Abstract
Alu elements are the most abundant repetitive elements in the human genome; they emerged 65 million years ago from a 5′ to 3′ fusion of the 7SL RNA gene and amplified throughout the human genome by retrotransposition to reach the present number of more than one million copies. Over the last years, several lines of evidence demonstrated that these elements modulate gene expression at the post-transcriptional level in at least three independent manners. They have been shown to be involved in alternative splicing, RNA editing and translation regulation. These findings highlight how the genome adapted to these repetitive elements by assigning them important functions in regulation of gene expression. Alu elements should therefore be considered as a large reservoir of potential regulatory functions that have been actively participating in primate evolution.
INTRODUCTION
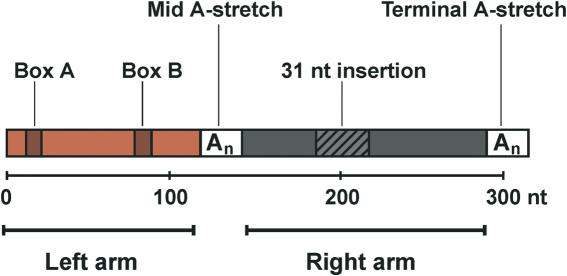
The initial sequencing of the human genome revealed that 55% of its nucleotide sequence is composed of repetitive elements (1). Among different families of repetitive elements, Alu elements are the most abundant in the human genome. They are present in more than one million copies, which altogether represent 10% of the whole genome mass. Alu belong to the SINE family (Short Interspersed Nuclear Elements) of repetitive elements; they emerged 55 millions years ago with the radiation of primates by a fusion of the 5′ and 3′ ends of the 7SL RNA gene, which encodes the RNA moiety of the signal recognition particle (SRP). The first fossil Alu monomers (FAMs) arose from this fusion (2); they were ∼160 bp long and are poorly represented in the human genome (2). According to the current model, modern Alu elements emerged from a head to tail fusion of two distinct FAMs (3) that gave rise to a dimeric structure composed of two similar but distinct monomers (left and right arms) joined by an A-rich linker (Figure 1). Modern Alu elements are ∼300 bp in length and are classified into subfamilies according to their relative ages [for a review see Ref. (4)]. Dimeric Alu elements are unique to primates; they amplified throughout the primate genomes via RNA intermediates by a mechanism of retrotransposition that remains to be elucidated. Their amplification has been dependent on the transposition machinery of other retrotransposons, since they do not encode any protein. It has recently been shown that they could use LINE-1 (long interspersed nuclear elements) elements for this purpose (5).
Figure 1.
Architecture of Alu elements. Alu elements are about 300 nt long; they have a dimeric structure composed of two related but not equivalent monomers (left and right arms). The right arm contains a 31 nt insertion as compared to the left arm. Left and right arms are separated by an A-rich region (Mid A-stretch) and followed by a short poly(A) tail (Terminal A-stretch). The left arm contains functional, but weak, A and B boxes of the RNA polymerase III internal promoter.
Alu elements, as well as other repetitive elements, were at the origin considered as parasites of the genome that had no major effect on its stability and genic expression. They were thought to be ‘selfish’ or ‘junk’ DNA (6,7), but nowadays, several lines of evidence show that the presence of repetitive elements and especially of Alu elements, had a great influence on the human genome, in particular on its evolution. These effects were both negative and positive. On one hand, integration into genic regions that caused gene inactivation might often have been deleterious for the organism. On the other hand, because of their extended sequence homology, Alu elements induced a considerable number of non-allelic recombinations that lead to both duplications and deletions of DNA segments, thereby accelerating evolution by several orders of magnitude. Another function frequently attributed to Alu elements is their ability to provide new regulatory elements to neighboring genes. It was, indeed, reported several times that Alu elements became effectors of gene transcription by providing new enhancers, promoters and polyadenylation signals to many genes [for reviews see Ref. (8,9)].
In this review, we focuse on more recently discovered effects of Alu elements on gene expression, at the post-transcriptional level. We describe their influence on mRNA splicing, on RNA editing and on protein translation.
Alu elements and alternative splicing
Alternative splicing is a mechanism by which the use of alternative splice sites in pre-mRNAs generates multiple variants of proteins from a single gene. This type of variation in splicing patterns is a major source of protein diversity in the genome and it has been estimated that 30–60% of all human genes produce alternative exons (10). This mechanism could expand the proteome diversity by several orders of magnitude and, in an extreme example, the Drosophila axon guidance receptor gene, Dscam, may potentially generate 38 000 DSCAM isoforms by alternative splicing (11). There are several ways by which a gene can acquire new alternative splice sites; one of them is the mutation of pre-existing intronic sequences that result in the recruitment of intronic sequences into coding regions of mRNAs. This process is called exonization.
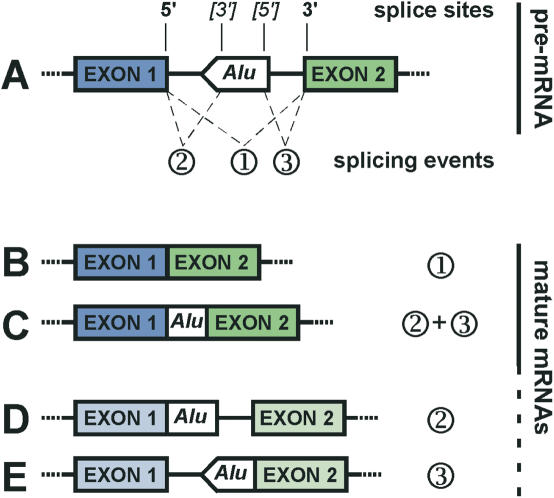
Of the thousands of Alu elements that are found in introns of the human genome, a certain number of complete or partial Alu sequences are also present in the coding regions of mature mRNAs (12). The presence of several potential splice sites in the Alu consensus sequence (13,14) strongly suggested that they were recruited in the coding region through exonization (Figure 2). Recently, genome-scale computational studies confirmed this hypothesis. By comparing the human genome to cDNA and EST databases (14), Sorek and colleagues identified a subset of alternatively spliced internal exons of which 5% were derived from Alu elements. As the same analysis performed on constitutive exons failed to identify any Alu element, they concluded that all Alu containing exons are alternatively spliced.
Figure 2.
Exonization of intronic Alu elements. Hypothetical Alu element inserted in an orientation that opposes the sense of transcription in an intronic region. This element has a major 3′ splice site near position 275 and a major 5′ splice site near position 158 as described in the text. The use of alternative splice sites of the Alu element can lead to a variety of mature mRNAs. (A) Pre-mRNA with possible splicing events (1; 2; 3). 5′ and 3′splice sites are indicated. Alternative splice sites of the Alu element are under brackets. (B) Regular splicing without Alu exonization resulting from splicing event 1. (C) Exonization of the intronic Alu element by use of its 5′ and 3′ splice sites together with the 5′ splice site of exon 1 and 3′ splice site of exon 2; result of the splicing event 2 and 3. Only the Alu element is exonized. (D) Exonization of the intronic Alu element by use of its 3′ splice site together with the 5′ splice site of exon 1; result of the splicing event 2. The Alu element and the 3′ end of the intron are exonized. (E) Exonization of the intronic Alu element by use of its 5′ splice site together with the 3′ splice site of exon 2; result of the splicing event 3. The Alu element and the 5′ end of the intron are exonized. Notably, (D and E) represent theoretical possibilities, which have so far never been observed in vivo. Because of exon length restriction (68), they are unlikely to occur. However, they might have existed as rare splice variants of evolutionary intermediates of (A).
The Alu consensus contains 9 potential 5′ splice sites and 14 potential 3′ splice sites (14). Along the consensus, 19 out of 23 potential splice sites are present on the minus strand; this means that an Alu element has more chances to be exonized when inserted in the sense that opposes the one of transcription. It was indeed confirmed by the same study that 85% of Alu containing exons derive from antisense Alu elements (14).
Sequence comparison of Alu containing exons revealed that all potential splice sites were not used at the same frequency (14); the most favored sites in the antisense Alu consensus were positions 275 and 279 used as 3′ splice site (referred as proximal and distal splice sites, respectively) and position 158 used as 5′ splice site. Further sequence comparison of nucleotides surrounding the alternative 3′ splice sites helped to determine mutations in the Alu consensus that govern the selection of this site. The results of sequence analysis were confirmed in vivo by experiments using an ADAR2 minigene. The ADAR2 gene is known to contain an alternative Alu exon that adds 40 amino acids to the open reading frame (ORF) of the protein (15). By site-directed mutagenesis of the ADAR2 minigene, Lev-Maor et al. (16) discovered a delicate interplay between the proximal and distal 3′ splice sites; when the G immediately upstream the distal site is mutated to any other nucleotide, the Alu exon becomes constitutively spliced and uses the proximal 3′ splice site. A similar computational and mutational approach determined the minimal base substitutions required in an antisense intronic Alu element to form a 5′ splice site at position 158. The base compositions at positions 2 and 5 of the intron were found to be critical for maintaining a fine balance of base pairing between the 5′ splice site and U1 snRNA, which determines the level of alternative splicing (17).
Most striking was the fact that single mutations could turn the splicing from alternative to constitutive (16,17). Several cases are known, such as the Alport (18) and the Sly syndromes (19), where constitutive inclusions of an Alu exon lead to genetic diseases. More recently, it was discovered that insertion of an alternative Alu exon could also lead to a genetic disease. A mutation in the intron 6 of the CTDP1 gene, which creates an alternatively spliced Alu exon, results in CCFDN (congenital cataracts, facial dysmorphism and neuropathy) syndrome (20).
Although, these studies provided the molecular bases of Alu elements exonization, the regulation of Alu exonization might be even more complex. Recent work on the intron 16 of the human angiotensin converting enzyme gene revealed that exonized antisense Alu elements contain several auxillary splicing enhancer sequences that facilitate their exonization (21). Similarly, it was determined by the same group that Alu elements inserted in sense orientation contain strong splicing silencer sequences that repress their exonization (22). The presence of such enhancers may provide an alternative explanation for the observation that 85% of exonized Alu elements are inserted in antisense orientation (14).
The evolutive process leading to exonization of a partial or complete Alu element is a complex series of successive mutations. Insertion of an Alu exon is likely to introduce a premature termination codon or a frame shift. Several mutations are therefore required to create an Alu exon. The precise scenario of sequential mutations that lead to exonization of an Alu element has recently been elucidated for five different loci by careful PCR analyses on different species among the primates lineage (23,24). For example, in the TNF receptor gene icp75TNRF, the alternative replacement of the first exon by an Alu exon, which lies upstream of exon 1, required three successive mutations. After integration of the Alu elements (58–40 mya ago), an A to G transition gave birth to a new ATG initiator codon before the divergence of platyrrhines. Following the New World Monkeys divergence (40–25 mya),a C to T transition then created a new 3′ splice site and a subsequent 7 nt deletion enabled the ORF by disabling a stop codon (23).
Taken altogether, these data showed that the million Alu elements present in the human genome could act as a very large reservoir of alternative exons. As all identified Alu exons were shown to be alternatively spliced (14), a strong selective pressure must exist to avoid the loss of the original form of the protein. As Alu exonization is prone to introduce premature termination by frame-shifts, the loss of the natural polypeptide would most likely be deleterious for the organism but these alternative Alu exons might have played an important role in the evolution of primates. For example, humans and mice almost have all of their respective genes in common. However, it has been estimated that although they share 70% of their constitutive splice sites, they only share 15% of their alternative splice sites (25). This difference in alternative splicing creates a species-specific pool of alternative exons and might therefore account for a significant part of the morphological and physiological differences between mice and humans.
Alu elements and A–I editing
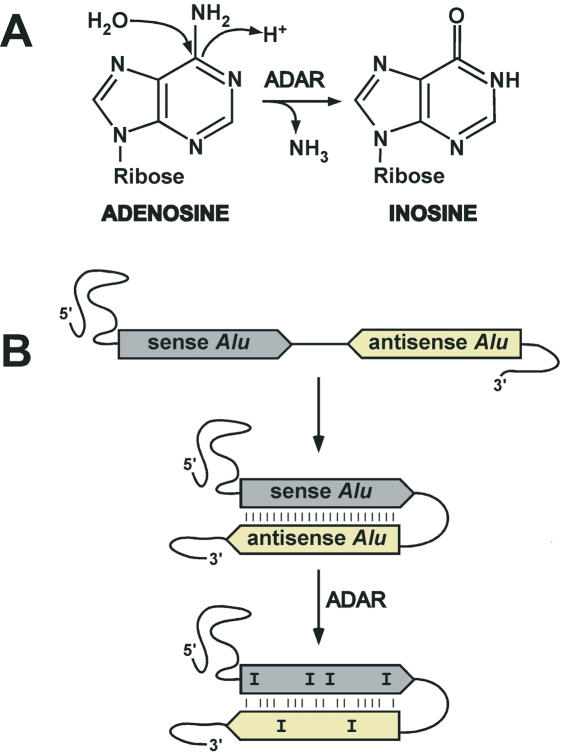
RNA editing is a process by which the nucleotide sequence of RNA molecules is changed co- or post-transcriptionally. The modifications in the RNA include nucleotide insertions, deletions or base modifications. Among these modifications, base conversions appear to be the major type of editing. The best-characterized base conversions are hydrolytic deamination reactions by which cytosine are converted to uracyl and adenosine (A) to inosine (I). The A–I editing reaction is catalyzed in vivo by members of the adenosine deaminase acting on RNA (ADAR) family of enzymes (Figure 3A), which preferentially edit adenosines located in double-stranded regions of RNA molecules [for reviews see Ref. (26,27)]. The precise role of A–I editing in cell metabolism is still unclear but it has been shown that it is required for normal life; the knockout ADAR1 is embryonically lethal by liver disintegration (28), while ADAR2−/− mice die young and are prone to seizures (29). Until recently, very few positions edited by ADAR were known in the human transcriptome. This was in contrast to the apparent mass of inosine estimated to one molecule per 17 000 bases in rat brain tissue and one molecule per 33 000 bases in heart tissue (30).
Figure 3.
A-to-I editing in Alu elements. (A) Deamination of adenosine by ADAR leading to the production of inosine. (B) Intramolecular base paring of a mRNA containing two Alu elements in opposite orientation. Base pairing between the two Alu elements leads to the formation of a long stable double-stranded RNA region in which ADAR performs A-to-I substitutions (marked as I).
The missing mass of inosine in the human transcriptome has recently been localized within Alu elements by three independent groups using genome-scale computational searches for A–I editing sites. These three groups identified editing sites by aligning mRNAs (EST and cDNA databases) to the human genome sequence and detecting A–G substitutions. As I is read as G by sequencing, the presence of A–G substitutions between genomic DNA and mRNA reflects the presence of an inosine edited site. To get rid of single nucleotide polymorphisms and sequencing errors, the three groups designed elaborated in silico filters allowing to select only A–I substitutions. Datasets of mRNAs and filters used are the major differences between these three studies. Using this approach, Kim et al. (31) identified 30 085 substitutions in 2674 different transcripts, Levanon et al. (32) identified 12 723 substitutions in 1637 different transcripts, and Athanasiadis et al. (33) found 14 500 substitutions in 1445 mRNAs.
Together, these three studies demonstrated that A–I editing is a widespread mechanism and, more surprisingly, that >90% of all A–I substitutions occur within Alu elements contained in mRNAs. Further investigations revealed that 54% of all identified editing events occurred in the 3′-untranslated region (3′-UTR) of mRNAs, 12% occurred in 5′-UTRs and 33% in introns (32); this clearly demonstrated that A–I editing preferentially occurs in non-coding regions of mRNAs. In order to assess the accuracy of the in silico analysis, 26 previously unknown editing sites were confirmed experimentally (32).
ADAR enzymes have no strict sequence requirements at the editing sites; a double-stranded RNA region seems to be sufficient. By comparing edited mRNAs, hot spots of editing along the Alu consensus sequence were identified. Adenosines 27, 28, 136 and 162 of the Alu consensus were more prone to editing than the others (32). Neighboring bases might also influence the editing frequency; T is over-represented at the position preceding the edited adenosine, whereas G is under-represented at the same position (31).
At first, the preferential editing of Alu sequences inside mRNAs might have been attributed to the secondary structure of Alu RNA that contains long double-stranded regions (34) (Figure 4). However, Athanasiadis et al. (33) determined an interesting correlation between adenosine editing inside an Alu element and the presence of an inverted Alu element in close proximity. They demonstrated that editing is favored when a distance <2 kb separates two Alu elements in opposite orientations. These data defined a model in which two closely inserted Alu elements base pair and become an ideal substrate for ADAR (Figure 3B). This model was recently confirmed by the study of the editing patterns of cyclin M3 intron 2 and NFκB1 intron 16 showing that the base pairing between two Alu elements occurs intramolecularly, and not intermolecularly (35), and therefore confirmed this model.
Figure 4.
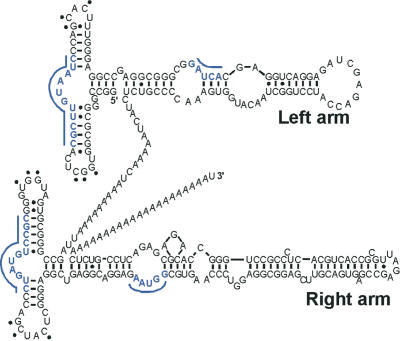
AluRNA secondary structure. Secondary structure of a Pol.III-transcribed Alu RNA drawn based on a previously determined secondary structure (34) and adapted to the sequence of the Alu element of intron 4 of the α-Fetoprotein gene (Alu Y) (69). Underlined blue letters and dots indicate the binding sites of SRP9/14 and the tertiary base pairing between the two loops, respectively, by analogy to SRP RNA (52,70).
Interestingly, the three genome-scale studies failed to detect any known editing sites in the coding region of glutamate (36) and serotonin receptor (37) mRNAs; this might be explained by the high stringency of the in silico filters used. Moreover, it strongly suggests that not all editing sites of the human transcriptome have been uncovered.
Similar sequence comparisons carried out in mouse and fly transcriptomes revealed that the high levels of editing are specific to primates (38). As Alu elements exist only in primates, the correlation between their presence and abundant A–I editing is striking. This logically led to the speculation that appearance of Alu elements in the primate lineage lead to widespread editing, which most likely played a role in primate evolution (38).
In summary, Alu elements play an important role in editing of the human transcriptome by providing ideal templates to the ADAR family of enzymes. The large number of Alu elements present in mRNAs and their relatively low divergence explains why they are more prone to be edited than other sequences, and why widespread editing is specific to primates (39). Although the precise role of RNA editing is still speculative, it might affect gene expression at several steps. As inosine does not base pair with uracyl but with cytosine, editing might influence the stability of RNA molecules by creating and disrupting secondary structures. At another level, as inosine is recognized as guanosine by the translation and splicing machineries, A–I editing could lead to amino acid substitutions in the coding sequence, or to modification of splice sites in introns that could induce premature termination or frame-shifts. Knowing that aberrant editing is found in several neurological disorders (40,41), it is highly probable that the phenomenon is of great physiological importance.
Alu elements and protein translation
It has been known for a long time that Alu RNAs, transcribed from Alu elements, are present in the cytosol of primate cells. As mentioned previously, Alu elements contain the internal A and B boxes of the RNA polymerase III promoter from the 7SL RNA gene (Figure 1). These internal promoter elements significantly diverge from the consensus (42) and are too weak to drive efficient transcription of Alu elements, which is then dependent on sequences flanking their site of insertion (43). In normal growth conditions, Alu RNAs are present at very low levels in the cytosol (103–104 molecules per cell) but numerous stress conditions, such as viral infection, cycloheximide exposure or heat shock, transiently increase their level of expression (44), which rapidly decreases upon recovery. This precisely controlled regulation raised the attractive possibility that Alu RNAs may serve a specific function in cell metabolism, which is required during stress conditions. This hypothesis was supported by two independent studies showing that an overexpressed Alu RNA was able to stimulate translation of a co-transfected reporter gene in mammalian cells (45,46). These data suggested for the first time that transcribed Alu elements may serve a specific function in translation regulation. Alu RNAs were initially thought to act as inhibitors of the double-stranded RNA-dependent protein kinase, PKR (45). They were proposed to bind PKR and prevent its autophosphorylation; this, in turn, would avoid eIF2α phosphorylation that would itself inhibit translation initiation. By preventing inhibition of translation initiation, Alu RNA would then behave as translational activators. However, it has been discovered that Alu RNAs can activate PKR when they are present at low concentrations and can inhibit it at higher concentrations (47). Knowing that Alu RNA stimulates translation of reporter genes in PKR knockout cells (46), the involvement of PKR in this mechanism is questionable, even though it cannot be excluded.
Alu RNAs transcribed from Alu elements are highly structured RNAs that maintained strong structural similarities with their ancestor, SRP RNA. The typical Alu RNA is a dimer of related but non-equivalent arms that are joined by an A-rich linker and followed by a short poly(A) stretch (Figure 4). Each arm is related to the Alu domain of SRP RNA in terms of sequence and secondary structure and can bind the cognate SRP protein SRP9/14 in vitro (48) and in vivo (49). However, the left arm shows a higher affinity for these proteins than the right one (48). Recent results from our group showed that synthetic Alu RNPs composed of Alu RNA in complex with recombinant SRP9/14 have a different effect on protein translation than naked Alu RNA. While Alu RNA stimulates the translation of all reporter mRNAs in a cell free translation system, Alu RNP acts as a general inhibitor of protein translation (50). Such opposite activities of Alu RNP and Alu RNA were at first quite surprising; however, these data could easily be explained by conformational changes of Alu RNA upon SRP9/14 binding. It was previously observed in SRP that the RNA is in a loosely folded state in the absence of SRP9/14 while it assumes a very compact structure in its presence (51,52).
As it is known that SRP9/14 are present in a large excess over SRP in mammalian cells (53), the occurrence of Alu RNP in vivo becomes very likely. Moreover, it has been shown that Alu RNA sediments in high molecular weight complex (53,54) and that Alu RNAs expressed in response to adenovirus infection are assembled in SRP9/14-containing RNPs (49). These data make the occurrence of naked Alu RNA in the cytosol questionable; however, they do not exclude a mechanism by which the activity of Alu RNA would be modulated by the binding of SRP9/14.
Further investigation about the mechanism by which Alu RNP and Alu RNA influence protein translation showed that both of them act at the level of translation initiation (50). These results unexpectedly demonstrated that despite strong structural similarities, Alu RNP and SRP influence translation in very different ways; while SRP mediates a transient delay in translation by blocking the elongation step, Alu RNP inhibits translation by reducing initiation.
Alu elements are also frequently found in UTR of several transcripts (55,56). They are transcribed by RNA polymerase II as part of mRNAs. As 5′- and 3′-UTRs are hot spots of regulation of translation initiation, these elements are suitably located to modulate translation initiation. Several independent studies described a role for Alu elements present in UTRs in regulating translation initiation. For example, it was suggested that an Alu element in the 5′-UTR of human growth hormone receptor (hGRH) could regulate the translation of this mRNA (57). Similarly, an antisense Alu element in the 3′-UTR of the manganese superoxide dismutase (MnSOD) acts as a translation inhibitor (58). In the case of ZNF177 mRNA, a zinc finger protein of unknown function, an inverted partial Alu element in the 5′-UTR has been shown to strongly decrease the translation efficiency of the mRNA (56). The best-characterized example of translation regulation by an Alu element in the UTR is BRCA1. BRCA1 is a DNA repair protein whose mutation is associated with breast cancer. The 80 kb genomic sequence of this gene is composed at 40% of Alu elements (59). BRCA1 mRNA exists in two forms that differ in their leader sequences and in their patterns of expression (60). These two transcripts are formed by selective use of different promoters (61). The isoform with a short 5′-UTR is expressed in normal and cancerous mammary tissue whereas the isoform with a longer 5′-UTR is expressed only in breast cancer tissue (62). The latter mRNA is much less efficiently translated than the other one and this translational defect has been shown to be due to an Alu element in the 5′-UTR of this transcript (62). This Alu element has a 60 nt deletion in the left arm but the right one is intact and forms the stable secondary structure that partially prevents translation initiation. Deregulation of BRCA1 transcription in cancer then results in a higher proportion of translationally inhibited mRNA, which contributes to a decrease in the BRCA1 protein level leading to accumulation of defects and mutations, and ultimately to cancer.
Altogether, these data show that Alu elements modulate protein translation in at least two different manners; they can act as trans regulatory factors when transcribed by Pol.III and assembled in Alu RNP, and act as cis regulatory elements when transcribed by Pol.II in 5′- and 3′-UTRs. Unpublished data from our group show that Alu elements in UTRs of some mRNAs are able to bind SRP9/14 in vitro. This observation suggests new potential roles for Alu elements in UTRs, for example a role in stabilization of the mRNA by SRP9/14 binding. The impact of Alu elements on protein translation is most likely only partially uncovered and further work will be required fully to understand their impact on gene expression.
CONCLUDING REMARKS
Together with the fact that Alu elements have a high potential to modulate gene transcription by binding several transcription factors (63), the findings reported here demonstrate that ‘junk RNA’, transcribed from Alu elements, is useful for many purposes in cell metabolism. Alu elements were probably no more than selfish DNA at the origin, but the genome clearly adapted to their presence by assigning them some important function in regulation of gene expression. This gain of regulatory function, known as exaptation (64), most likely participated in primate evolution and probably helped in their divergence from other mammals. Alu elements should then rather be considered as a huge reservoir of potential regulatory functions that are expressed or not, at the mercy of point mutations occurring randomly over the time, as detailed previously for alternative splice site formation (23,24).
Other families of retrotransposons were already suggested to affect gene expression. LINE-1, for example, have been shown to influence gene transcription and to introduce polyadenylation signals in several reports [reviewed in Refs. (65,66)]. However, the effects of LINE-1 elements on gene expression have been less studied, and roles, such as described for Alu elements are still speculative.
We believe that the amplitude of the ‘Alu phenomenon’ in both human genome and transcriptome has been only partially uncovered. Several other potential functions of Alu elements might be unsuspected yet; it was indeed recently reported that Alu elements in 3′-UTRs of mRNAs are probable microRNA targets (67).
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the Canton of Geneva.
Conflict of interest statement. None declared.
REFERENCES
- 1.International-Human-Genome-Sequencing-Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Quentin Y. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res. 1992;20:3397–3401. doi: 10.1093/nar/20.13.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quentin Y. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 1992;20:487–493. doi: 10.1093/nar/20.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nature Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 5.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 6.Orgel L.E., Crick F.H. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 7.Orgel L.E., Crick F.H., Sapienza C. Selfish DNA. Nature. 1980;288:645–646. doi: 10.1038/288645a0. [DOI] [PubMed] [Google Scholar]
- 8.Tomilin N.V. Control of genes by mammalian retroposons. Int. Rev. Cytol. 1999;186:1–48. doi: 10.1016/s0074-7696(08)61050-5. [DOI] [PubMed] [Google Scholar]
- 9.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 10.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 11.Schmucker D., Clemens J.C., Shu H., Worby C.A., Xiao J., Muda M., Dixon J.E., Zipursky S.L. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 12.Nekrutenko A., Li W.H. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 13.Makalowski W., Mitchell G.A., Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 14.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai F., Chen C.X., Carter K.C., Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 17.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 18.Knebelmann B., Forestier L., Drouot L., Quinones S., Chuet C., Benessy F., Saus J., Antignac C. Splice-mediated insertion of an Alu sequence in the COL4A3 mRNA causing autosomal recessive Alport syndrome. Hum. Mol. Genet. 1995;4:675–679. doi: 10.1093/hmg/4.4.675. [DOI] [PubMed] [Google Scholar]
- 19.Vervoort R., Gitzelmann R., Lissens W., Liebaers I. A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- 20.Varon R., Gooding R., Steglich C., Marns L., Tang H., Angelicheva D., Yong K.K., Ambrugger P., Reinhold A., Morar B., et al. Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nature Genet. 2003;35:185–189. doi: 10.1038/ng1243. [DOI] [PubMed] [Google Scholar]
- 21.Lei H., Day I.N., Vorechovsky I. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 2005;33:3897–3906. doi: 10.1093/nar/gki707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei H., Vorechovsky I. Identification of splicing silencers and enhancers in sense Alus: a role for pseudoacceptors in splice site repression. Mol. Cell. Biol. 2005;25:6912–6920. doi: 10.1128/MCB.25.16.6912-6920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer S.S., Mannel D.N., Hehlgans T., Brosius J., Schmitz J. From ‘junk’ to gene: curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 24.Krull M., Brosius J., Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 25.Thanaraj T.A., Clark F., Muilu J. Conservation of human alternative splice events in mouse. Nucleic Acids Res. 2003;31:2544–2552. doi: 10.1093/nar/gkg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaub M., Keller W. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie. 2002;84:791–803. doi: 10.1016/s0300-9084(02)01446-3. [DOI] [PubMed] [Google Scholar]
- 27.Valente L., Nishikura K. ADAR gene family and A–I RNA editing: diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- 28.Hartner J.C., Schmittwolf C., Kispert A., Muller A.M., Higuchi M., Seeburg P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 29.Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 30.Paul M.S., Bass B.L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., et al. Systematic identification of abundant A–I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 33.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinnett D., Richer C., Deragon J.M., Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J. Biol. Chem. 1991;266:8675–8678. [PubMed] [Google Scholar]
- 35.Kawahara Y., Nishikura K. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeburg P.H. The role of RNA editing in controlling glutamate receptor channel properties. J. Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- 37.Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 38.Eisenberg E., Nemzer S., Kinar Y., Sorek R., Rechavi G., Levanon E.Y. Is abundant A–I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Neeman Y., Levanon E.Y., Jantsch M.F., Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006 doi: 10.1261/rna.165106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palladino M.J., Keegan L.P., O'Connell M.A., Reenan R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 41.Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 42.Muller J., Benecke B.J. Analysis of transcription factors binding to the human 7SL RNA gene promoter. Biochem. Cell. Biol. 1999;77:431–438. [PubMed] [Google Scholar]
- 43.Li T.H., Schmid C.W. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 44.Liu W.M., Chu W.M., Choudary P.V., Schmid C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu W.M., Ballard R., Carpick B.W., Williams B.R., Schmid C.W. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubin C.M., Kimura R.H., Schmid C.W. Selective stimulation of translational expression by Alu RNA. Nucleic Acids Res. 2002;30:3253–3261. doi: 10.1093/nar/gkf419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams B.R. PKR: a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 48.Bovia F., Wolff N., Ryser S., Strub K. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res. 1997;25:318–326. doi: 10.1093/nar/25.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang D.Y., Hsu K., Maraia R.J. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasler J., Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006;34:2374–2385. doi: 10.1093/nar/gkl246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weichenrieder O., Stehlin C., Kapp U., Birse D.E., Timmins P.A., Strub K., Cusack S. Hierarchical assembly of the Alu domain of the mammalian signal recognition particle. RNA. 2001;7:731–740. doi: 10.1017/s1355838201010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weichenrieder O., Wild K., Strub K., Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- 53.Bovia F., Fornallaz M., Leffers H., Strub K. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol. Biol. Cell. 1995;6:471–484. doi: 10.1091/mbc.6.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W.M., Maraia R.J., Rubin C.M., Schmid C.W. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yulug I.G., Yulug A., Fisher E.M. The frequency and position of Alu repeats in cDNAs, as determined by database searching. Genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 56.Landry J.R., Medstrand P., Mager D.L. Repetitive elements in the 5′-untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–116. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- 57.Goodyer C.G., Zheng H., Hendy G.N. Alu elements in human growth hormone receptor gene 5′-untranslated region exons. J. Mol. Endocrinol. 2001;27:357–366. doi: 10.1677/jme.0.0270357. [DOI] [PubMed] [Google Scholar]
- 58.Stuart J.J., Egry L.A., Wong G.H., Kaspar R.L. The 3′-UTR of human MnSOD mRNA hybridizes to a small cytoplasmic RNA and inhibits gene expression. Biochem. Biophys. Res. Commun. 2000;274:641–648. doi: 10.1006/bbrc.2000.3189. [DOI] [PubMed] [Google Scholar]
- 59.Smith T.M., Lee M.K., Szabo C.I., Jerome N., McEuen M., Taylor M., Hood L., King M.C. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 60.Xu C.F., Brown M.A., Chambers J.A., Griffiths B., Nicolai H., Solomon E. Distinct transcription start sites generate two forms of BRCA1 mRNA. Hum. Mol. Genet. 1995;4:2259–2264. doi: 10.1093/hmg/4.12.2259. [DOI] [PubMed] [Google Scholar]
- 61.Xu C.F., Chambers J.A., Solomon E. Complex regulation of the BRCA1 gene. J. Biol. Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- 62.Sobczak K., Krzyzosiak W.J. Structural determinants of BRCA1 translational regulation. J. Biol. Chem. 2002;277:17349–17358. doi: 10.1074/jbc.M109162200. [DOI] [PubMed] [Google Scholar]
- 63.Polak P., Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brosius J., Gould S.J. On ‘genomenclature’: a comprehensive (and respectful) taxonomy for pseudogenes and other ‘junk DNA’. Proc. Natl Acad. Sci. USA. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han J.S., Boeke J.D. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 66.Kim D.S., Kim T.H., Huh J.W., Kim I.C., Kim S.W., Park H.S., Kim H.S. LINE FUSION GENES: a database of LINE expression within human genes. BMC Genomics. 2006;7:139. doi: 10.1186/1471-2164-7-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smalheiser N.R., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 69.Gibbs P.E., Zielinski R., Boyd C., Dugaiczyk A. Structure, polymorphism, and novel repeated DNA elements revealed by a complete sequence of the human alpha-fetoprotein gene. Biochemistry. 1987;26:1332–1343. doi: 10.1021/bi00379a020. [DOI] [PubMed] [Google Scholar]
- 70.Strub K., Moss J., Walter P. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol. Cell. Biol. 1991;11:3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]