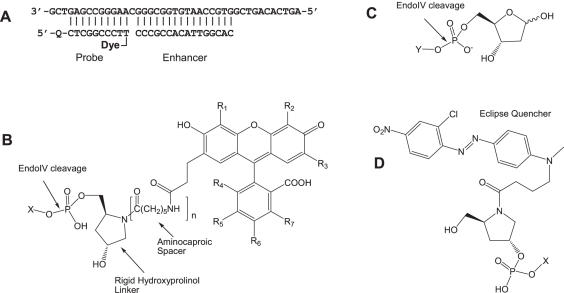
(A) Endonuclease IV model assay showing a target sequence, a 16mer enhancer (calculated Tm 50°C) and a 10mer Endo IV probe (calculated Tm 46.7°C). Q is Eclipse Dark Quencher and the Dyes are compounds from FL1 to FL4. (B) Dyes with hydroxyprolinol (n = 0) and extended (n = 1) hydroxyprolinol linkers. (C) Natural abasic substrate. (D) Eclipse Dark Quencher attached to the 5′ end of the probe. X shows the connection to the oligonucleotide probe. Y shows the connection to the single-stranded DNA.
ENDO IV ASSAY
PCR
PCR was conducted in 384-well plates on MJ Research PTC-200 Peltier Thermal Cycler (Waltman, MA). Fifty cycles of three-step PCR profile (95°C for 5 s, 58°C for 30 s, 72°C for 30 s) were run after an initial 2 min denaturation step at 95°C. Each 5 μl PCR contained 2.5 μl JumpStart Taq ReadyMix (Sigma, MI) and 5–10 ng lyophilized human genomic DNA from Coriell Biorepository (Camdem, NJ). Primer concentrations were 0.1 and 1.0 μM for forward and reverse primers, respectively. A Biomek® 2000 Laboratory Automation Station (Beckman Coulter, USA) was used to set up PCRs.
The Endo IV Master Mixture was prepared by mixing appropriate volumes of 5× Endo IV reaction buffer [100 mM Tris–HCl (pH 8.5), 25 mM MgCl2, 5× E.coli Endo IV enzyme solution, 50% glycerol, 50 mM Tris–HCl (pH 7.5), 0.1 M NaCl, 1 mM DTT, 0.1% Triton X-100, 0.02 U E.coli Endo IV enzyme and 20× of Endo IV Probes/Enhancer Master Mixture], containing each probe and enhancer at 20 μM concentration.
Post-PCR Endo IV reaction
The endo IV reaction was prepared by adding 5 μl Endo IV Master Mixture to 5 μl PCR mixture. Reaction was incubated at 50°C for 50–60 min. Fluorescent detection was performed on ABI 7700 or 7900 in the two channels typically used to detect FAM (for FL1) and VIC (for FL2, FL3 and FL4) utilizing the ABI software for Allelic Discrimination. Alternatively, the fluorescence could also be detected in a LightCycler® LC24 Real-Time PCR System. All assays unless otherwise indicated were performed at least in triplicate.
Tm determination
Thermodynamic parameters of duplex formation were derived by the van't Hoff analysis methods. The shape of each melting curve was fitted to the two-state model with linear base lines (22) using a nonlinear least-square program (23). Unless otherwise stated, Tm of DNA duplexes were measured at a concentration of 5 × 10−7 M in 1× PCR buffer containing 10 mM Tris–HCl (pH 8.5), 5 mM MgCl2 and 40 mM NaCl, as described earlier (23).
Primers, enhancer and probe design
The MGB Eclipse® Design Software 3.0 (http://www.nanogen.com/products/customassays/mgb_ecliplse/) was used to design the primers for PCR. It was also used to calculate the Tm of the enhancer and probe.
RESULTS
Endo IV assay
An example of the new Endo IV-based assay is shown in Figure 2. A probe and an enhancer are hybridized to a complementary synthetic target with a one base gap between them.
The signal is substantially diminished in the absence of the enhancer. A rigid hydroxyprolinol linker (Figure 3B) between the oligonucleotide and fluorophore FL1, critical for sensitivity and specificity, was used in this experiment. The results in Figure 2 illustrate the importance of all components used in the assay. The cleavage of the fluorophore is highly target dependent and its rate is amplified in the presence of the enhancer oligonucleotide. Very low non-target-dependent cleavage is an important feature of the Endo IV assay.
Optimization of Endo IV enzyme activity
Effects of pH and cations on E.coli Endo IV activity was determined using the probe, enhancer and target (Figure 3A). It was shown that Endo IV cleavage of the probe has a relatively flat pH optimum in the range between pH 8.5 and 9.5. For compatibility with post-PCR Endo IV amplification the assay buffer with pH 8.6 was chosen, similar to that of the PCR master mixture. Inclusion of the divalent magnesium cation in the buffer ranging from 0 to 20 mM, resulted in more than 2-fold increase in cleavage rate, the optimum range was ∼5–8 mM. A concentration of 5 mM was chosen for the Endo IV assay. It was determined that the monovalent cations, such as Li, Na, K and Rb, inhibit Endo IV cleavage activity for the concentrations studied in the range from 0 to 100 mM. An inhibition of ∼30% was observed at 20 mM monovalent cation concentration, which increased to ∼60–70% at 100 mM concentration. The optimum probe concentration was determined to be ∼600 nM, please see Supplementary Figures 1–3.
Optimization of fluorophore and linker structures
Figure 3A shows a model Endo IV assay that requires a target complementary to an enhancer and a probe labeled at the 5′ end with an Eclipse Dark Quencher and a fluorescent dye at the 3′ end. As shown, the probe (calculated Tm = 46.7°C) and enhancer (calculated Tm = 50.2°C) are separated by one base to mimic a natural abasic site. Four probes with different fluorescent dyes and linkers (Figure 3B) were evaluated for their performance in the new assay. Fluorophores FL1, FL2 and FL3 of the probes I, II and III (with similar quantum yields) contained a hydroxyprolinol linker between the 3′-phosphate of the probe sequence and the core of the dyes. Fluorophore FL4 of probe IV was analogous to FL3 of probe III, with an additional aminocaproic spacer introduced between the hydroxylprolinol moiety and the dye; the purpose of this spacer was to distance the fluorophore from the cleavage site.
The probe specificity was evaluated by comparing the cleavage rates in the presence and absence of target. Target-dependent (specific) and target-independent (non-specific) cleavage rates of the probe were measured as the percent target cleaved per minute. The results are summarized in Table 1. Probes I–III demonstrated similar specific cleavage. A 2- to 3-fold decrease in the rate of non-specific cleavage for probes II and III versus probe I was observed. These low, non-specific cleavage rates translate into high-specific to non-specific cleavage ratios. It appears that chloro- or chloro/methyl substitutions in the fluorophores of probes II and III, respectively, are beneficial for the reduction of non-specific cleavage. One possible explanation for this reduction may be an effect of increased hydrophobicity of FL2 and FL3 on enzyme activity. In the case of probe IV, with the extended hydroxyprolinol linker, the rate of specific cleavage almost doubled. However, the corresponding increase in the rate of non-specific cleavage was ∼20 times higher than that observed in probe III, substantially compromising the ratio of specific to non-specific cleavage rates. Once again, this result implies the existence of inhibiting interactions between Endo IV enzymatic activity and fluorophore which can be adjusted by moving the fluorophore away from the active center.
Table 1.
Effects of dyes and linkers on the rates and ratios of specific and non-specific cleavage
| Probe nos | Dye | Substitution on Figure 3B | Cleavage rate | Specific/non-specific rate ratios | |
|---|---|---|---|---|---|
| Target-specific %/min | Target-non-specific %/min | ||||
| I | FL1 | R1–R7 = H; n = 0 | 0.97 | 0.0031 | 313 |
| II | FL2 | R1 = R3 = R4 = R5 = R6 = R7 = Cl; R2 = H; n = 0 | 0.93 | 0.0012 | 775 |
| III | FL3 | R1 = R3 = R5 = R6 = Cl; R4 = R7 = H; R2 = CH3; n = 0 | 1.04 | 0.0010 | 1040 |
| IV | FL4 | R1 = R3 = R5 = R6 = Cl; R4 = R7 = H; R2 = CH3; n = 1 | 1.90 | 0.0260 | 73 |
The rigid hydroxyprolinol linker with low non-specific cleavage rates utilized in probes I–III was identified as the linker of choice in the Endo IV assays.
The use of a flexible, straight chain linker (C6) instead of hydroxyprolinol-based linker increased the rates of non-specific cleavage to even higher degree than the extended hydroxyprolinol linker and reduced the specific to non-specific ratios to an unacceptable level (data not shown).
It appears that both the structure of the sterically constrained hydroxyprolinol linker and proximity of the bulky dye moiety to the cleavage site moderate cleavability of the 3′-terminal phosphodiester bond and practically eliminate non-template-dependent cleavage.
Enhancer optimization
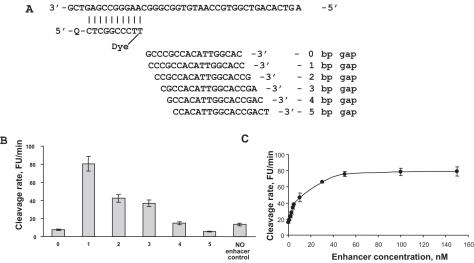
Figure 4A shows the target, probe and the enhancer aligned with the target to indicate different gaps. The Tm of the enhancer is typically chosen to be at least 5°C higher than that of the probe. The cleavage rates in the presence of enhancer are shown with 1–5 base gaps (Figure 4B). As expected, based on the natural abasic substrate requirement shown in Figure 1B, a one base gap gave the highest cleavage rate. The probe in the absence of enhancer showed a cleavage rate nearly equivalent to that of a 4 base gap in the presence of enhancer. As shown in Figure 4C, the maximum cleavage rate is achieved at a concentration of enhancer greater than ∼80 nM. In practice, the concentration of the enhancer is typically at least equivalent or larger than that of the probe. Some cleavage is observed with no enhancer present.
Figure 4.
(A) Endo IV assay investigating a gap of 0–5 bases between the probe and primer. (B) Shows the cleavage rate dependence on the gap between the probe and primer in reference to the cleavage rate when no enhancer is present. (C) Shows the cleavage rate in the presence of different concentrations of a 16mer enhancer. Experiments were performed in quintiplicate.
Probe Tm optimization
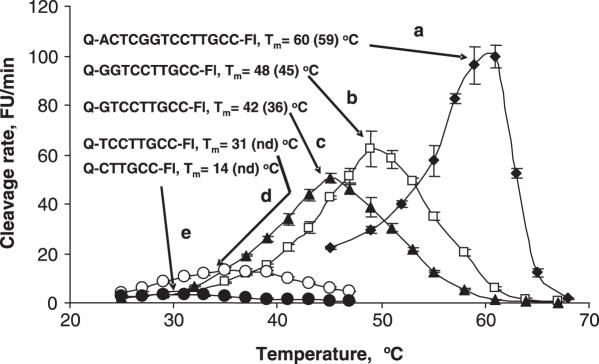
Figure 5 shows how the cleavage rate of probes with different Tms changes with temperature. Probe lengths ranging from a 6mer to a 14mer were investigated and are shown in Figure 5 with calculated Tms of between 14 and 60°C. All the probes showed a bell-shaped relationship for cleavage rate with temperature. As expected, the cleavage rate for all probes increased with increased temperature. A relatively sharp cleavage rate optimum was observed for the probes a, b and c with calculated Tms of 60, 48 and 42, respectively. It appears that the best probe cleavage occurs close to the calculated Tm or slightly above it. Performing the assay at a temperature slightly higher than the Tm (Figure 5) showed not only optimum activity but also appears to allow probe cleavage cycling, an important feature of this assay.
Figure 5.
The effect of temperature on the cleavage rate of probes with different length and calculated Tm. The numbers in parentheses are the determined Tms. ‘nd’ is not determined. The target and enhancers sequences are, respectively, 5′-AGTCACAGTCGGTGCCAATGTGGCGGGCAAGGACCGAGTCG-3′ and 3′-CAGCCACGGTTACACCG-5′.
Determination of assay sensitivity
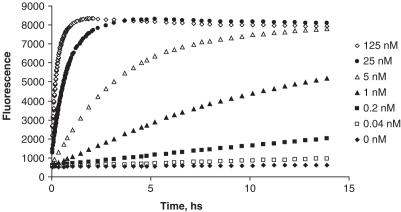
The ability of the Endo IV assay to detect different target concentrations is shown in Figure 6. It was observed that in the case of 125 nM target concentration, the probe-based fluorescence plateaued in ∼1 h, while in the presence of 25 nM target a fluorescence plateau was reached in ∼3 h. The limit of detection of this assay is 0.04 nM, clearly distinguishable from the stable background (Figure 6). This stable background, a characteristic feature of the assay, is maintained over a period of 15 h. This property is also observed in the assay with other nucleic acid targets.
Figure 6.
Dependence of rates of fluorescence generation on target concentration. Probe, enhancer and target sequences are shown in Figure 3. The concentration of reagents were 150 nM probe, 150 nM enhancer, 0.2 U/μl enzyme, 5 mM MgCl2 in 20 mM Tris–HCl buffer, pH 8.5. The reaction was performed at 45°C.
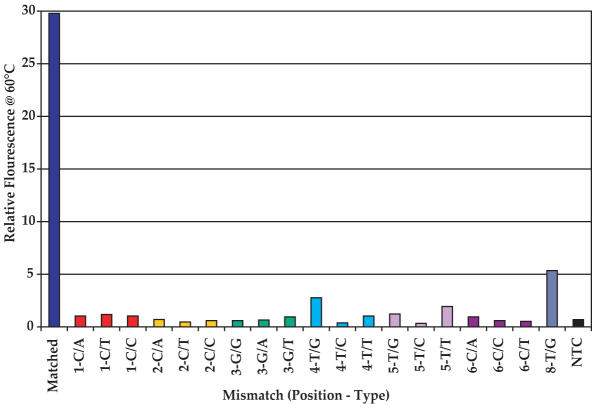
Mismatch discrimination
The ability of the Endo IV assay to discriminate mismatches at different positions of a 14mer probe is illustrated in Figure 7. Mismatches were introduced in the probe one at a time, from positions 1 to 8. The Endo IV assay shows excellent specificity for the different mismatches from base 1 to base 6 with large match/mismatch signal ratios. As shown in Figure 7, the mismatch signal is often about the same as that of the NTC or slightly higher. The exceptions are the difficult 4-G/T- and 8-T/G-mismatches (24) where match/mismatch ratios of ∼16 and 7 were observed, respectively. These ratios are quite adequate to differentiate matched and mismatched sequences. It appears (data not shown) that the discrimination in positions 1 and 2 is largely determined by substrate requirements while discrimination in other positions is determined by thermodynamic considerations. Experiments with a large number of assays have also indicated that, with 10mer to 12mer probes, satisfactory discrimination is observed with the mismatch position in all except the last three bases at the 5′ end.
Figure 7.
The comparison of the change in relative signal fluorescence of match and different mismatches at different positions in a 14mer probe in an Endo IV assay run at 60°C. The probe sequence of the matched probe and target sequence are, respectively, 5′-Q-ACTCGGTCCTTGCC-FL-3′ and 5′-AGTCACAGTCGGTGCCAATGTGGCGGGCAAGGACCGAGTCG-3′. All the complementary sequences indicating the different mismatches are shown in the Supplementary Data. NTC is the no template control.
The exquisite specificity of the Endo IV assay appears to be the result of at least two factors. The first is the strict intrinsic substrate specificity requirement of the Endo IV enzyme which appears to require the rigid prolinol linker (Figure 3B, n = 0) used in the probes I–III. Second, the use of short probes (10mer to 12mer) gives the Endo IV a distinct advantage over most other assays where probes are typically much longer (1,2,6,25).
Post-PCR Endo IV genotyping
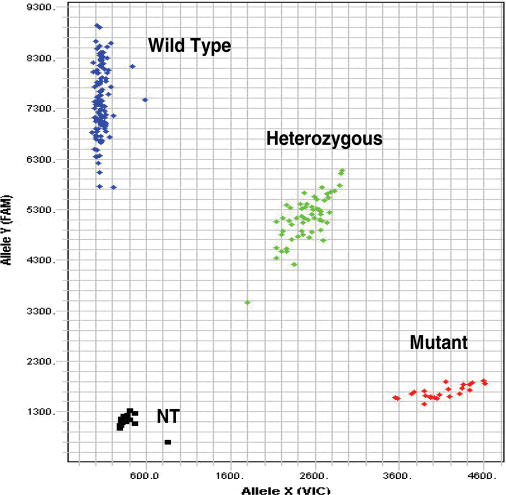
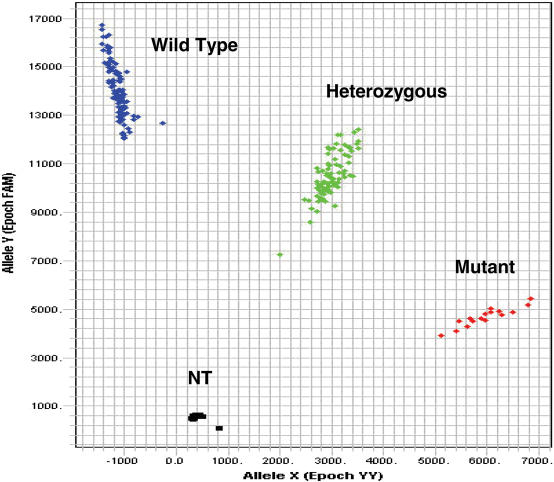
The application of the Endo IV cleavage as a post-PCR detection system is shown in Figures 8 and 9. Following an asymmetric PCR using primers to produce predominantly single-stranded amplicon, two allele-specific probes, an enhancer and Endo IV enzyme are added to the amplification mixture and incubated isothermally for about an hour. The cleaved fluorescence from the two allele-specific probes, each labeled with a different fluorescent dye, is then plotted in a scatter diagram as shown in Figures 8 and 9 for the genotyping analysis of two single nucleotide polymorphisms (SNPs) which are of immediate importance to in cancer (http://snp500cancer.nci.nih.gov). The scatter diagrams of two polymorphisms, namely agouti signal molecular epidemiology studies protein (ASIP-01) and adenomatous polyposis coli (APC-03) (http://snp500cancer.nci.nih.gov) are shown in Figures 8 and 9, respectively. In both cases, a probe specific to the wild-type allele is labeled with FL1 and a probe specific for the mutant allele labeled with FL3 is used.
Figure 8.
A scatter plot of the genotyping of an agouti signal protein (ASIP-01) mutation. The probe sequences for the wild-type and mutant alleles are, respectively, 5′-Q-ACCTTCATTCCC-FL1-3′ and 5′-Q-GACCTTCATTCTC-FL3-3′. The mismatch is underlined. NT is no template control and Q is the Eclipse Quencher. The primer sequences are TCCAGTGCCCCTCATATTTTCC and CTCCTGTCAACAGTAACAAACTTC.
Figure 9.
A scatter plot of the genotyping of an adenomatous polyposis coli (APC-03) mutation. The probe sequences for the wild-type and mutant alleles are, respectively, 5′-Q-TACTTTCTGTGGC-FL1-3′ and 5′-Q-CTTTTCCGTGGC-FL3-3′. The mismatch is underlined. NT is no template control and Q is the Eclipse Dark Quencher. Primer sequences are AGCACTCAGGCTGGATGAACAA and GTTCAGAGGGTCCAGGTTCTT.
The probe sets, designed to analyze ASIP-01 and APC-03 polymorphisms, were used to assay 102 unrelated human DNA samples obtained from the Coriell Institute. In the case of the ASIP-01 target the mismatch was situated two bases from the 3′ end of the probe, while in the case of APC-03, the mismatch was six bases from the 3′ end of the probe.
From our experience, the spacing of the allele clusters in Figures 8 and 9 are comparable or better than those scatter plots observed in other methods (2,26,27). The exquisite specificity achieved with the Endo IV enzyme, combined with the shorter probes used and the high signal accumulation, gives this assay an advantage over other SNP assays. In comparison with other signal accumulating assays (2), the Endo IV assay has similar levels of high signal accumulation, but uses shorter probes with higher specificity than those assays. In the case of hybridization-based assays (5,25), the Endo IV assay has not only a signal accumulation advantage, but also uses shorter probes with a specificity advantage. In addition, the shorter probes allow much more flexibility in assay design compared to other methods with longer probes.
CONCLUSIONS
We disclosed a novel nucleic acid signal detection system based on the cleavage of a phosphodiester linkage in a dual-dye-labeled oligonucleotide probe by the Endo IV enzyme from E.coli. The parameters influencing the activity of the Endo IV enzyme were optimized. This optimized Endo IV detection assay requires, in addition to an oligonucleotide probe substrate, a fluorophore attached through a relatively rigid linker to the oligonucleotide and a helper oligonucleotide. The rigid linker conveys the exquisite specificity of Endo IV cleavage of the phosphodiester linkage in the probe and shows high-specific/non-specific rate ratios. The synthesis of fluorophore analogs coupled to a rigid linker and solid support, used in the synthesis of labeled oligonucleotides, was also disclosed. Excellent mismatch discrimination is observed from positions 1 to 6 in a 12mer probe. The ability to put the mismatch in any one of positions 1–6, allows for flexibility in probe design for genotyping assays where sequence and secondary structural constraints are considered. This method allows the use of probes shorter than those typically used in other methods (28) contributing to the improved mismatch discrimination.
The combination of the Endo IV assay with a PCR as a post-amplification detection system has been demonstrated. Post-PCR Endo IV genotyping shows excellent spacing of scatter plot endpoint signals of no template controls, wild-type, mutant and heterozygous samples allowing clear allele differentiation. In contrast, other endpoint genotyping methods may suffer from overlapping endpoint signals due to PCR efficiency issues (25,29). This Endo IV post-PCR genotyping has been successfully used in an industry setting for >5000 genotype assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
We thank Alan Mills for his excellent technical assistance in the preparation of figures. Funding to pay the Open Access publication charges for this article was provided by Nanogen, Inc., San Diego, CA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holland P.M., Abramson R.D., Watson R., Gelfand D.H. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus. Proc. Natl Acad. Sci., USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livak K.J. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Gen. Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y.-W., Procop G.W., Persing D.H. Molecular diagnostics of infectious diseases. Clin. Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 4.Livak K.J., Flood S.J., Marmaro J., Giusti W., Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 5.Marras S.A., Kramer F.R., Tyagi S. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyamichev V., Mast A.L., Hall J.G., Prudent J.R., Kaiser M.W., Takova T., Kwiatkowski R.W., Sander T.J., de Arruda M., Arco D.A., et al. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- 7.Todd A.V., Fuery C.J., Impey H.L., Applegate T.L., Haughton M.A. DzyNA-PCR: use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format. Clin. Chem. 2000;46:625–630. [PubMed] [Google Scholar]
- 8.Guatelli J.C., Whitfield K.M., Kwoh D.Y., Barringer K.J., Richman D.D., Gingeras T.R. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc. Natl Acad. Sci. USA. 1990;87:1874–1878. doi: 10.1073/pnas.87.5.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modrusan Z., Bekkaoui F., Duck P. Spermine-mediated improvement of cycling probe reaction. Mol. Cell Probes. 1998;12:107–116. doi: 10.1006/mcpr.1998.0156. [DOI] [PubMed] [Google Scholar]
- 10.Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 11.Loeb L.A., Preston B.D. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 12.Wilson D.M., III, Takeshita M., Grollman A.P., Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 13.Strauss P.R., O'Regan N.E. Abasic site repair in higher eukaryotes. In: Nickoloff J.A., Hoekstra M.F., editors. DNA Damage and Repair. Vol. III. Totowa: Humana Press; 2001. pp. 43–86. [Google Scholar]
- 14.Levin J.D., Johnson A.W., Dempe B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J. Biol. Chem. 1988;263:8066–8071. [PubMed] [Google Scholar]
- 15.Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem. 1977;252:2808–2814. [PubMed] [Google Scholar]
- 16.Haas B.J., Sandigursky M., Tainer J.A., Franklin W.A., Cunningham R.P. Purification and characterization of Thermotoga maritima endonuclease IV, a thermostable apurinic/apyrimidinic endonuclease and 3′-repair diesterase. J. Bacteriol. 1999;181:2834–2839. doi: 10.1128/jb.181.9.2834-2839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantor C.R., Warshow M.M., Shapiro H. Oligonucleotide interaction III. Circular dicroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 18.Herbert N. Oligomeric aminodiol-containing compounds, libraries thereof, and process of preparing the same. 2004. US Patent 6,828,427.
- 19.Haralambidis J., Duncan L., Angus K., Tregear G.W. The synthesis of polyamide–oligonucleotide conjugate molecules. Nucleic Acids Res. 1990;18:493–499. doi: 10.1093/nar/18.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plattner J.J., Fung A.K.L., Parks J.A., Pariza R.J., Crowley S.R., Pernet A.G., Bunnell P.R., Dodge P.W. Substituted 5,6-dihydrofuro[3,2-f]-1,2-benzisoxazole-6-carboxylic acids: high-ceiling diuretics with uricosuric activity. J. Med. Chem. 1984;27:1016–1026. doi: 10.1021/jm00374a014. [DOI] [PubMed] [Google Scholar]
- 21.Hebert N., Davis P.W., DeBaets E.L., Acevedo O.L. Synthesis of N-substituted hydroxyprolinol phosphoramidites for the preparation of combinatorial libraries. Tetrahedron Lett. 1994;35:9509–9512. [Google Scholar]
- 22.Xia T., SantaLucia J., Jr, Burkard M.E., Kierzek R., Schroeder S.J., Jiao X., Cox C., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 23.Lohkov S.G., Pyshnyi D.V. Thermodynamic and spectral properties of DNA miniduplexes with the terminal G:A mispairs and 3′ or 5′ dangling bases. FEBS Lett. 1997;420:134–138. doi: 10.1016/s0014-5793(97)01502-0. [DOI] [PubMed] [Google Scholar]
- 24.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 25.Afonina I.A., Reed M.W., Lusby E., Shishkina I.G., Belousov Y.S. Minor groove binder-conjugated DNA probes for quantitative DNA detection by hybridization-triggered fluorescence. Biotechniques. 2002;32:940–944. doi: 10.2144/02324pf01. [DOI] [PubMed] [Google Scholar]
- 26.Hampe J., Wollstein A., Lu T., Frevel H.J., Will M., Manaster C., Schreiber S. An integrated system for high throughput TaqMan based SNP genotyping. Bioinformatics. 2001;17:654–655. doi: 10.1093/bioinformatics/17.7.654. [DOI] [PubMed] [Google Scholar]
- 27.Hsu T.M., Law S.M., Duan S., Neri B.P., Kwo P.-Y. Genotyping single-nucleotide polymorphisms by the invader assay with dual-color fluorescence polarization detection. Clin. Chem. 2001;47:373–377. [PubMed] [Google Scholar]
- 28.Chantratita W., Pongtanapisit W., Piroj W., Srichunrasmi C., Seesuai S. Development and comparison of the real-time amplification based methods—NASBA-Beacon, RT–PCR Taqman and RT–PCR hybridization probe assays—for the qualitative detection of SARS coronavirus. Southeast Asian J. Trop. Med. Public Health. 2004;35:623–629. [PubMed] [Google Scholar]
- 29.Afonina I.A., Belousov Y.S., Metcalf M., Mills A., Sanders S., Kutyavin I., Walburger D., Gorn V., Shishkina I., Adams A., et al. Single nucleotide detection with MGB Eclipse™ assays. J. Clin. Ligand Assay. 2002;25:268–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.