Abstract
Linkage analysis is a successful procedure to associate diseases with specific genomic regions. These regions are often large, containing hundreds of genes, which make experimental methods employed to identify the disease gene arduous and expensive. We present two methods to prioritize candidates for further experimental study: Common Pathway Scanning (CPS) and Common Module Profiling (CMP). CPS is based on the assumption that common phenotypes are associated with dysfunction in proteins that participate in the same complex or pathway. CPS applies network data derived from protein–protein interaction (PPI) and pathway databases to identify relationships between genes. CMP identifies likely candidates using a domain-dependent sequence similarity approach, based on the hypothesis that disruption of genes of similar function will lead to the same phenotype. Both algorithms use two forms of input data: known disease genes or multiple disease loci. When using known disease genes as input, our combined methods have a sensitivity of 0.52 and a specificity of 0.97 and reduce the candidate list by 13-fold. Using multiple loci, our methods successfully identify disease genes for all benchmark diseases with a sensitivity of 0.84 and a specificity of 0.63. Our combined approach prioritizes good candidates and will accelerate the disease gene discovery process.
INTRODUCTION
The identification of genes responsible for human disease is critical to gain an understanding of disease mechanisms and is essential for the development of new diagnostics and therapeutics. Genetic linkage analysis has been used successfully to identify chromosomal loci. Unfortunately, isolating the disease-causing gene(s) within these loci can be difficult: genomic regions are often large, containing hundreds of possible candidate genes, making experimental methods time-consuming and expensive. Furthermore, searches for single nucleotide polymorphisms (SNPs) in the genomes of individual patients from clinical studies will produce a large number of potential gene candidates (1,2). Clearly, these high-throughput analyses will require computational approaches to identify the best candidates for further study.
The completion of the human genome sequencing project has stimulated the development of new genome-scale bioinformatics approaches to understand disease. While some progress has been made in candidate gene prediction, these systems can, at best, only claim modest pruning of the genes in a disease interval (3).
Previous candidate gene prediction systems have largely been based on keyword similarity to known disease genes or phenotypes. For example, the G2D system (4,5) is based on biomedical literature searches and associates pathological conditions with gene ontology (GO) terms (6). Candidate genes are then identified by homology to GO-annotated and disease-associated genes. POCUS (3) finds candidate genes by identifying an enrichment of keywords associated with GO, shared InterPro domains (7) and expression profiles among a given set of susceptibility loci relative to the genome at large. The method by Tiffin et al. (8) selects candidates according to their expression profiles within tissues associated with disease, and relationships between clinical and molecular data are identified using the eVOC anatomy ontology (9). The recent method SUSPECTS (10) again compares GO, InterPro and expression libraries of putative disease genes with those known to be involved with the same disease. Similarly, GeneSeeker (11) integrates keyword data based on mapping, expression and phenotypic databases from human and mouse studies. Finally, the method by Freudenberg and Propping (12) is based on a measure of phenotypic similarity between diseases and produces clusters of disease genes using keywords derived from OMIM (13).
Some of these methods have been incorporated into a consensus approach that has been applied to select candidates for the complex diseases type 2 diabetes and obesity (14). Using a consensus of combined methods appears to be effective for ranking predicted candidate disease genes.
Here, we present Gentrepid, a system which improves on these early methods by using a combined bioinformatics approach encompassing methods of domain comparison and protein pathway and interaction data analysis. The system combines two methods for the automated prediction of disease genes within known disease intervals. The first, Common Pathway Scanning (CPS), is based on the assumption that common phenotypes are generally associated with disruption in proteins that participate in the same complex or pathway (15). Recently, Gandhi et al. (16) showed that disease genes preferentially interact with other disease-causing genes and a study by Oti et al. (17) predicted that 10% of proteins interacting with a disease gene product are likely to participate in the same disease. Franke et al. (18) described a system, PRIORITIZER, based on predicted protein–protein interactions (PPIs), whereby disease genes are identified through common interactions of proteins in multiple disease intervals that have common phenotypes.
Our second method, Common Module Profiling (CMP), is based on the principle that candidate genes may have similar functions to disease genes that have already been determined (19). CMP is similar in concept to methods using functional annotations, but many human proteins lack annotation (20) and, therefore, similarities would be missed when comparing keywords alone. For example, only 10 000 human proteins, ∼25% of the human proteome, have manually curated GO-terms.
CMP uses a domain-based comparative sequence analysis to identify those proteins with potential functional similarity. Domain-based sequence comparison searches have been shown to be more accurate than full-sequence searches (21) as commonly applied in BLAST or PSI-BLAST database searches (22). Unlike the keyword systems, CMP calculates a measure of domain-based similarity to known disease genes rather than making a binary comparison.
Both methods use two sources of input for disease gene prediction. First, known disease genes are used to predict novel disease genes in chromosomal intervals associated with the same disease. Second, without knowledge of the disease genes, candidate disease genes are predicted by comparing all the genes in the multiple intervals associated with the same disease to find relationships between proteins linking the intervals. The proteins may be related via a common pathway or shared domains.
MATERIALS AND METHODS
Annotation pipeline
All biological data were combined into a relational database. Human disease gene information was extracted from the OMIM database and lists of genes flanking the disease genes were obtained from EntrezGene (build 35) (23). Protein sequence data were taken from GenBank (24) and complete protein domain annotation was performed on all protein sequences using Pfam Hidden Markov models (25). Finally, all genes were mapped to the latest pathway and PPI data.
There are currently over 200 biological pathway and network resources available (26). Here, we utilize data from BioCarta (www.biocarta.com), KEGG (27) and OPHID (28), the most comprehensive databases of their type. BioCarta and KEGG are chiefly pathway databases with BioCarta specializing in signalling pathways and KEGG in metabolic pathways. OPHID is a secondary PPI database containing literature-derived interaction data from BIND (29), MINT (30) and HPRD (31), as well as data from recent high-throughput experimentation (32–35). OPHID also contains transferred interactions from orthologous proteins in model organisms.
CPS
Potential disease genes were predicted by identifying all proteins within a disease interval that are part of a pathway, described in BioCarta and KEGG. PPI data from OPHID was used to identify novel disease genes by finding the interaction partners of known disease genes in a disease interval. Three levels of interactions were tested for potential disease genes, based on the shortest path length to a known disease gene. Unless stated otherwise, result summaries for all the combined methodologies are presented using OPHID interactions at a distance of one to the nearest disease gene.
When CPS is applied across multiple intervals, i.e. in the absence of known disease genes, all interaction partners and pathways associated with the genes in each interval are compared across intervals. Disease genes are predicted by identifying common pathways or interaction partners shared by the intervals associated with a specific phenotype.
CMP
CMP compares the Pfam domain content of each protein within a disease interval to identify putative disease genes. Different calculations are performed depending on whether CMP uses known disease genes or multiple intervals as input.
When known disease genes are used as input, a protein (candidate) observed to have disease-like domains is assigned a score (S). Scores are based on the similarity between the protein's domains (j) and the domains (i) in the known disease gene (dg) using SSEARCH (36) bit scores (s). SSEARCH is an implementation of the Smith and Waterman local alignment algorithm (37). Scores are normalized by matching the equivalent region of the disease gene against itself on a domain by domain basis (Equation 1).
| 1 |
If a protein has multiple domains of the same type, the highest scoring matching domain is used.
When CMP is used across multiple intervals, a census of all domains in every interval associated with the disease is taken. Disease genes are predicted based on the similarity of their domain content to genes from other intervals associated with the phenotype. The domain combination is tested for over-representation in the intervals compared to the genome as a whole. A similarity score based on the numerator of Equation 1 is calculated as well as two measures of statistical significance. In the first calculation of significance, domains in a sequence are assumed to be completely uncorrelated. This represents an upper limit of significance. The expected (ea) number of genes containing those domains is calculated by:
| 2 |
where m is the number of intervals containing the domains of interest, n is the number of genes in the interval and f is a form factor related to the average number of domains per gene. The probability of encountering domain i is given by:
| 3 |
where N is all domain types. These numbers are determined from a census of all domains across the genome. For the second calculation of significance, domains are assumed to be completely correlated. This represents a lower limit of significance. The expectation (eb) is based on the prevalence of the rarest domain:
| 4 |
Two χ2 tests ( and ) are then calculated in the usual manner using the two expectation values at a significance level of 0.995. Clusters of genes containing the same domains are then ranked according to the two alternative χ2 values.
Benchmarking
We validated the CPS and CMP methods using data from previously determined disease phenotypes where at least three disease genes have been identified (3). Our benchmark disease set had 170 disease genes. This same set contained 163 disease genes in 2003 when used in the analysis of POCUS. The disease genes may have been identified via linkage analysis or through a candidate gene approach. For the purpose of benchmarking, the disease genes are used to generate pseudo-intervals to simulate an interval derived by linkage analysis. Three pseudo-interval sizes are used that encompass 50, 100 and 150 genes around the known disease genes. Hereafter, the term interval will be used to refer to pseudo-intervals in the test set.
When the disease genes are used as the input, the predictive power of each method is tested on each disease gene using leave-one-out cross validation. In this method one of the disease genes is disregarded and the remaining known disease genes are used to identify the omitted disease gene in its interval. When using multiple intervals, all genes in the intervals sharing a phenotype are used to identify links between the intervals via common protein relationships. The multiple interval technique is useful for phenotypes were no disease genes are known.
Several measures of predictive power were used: sensitivity, the probability of finding a disease gene among disease genes [TP/(TP + FN)]; and specificity, the probability of not finding a disease gene among non-disease genes [TN/(TN + FP)]; where TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives and FN is the number of false negatives. An enrichment ratio (ER) is also calculated for each disease from the proportion of disease genes predicted by the methods divided by the proportion of disease genes within the disease intervals (Equation 5). Enrichment is a measure of how well the system prunes a list of genes in a disease interval to a list of final candidate disease genes.
| 5 |
CPS and CMP predictions were compared with a random selection of candidate genes within a disease interval. The number of random assignments made is based on the number of predictions made by each method. Random selections were performed 1000 times for each disease, from which an average number of correctly identified disease genes is calculated.
RESULTS
Known disease gene input
Table 1 shows the results of candidate gene prediction for each of our methods on 170 disease genes for 29 diseases as used by Turner et al. in their analysis of POCUS. When using known disease genes as input, our methods make predictions for all 29 diseases in each of the 50, 100 and 150 gene intervals and correctly predict a disease gene in 20 diseases. In comparison, POCUS made candidate predictions for eight of the 29 diseases and only five of the diseases had a disease gene correctly identified.
Table 1.
Number of correctly predicted disease genes by each method using known disease genes
| Disease | Known Disease Genes | Successful Automated Predictions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CMP | CPS | CPS | CPS | CPS | CPS | CPS | Random | |||||
| BioCarta | KEGG | OPHID | OPHIDh | OPHIDlit+ | OPHIDlit− | Total | 50 | 100 | 150 | |||
| aan | 4 | 0 | 0 | 0 | 3 | 3 | 3 | 2 | 3 | 0.1 | 0.1 | 0.1 |
| alz | 8 | 2 | 3 | 6 | 5 | 5 | 5 | 3 | 6 | 0.3 | 0.2 | 0.2 |
| aml | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.2 | 0.2 |
| bb | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 |
| bc | 9 | 0 | 4 | 0 | 6 | 6 | 6 | 0 | 6 | 0.5 | 0.5 | 0.5 |
| bcc | 4 | 1 | 1 | 2 | 3 | 3 | 3 | 0 | 3 | 0.1 | 0.0 | 0.1 |
| cchn | 6 | 5 | 0 | 0 | 5 | 4 | 4 | 4 | 5 | 0.4 | 0.3 | 0.3 |
| cf | 5 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 0.2 | 0.2 | 0.2 |
| cfh | 12 | 5 | 0 | 4 | 4 | 4 | 4 | 0 | 9 | 1.0 | 0.7 | 0.8 |
| cmt | 5 | 0 | 0 | 0 | 2 | 2 | 2 | 0 | 2 | 0.2 | 0.2 | 0.2 |
| ebl | 5 | 3 | 0 | 5 | 5 | 5 | 5 | 0 | 5 | 0.2 | 0.1 | 0.1 |
| ed | 7 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 5 | 0.4 | 0.3 | 0.2 |
| fap | 4 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | 0.2 | 0.2 | 0.1 |
| gc | 5 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 4 | 0.3 | 0.2 | 0.2 |
| h | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0.2 | 0.2 |
| ibd | 5 | 0 | 2 | 3 | 4 | 4 | 4 | 2 | 4 | 0.4 | 0.3 | 0.3 |
| joag | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0.1 | 0.1 |
| lca | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0.1 | 0.1 |
| lhscr | 5 | 0 | 0 | 2 | 2 | 2 | 2 | 0 | 4 | 0.2 | 0.3 | 0.3 |
| md | 6 | 2 | 0 | 0 | 2 | 2 | 2 | 0 | 3 | 0.1 | 0.1 | 0.1 |
| mf | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.2 | 0.2 |
| mody | 6 | 2 | 0 | 0 | 4 | 4 | 4 | 2 | 5 | 0.3 | 0.3 | 0.3 |
| niddm | 8 | 4 | 2 | 0 | 2 | 2 | 2 | 2 | 5 | 0.6 | 0.4 | 0.3 |
| oc | 4 | 0 | 0 | 4 | 2 | 2 | 2 | 2 | 4 | 0.3 | 0.3 | 0.3 |
| pc | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0.1 | 0.2 |
| pd | 3 | 0 | 0 | 3 | 2 | 2 | 2 | 0 | 3 | 0.1 | 0.0 | 0.0 |
| rp | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.2 | 0.2 |
| sle | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.1 | 0.2 |
| tcp | 13 | 3 | 0 | 2 | 4 | 4 | 4 | 0 | 7 | 0.9 | 0.8 | 0.8 |
| Total | 170 | 32 | 16 | 41 | 55 | 54 | 54 | 17 | 88 | 8.0 | 6.6 | 6.7 |
CMP results are based on a cut-off threshold of 0.1. CPS-interactions go to the 1st level of interaction only. CPS-OHPID contains all PPI data from OPHID. CPS-OPHIDh contains human data only. CPS-OPHIDlit+ contains data from literature databases only. CPS-OPHIDlit− does not contain PPI data from literature databases. Random is calculated on total predictions for the 50, 100 and 150 interval size. Disease abbreviations: aan, adrenoleukodystrophy, autosomal neonatal; alz, Alzheimer disease; aml, acute myeloid leukemia; bb, Bardet-Biedl syndrome; bc, breast cancer; bcc, basal cell carcinoma; cchn, colorectal cancer, hereditary nonpolyposis; cf, cystic fibrosis; cfh, cardiomyopathy, familial hypertrophic; cmt, Charcot-Marie-Tooth disease; ebl, epidermolysis bullosa letalis; ed, epiphyseal dysplasia, multiple types 1–5; fap, familial adenomatous polyposis; gc, gastric cancer; h, hypertension; ibd, inflammatory bowel disease; joag, juvenile-onset primary open angle glaucoma; lca, Leber congenital amaurosis; lhscr, long-segment Hirschsprung disease; md, muscular dystrophy, limb-girdle; mf, familial meningioma; mody, maturity-onset diabetes of the young; niddm, type 2 diabetes mellitus; oc, ovarian carcinom; pc, prostate cancer; pd, Parkinson disease; rp, retinitis pigmentosa; sle, systemic lupus erythematosus; tcp, thyroid carcinoma, papillary.
CPS benchmark performance: CPS identifies novel disease genes by finding proteins that are linked with the product of a known disease gene in the pathway and PPI databases. Results for CPS are divided into three datasets: pathway data from BioCarta, pathway data from KEGG and PPI data from OPHID. KEGG pathway data correctly predicts 41 disease genes in 13 diseases. For the 100 gene interval size, the probability of finding a disease gene (sensitivity) using KEGG data is 0.26, and the probability of not finding a disease gene among non-disease genes (specificity) by KEGG is 0.98. Overall data enrichment is 12-fold for the 100 gene interval size, reducing a list of 100 gene candidates to just eight genes.
BioCarta pathway data identifies 16 disease genes in seven diseases. BioCarta has a sensitivity of 0.15, a specificity of 0.99 and an enrichment of 16-fold for the 100 gene interval size. The complementary nature of these pathway databases is demonstrated by their unique results. BioCarta finds disease genes for two diseases, type 2 diabetes mellitus and breast cancer, where the KEGG data fails. KEGG finds disease genes for eight diseases where the BioCarta data fails.
The OPHID PPI dataset contains 48 321 interactions for 10 666 proteins representing 13% of the estimated complete human-interactome (38). Overall, OPHID has a sensitivity of 0.42, a specificity of 1.00 and an enrichment of 50-fold at the 100 gene interval size. These results appear much better than the pathway data, but the success of prediction might be influenced by PPI data derived from literature associations of well studied diseases. In an attempt to remove bias from literature PPIs and to assess the usefulness of orthology data, OPHID is further split into several overlapping sets: human-only data, i.e. the data does not contain transferred orthologous interactions (OPHIDh); PPI data derived from literature searches only, i.e. data from the BIND, HPRD and MINT databases (OPHIDlit+); and all PPIs except those from the literature databases (OPHIDlit−).
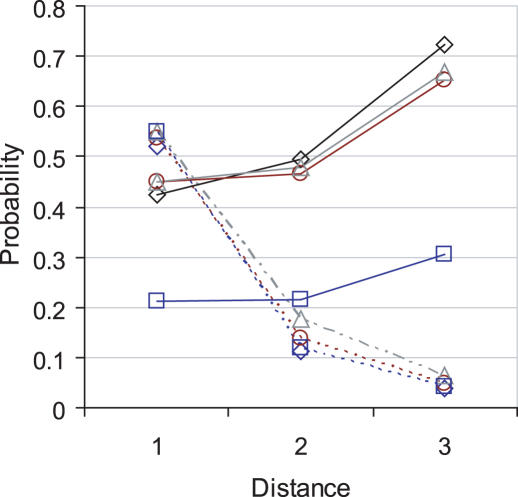
Using the orthology data leads to more false positives and the difference between correct predictions is negligible: OPHID finds one more disease gene than OPHIDh. Figure 1 shows the sensitivities for each of the datasets compared with the proportion of correct predictions at increasing path lengths for the 100 gene interval size. At the first level of interactions the majority of correct predictions, 54, is found using the OPHIDlit+ set, with a sensitivity of 0.45 and specificity of 1.00. The non-literature PPIs find 17 disease genes, with a sensitivity of 0.21 and a specificity of 1.00. While the probability of finding a disease gene is lower in the non-literature set, overall enrichment is the same, 53-fold, and the proportion of correct predictions is the same, 0.55. Therefore, it is the larger coverage of the literature data that gives it the advantage over the non-literature set and suggests that the experimental data and orthology data held in the OPHIDlit− set is of equal quality to the literature assignments.
Figure 1.
Sensitivity (continuous line) and proportion of predicted genes that are actually disease genes (dashed line) for OPHID (diamond), OPHIDh (circle), OPHIDlit+ (triangle) and OPHIDlit− (square) at three levels of interactions (Distance). Results are shown for the 100 interval size only.
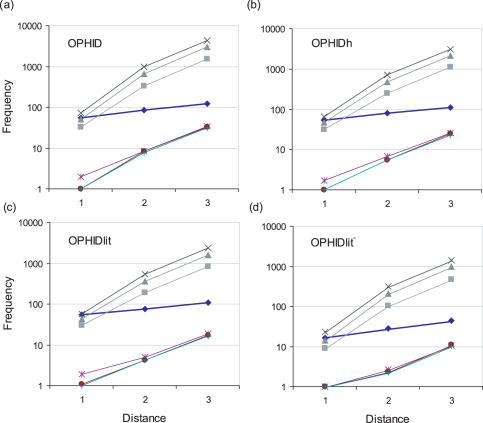
Figure 2 shows the number of false positives returned by the interaction data at increasing path lengths up to a distance of three interactions from the known disease genes. As the shortest path length increases, the sensitivity improves, but the number of false positives increases exponentially and reduces the specificity. At a distance of two interactions, the full OPHID set finds 84 disease genes with a sensitivity of 0.49, a specificity of 0.96 and an enrichment of 11-fold. Increasing the distance to three interactions, finds 123 disease genes, with a high sensitivity of 0.72, but a smaller specificity of 0.82 and a poor 4-fold enrichment.
Figure 2.
Performance of PPI data from (a) OPHID, (b) OPHIDh, (c) OPHIDlit+ and (d) OPHIDlit−. Results are shown for three levels of interaction using the shortest path length to a disease gene (Distance). Black diamonds represent the number of disease genes found. The number of non-disease genes returned are presented for the 50 gene interval (square), 100 gene interval (triangle) and 150 gene interval (x). The number of disease genes returned by random selection are presented for the 50 gene interval (*), 100 gene interval (circle) and 150 gene interval (+).
Combining the results from the full OPHID set (where the shortest path length is one) with the results from BioCarta and KEGG, CPS correctly identifies 78 disease genes for 20 diseases. Overall CPS performance has a sensitivity of 0.47 with a specificity of 0.98 and an enrichment of 17-fold at the 100 gene interval size. Less than 0.6% of proteins rejected will be disease genes.
CMP benchmark performance: CMP identifies disease genes using domain-based comparative sequence analysis. This is achieved by first using Pfam Hidden Markov models to annotate the domain content of known disease genes. Putative disease genes are then identified based on a shared domain content with the known disease genes. Several score thresholds were tested: the ratio of true positives to false positives is best at a threshold of 0.4. However, at a threshold of 0.1, CMP finds more disease genes and sensitivity is at its best. At this threshold, 7.5, 11.6 and 18.5% of predictions are disease-causing genes for the 50, 100 and 150 gene intervals, respectively. Less than 0.8% of proteins rejected will be disease genes.
Independently, CMP correctly predicts 32 disease genes for 10 diseases at a score threshold of 0.1 and has a sensitivity of 0.2 and a specificity of 0.98 for each interval size. Overall enrichment for all diseases was 11-fold at the 100 gene interval size.
Multiple interval input
For poorly characterized diseases no disease genes may have been identified, but several loci may have been isolated. In this case, a multiple interval comparison implementation of the two methods can be used which allows ab initio prediction of disease genes.
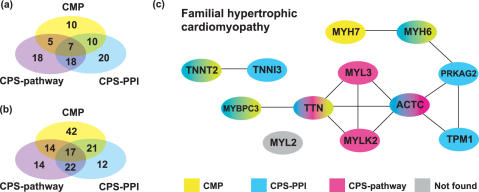
CPS benchmark performance: When multiple loci are used as the input to CPS the system found common pathways or PPIs for 100 disease genes in the 100 gene intervals. While sensitivity is high 0.59, more false positives are predicted compared to input from known disease genes. False positives occur because common pathway and PPIs are found for non-disease genes in intervals associated with each phenotype. This reduces specificity to 0.84 and the ER to 3.7-fold. The pathway and PPI data complement each other: CPS using pathway data alone finds 28 disease genes that are missed by the PPI data. Conversely, CPS using PPI data alone finds 33 disease genes that the pathway data misses and together they find the same 39 disease genes (Figure 3). In the absence of known disease genes, the use of network data on multiple disease loci is a powerful approach to identify disease genes. Table 2 shows the results for each of the individual methods.
Figure 3.
Combined prediction success. (a) Correct predictions based on known disease genes. (b) Correct predictions based on multiple intervals. (c) Combined CPS and CMP predictions for familial hypertrophic cardiomyopathy using known disease genes. Disease genes are represented by their HUGO-name. Gene-linking lines are predictions by CPS and CMP. For example, TNNT2 is found by the known disease gene TNNI3 using CPS-PPI and CMP predictions, and TNNI3 is found by the known disease gene TNNT2 using CPS-PPI predictions. PRKAG2 and TPM1 were found using PPI data at a distance of three, all other PPI predictions are at a distance of one.
Table 2.
Multiple interval benchmark results
| Method | 50 | 100 | 150 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | ER | Sensitivity | Specificity | ER | Sensitivity | Specificity | ER | |
| CPS-pathway | 0.35 | 0.90 | 3.4 | 0.39 | 0.89 | 3.4 | 0.41 | 0.88 | 3.2 |
| CPS-PPI | 0.39 | 0.95 | 7.3 | 0.42 | 0.93 | 6.1 | 0.47 | 0.92 | 5.6 |
| CPS | 0.54 | 0.87 | 4.0 | 0.59 | 0.84 | 3.7 | 0.62 | 0.82 | 3.5 |
| CMP ( multi) | 0.17 | 0.95 | 3.3 | 0.19 | 0.94 | 3.1 | 0.23 | 0.93 | 3.2 |
| CMP ( all) | 0.46 | 0.77 | 1.9 | 0.55 | 0.72 | 1.9 | 0.59 | 0.69 | 1.9 |
| CMP ( multi) | 0.16 | 0.95 | 3.2 | 0.18 | 0.94 | 3.1 | 0.22 | 0.94 | 3.3 |
| CMP ( all) | 0.46 | 0.77 | 2.0 | 0.55 | 0.72 | 1.9 | 0.58 | 0.69 | 1.9 |
| CPS-CMP ( all) | 0.74 | 0.69 | 2.3 | 0.84 | 0.63 | 2.2 | 0.87 | 0.59 | 2.1 |
, significance based on the assumption that domains in a gene are uncorrelated; , significance based on the assumption that domains in a gene are correlated; multi, genes that contain multiple Pfam domains only; all, genes that contain at least one Pfam domain. All χ2 tests are at a significance level of 0.995.
CMP benchmark performance: When multiple loci are used as the input to CMP, a census of the domain content of all genes in the specified loci is taken. The aim is to search for domain combinations that are over-represented in the loci associated with the phenotype. The tally of genes with a specific domain content is compared with the number of genes expected by chance based on the prevalence of those domains in the genome (see Materials and Methods). Clusters of genes with similar domain content are ranked based on two estimates of significance: the first assumes that the domain content of the cluster is completely uncorrelated and is an upper estimate of the significance (); the second assumes the domains are highly correlated and the prevalence is determined by the rarest domain (). These two values are the same for single domain proteins.
Comparison of the CMP results are shown in Table 2. Results have been split into subgroups: those that contain multiple Pfam domains (multi) and those that contain at least one Pfam domain (all). Sensitivity is low for the multi-domain method because disease genes with zero or one Pfam domain are included in the false negatives. However, the specificity is very high indicating that if the target disease genes are multiple domain proteins, the method is very effective.
As the method is essentially the same as CMP using known disease genes correct predictions are fairly similar. The 36 disease genes potentially identifiable by CMP, based on their domain similarity, can be divided into 16 clusters, containing two or more disease genes. Of these genes, 32 were identified by CMP using known disease genes as a starting point, while four fell below the 0.1 threshold similarity. Using multiple intervals as input, two clusters containing four genes were not found as determined by significance. For example, genes RET and NTRK1 involved in thyroid carcinoma have a protein kinase domain in common, but protein kinase domains are very common in the genome and thus lowered the significance of the shared domain.
Of the 14 successfully identified gene clusters, 11 were ranked in the top 10 for that disease based on either score of significance and 13 were in the top 20. The test favours multi-domain proteins whereas disease genes that are single domain proteins have a better chance of being detected with .
Success of combined methods
While both methods successfully identify disease-causing genes, performance is improved when the methods are combined. The methods tend to be complementary, with one finding disease genes where the other method fails (Figure 3).
An example of the success of the combined methods can be seen for familial hypertrophic cardiomyopathy (Figure 3c). For the 12 known disease genes, 9 were found by the standard implementation of CPS and CMP and a further 2 genes were found by CPS-PPI data using a distance of three interactions to a known disease gene. Both CPS-PPI data and CMP identify disease genes through relationships between titin (TTN) and myosin-binding protein C (MYBPC3), and between troponin I type 3 (TNNI3) and troponin T2 (TNNT2). CMP exclusively links disease genes myosin heavy polypeptide 6 (MYH6) and myosin heavy polypeptide 7 (MYH7). The CPS-pathway-data from KEGG links actin (ACTC), myosin light polypeptide kinase 2 (MYLK2), myosin light polypeptide 3 (MYL3) and titin through the ‘regulation of actin cytoskeleton’ pathway.
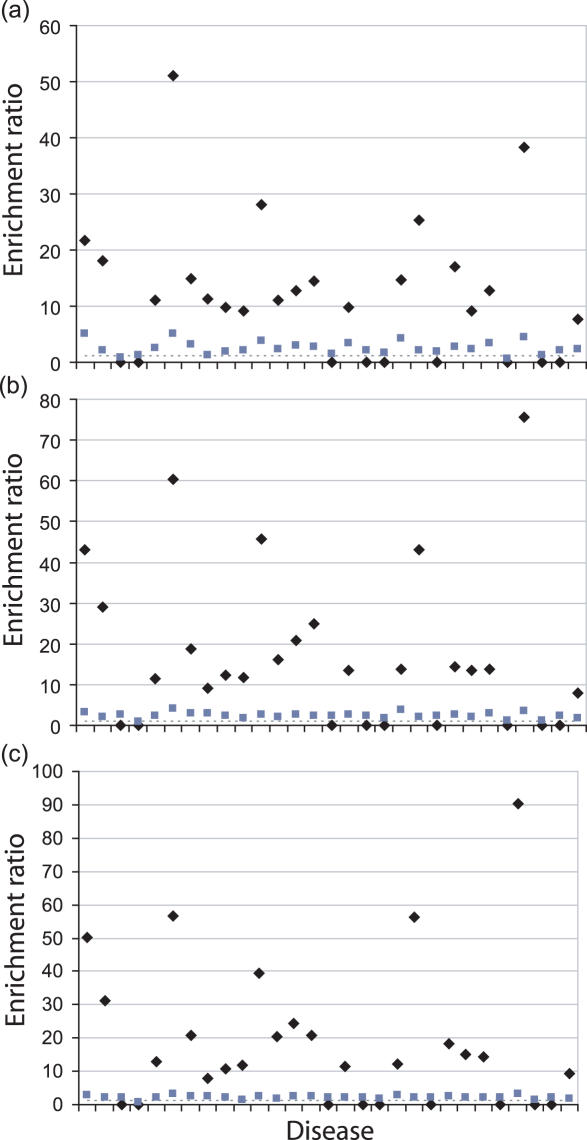
The probability of finding a disease gene increases when combining the results from the two methods: sensitivity increases to 0.51 with a specificity of 0.97 for the 50, 100 and 150 gene intervals when using known disease genes as input. Of the rejected genes, only 0.5% will be disease genes. Overall enrichment is 11-fold in the 50 gene interval and 13-fold in the 100 and 150 gene intervals. Figure 4 shows the enrichment scores for each disease using the combined methodology. The combined methods are only worse than random when no correct predictions are made.
Figure 4.
Candidate gene enrichment for the 50 (a), 100 (b) and 150 (c) gene interval sizes using the combined methods. Enrichment values are on the y-axis and diseases are listed alphabetically from left to right on the x-axis, as in Table 1. Black diamonds represent enrichment of data using known disease genes. Grey squares represent enrichment of data using multiple intervals. The dashed line represents data enrichment by random selection.
Removing the literature-derived PPI data, but still using known disease genes, only slightly reduces overall performance: sensitivity is 0.42, specificity is 0.97 and enrichment is 11-fold at the 100 gene interval. When extending the OPHID interaction data to the second level of interaction, overall sensitivity increases to 0.59, but with a reduction in both specificity, 0.93, and enrichment, 8-fold, for each interval size.
For the combined multiple interval predictions at the 100 gene interval size, sensitivity greatly improves to 0.84, however, the increase in false positives from CMP ( all) causes specificity and enrichment to fall to 0.63 and 2.2-fold, respectively.
Failed predictions
While our methods found disease genes for most of the diseases, all methods failed for nine diseases when using known disease genes as input (Table 1). For six of these diseases, disease genes are correctly predicted when the PPI interaction data are extended to a distance of two and three interactions to the nearest known disease gene. This leaves three diseases without successful predictions: Bardet-Biedl syndrome; juvenile-onset primary open angle glaucoma and Leber congenital amaurosis. Each of the genes involved in these diseases have distinct Pfam domains. For this reason they cannot be identified by the CMP method. CPS fails because interaction and pathway data are not available for these genes. However, it is likely that these genes perform their functions as part of the same biochemical pathway. This has recently become apparent for Bardet-Biedl syndrome where defects in ciliary proteins have been delineated (39). Our current knowledge of pathways is incomplete, but as more data becomes available predictions will improve.
DISCUSSION
Two methods for candidate disease gene prediction have been presented in this study. CPS hypothesizes that novel disease genes reside in the same pathways as those of known disease genes and CMP assumes that novel disease-causing genes that produce the same phenotype as known disease genes are likely to have similar functions. The genes in the novel interval of interest are then tested for relationships to known disease genes or genes in other characterized disease intervals. Both CPS and CMP can effectively recover known disease genes for a broad array of diseases.
Many previous candidate gene prediction methods have relied on functional annotation, such as GO terms, which can be general or absent. Only 25% of human proteins have manually annotated GO terms. Many more human proteins have predicted annotations, but 35% have no annotation at all. Furthermore, these systems will be biased to well studied and well annotated diseases and may not be useful in the analysis of uncharacterized diseases.
Our methods are based directly on biological data, and differ from earlier candidate gene prediction techniques, which use blanket systems based on descriptive keywords to cover all aspects of disease. Such methods include POCUS (3), G2D (4,5) and SUSPECTS (10). New systems biology approaches to candidate gene prediction, which are based directly on biological data, mine PPI and pathway databases. Those described by Franke et al. (18) and Oti et al. (17) as well as our own CPS fall into this category. Our CMP method is quite different to any other method described previously, in that it tries to associate particular protein modules with specific diseases. Not only does this technique represent a more powerful way of finding homologs than BLAST searches but it also has the potential to find otherwise unrelated proteins that engage in homophilic interactions (e.g. through EGF domains) or share a common functional unit but are otherwise unrelated, e.g. the protein kinase domains found in thyroid carcinoma.
Comparison with other methods is difficult as benchmark datasets are different and some methods merely rank candidates without applying a cut-off. In an attempt to fairly assess our methods compared to others, we have used the disease set as applied in the analysis of POCUS. Turner et al. (3) previously compared other methods against POCUS by calculating and comparing ERs: van Driel et al. (11) studied eight diseases and reduced an average 163 genes to 22, producing a 7-fold enrichment. Freudenberg and Propping (12) found two-third of the disease genes in the top 15% of candidates, giving a 7-fold enrichment. Generally, these keyword methods have been shown to provide a 7- to 10-fold enrichment (3). The updated G2D method is the most successful of these methods, correctly identifying disease genes for 47% of diseases within their ranked top eight predictions, which is below our performance. Using known disease genes as input, we correctly predicted disease genes for 69% of diseases with an average success rate of one in seven (14%) gene predictions and a 13-fold enrichment. A 13-fold enrichment prunes a list of 100 gene candidates to just eight genes, and significantly reduces the time and cost of experimental studies.
There are three other methods, POCUS, PRIORITIZER (18) and the method by Tiffin et al. (8), that attempt the more ambitious task of ab initio predictions in the absence of known disease genes. The ability to perform this task is particularly useful for phenotypes were no disease genes are known or where the known disease genes account for only a small percentage of cases presenting with the disease. While POCUS makes very few predictions, for the eight diseases that it does make predictions (28%), the quality of prediction is high with a one in four success rate and 23-fold enrichment. The PRIORITIZER method by Franke et al. (18) correctly identified disease genes for 64% of diseases with a success rate of one in eight predictions and a 2.8-fold enrichment. The method by Tiffin et al. correctly identified disease genes for 88% of diseases with a 1.6-fold enrichment. Our combined methods make correct predictions for all diseases with a 2.2-fold enrichment. Another consideration when comparing these results is the range of interval sizes used in the benchmark. POCUS used intervals based on keyword densities and sizes ranged from 2 to 19 Mb, which are small and more typical of monogenic diseases. Franke et al. (18) used intervals of 50, 100 and 150 genes, but only included those genes that had predicted interactions. Our benchmark intervals range from 50 genes (from 1 Mb) to 150 genes (up to 51 Mb). The larger interval sizes are realistic for complex diseases (40) and include all genes.
Our side-by-side use of two prediction systems based directly on independent biological data shows the value of this approach. Recently several prediction systems were benchmarked against each other using obesity and type 2 diabetes phenotypes (14). A meta-analysis was then used to choose the best candidates based on consensus. The complementarity of data predicted by our two systems (Figure 3) show that a consensus method is not always appropriate. Had we used this approach far fewer disease genes would have been found. Clearly the independence of data sources needs to be considered before applying consensus approaches. On the other hand, the type of relationships flagged by CMP is clearly related to pathway data. Pathways may expand by gene duplication and subsequent specialization of the daughters, possibly in association with discrete tissue expression. Similarly, protein complexes consisting of homo-oligomers may differentiate by duplication and specialization of genes encoding similar subunits. If pathway and interaction data were comprehensive then the alternative predictions provided by CMP may not be necessary, but clearly this is not yet the case.
Given that several systems biology approaches have now been published, it is worthwhile examining the caveats associated with these methodologies. CPS with PPI data alone found the majority of disease genes in the benchmark tests. But, some of the interaction data is likely to be dubious, because high-throughput experiments, such as yeast two-hybrid and TAP systems will associate proteins that would otherwise never be present in the same cell or subcellular compartment (41). Furthermore, the various PPIs curated from computational searches of the literature have limited overlap with each other (38), which may be indicative of a high false positive rate. While there is strong evidence to suggest that PPIs are conserved through evolution (42), errors in the source data will perpetuate through the databases. These caveats make predicted interactions, such as the Bayesian approach applied by Franke et al. (18), inaccurate. As more evidence for PPIs are collected, the performance of CPS and other similar methods will improve. The results using PPI data alone are already very encouraging: the full OPHID dataset enriches the candidate list by 50-fold, far better than any other reported method.
Finally, our methods were able to make predictions for both Mendelian and complex diseases. Identifying the disease genes for complex diseases is inherently difficult. Our methods scored well on our test set, but we intend to further investigate the performance on a larger dataset of diseases. In addition, although some of the predicted disease genes are not currently known to be involved in the disease, which are counted as false positives in this study, it is possible that they may be uncharacterized disease genes. Our benchmark results are available at our web site, www.gentrepid.org, for further analysis. Our methods are also available to identify potential disease genes in user-specified intervals.
A new era of genomics and bioinformatics has permitted a genome-scale perspective of disease and is enabling new technologies to identify disease-causing systems. Our methods will accelerate the disease gene discovery process by gathering and sifting through all knowledge of each candidate gene including its homologs and interaction partners. In addition, it will significantly reduce the cost of expensive experimental studies. Identification of the disease gene enables targeted research on how mutations in the gene contribute to disease and provides specific leads towards cures. The results presented here are better than other reported methods for disease gene prediction. CPS and CMP utilize information from protein sequence and interaction databases, enabling accurate disease gene identification. In the multiple interval input mode, our methods do not require a priori knowledge of the disease or disease genes. They will, therefore, be a powerful tool in candidate disease gene prediction for poorly characterized diseases.
Acknowledgments
The authors wish to thank Michael Wouters and Robyn Otway for helpful discussions. We acknowledge funding from the University of New South Wales, The Ronald Geoffrey Arnott Foundation and St Vincent's Clinic Foundation. D.F. acknowledges the Sylvia and Charles Viertel Charitable Foundation and the NHMRC for support. Funding to pay the Open Access publication charges for this article was provided by the Victor Chang Cardiac Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rudd M.F., Webb E.L., Matakidou A., Sellick G.S., Williams R.D., Bridle H., Eisen T., Houlston R.S. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006;16:693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth D.J., Cooper J.D., Bailey R., Field S., Burren O., Smink L.J., Guja C., Ionescu-Tirgoviste C., Widmer B., Dunger D.B., et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 3.Turner F.S., Clutterbuck D.R., Semple C.A. POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol. 2003;4:R75. doi: 10.1186/gb-2003-4-11-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Iratxeta C., Bork P., Andrade M.A. Association of genes to genetically inherited diseases using data mining. Nature Genet. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Iratxeta C., Wjst M., Bork P., Andrade M.A. G2D: a tool for mining genes associated with disease. BMC Genet. 2005;6:45. doi: 10.1186/1471-2156-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiffin N., Kelso J.F., Powell A.R., Pan H., Bajic V.B., Hide W.A. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res. 2005;33:1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelso J., Visagie J., Theiler G., Christoffels A., Bardien S., Smedley D., Otgaar D., Greyling G., Jongeneel C.V., McCarthy M.I., et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adie E.A., Adams R.R., Evans K.L., Porteous D.J., Pickard B.S. SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics. 2006;22:773–774. doi: 10.1093/bioinformatics/btk031. [DOI] [PubMed] [Google Scholar]
- 11.van Driel M.A., Cuelenaere K., Kemmeren P.P., Leunissen J.A., Brunner H.G. A new web-based data mining tool for the identification of candidate genes for human genetic disorders. Eur. J. Hum. Genet. 2003;11:57–63. doi: 10.1038/sj.ejhg.5200918. [DOI] [PubMed] [Google Scholar]
- 12.Freudenberg J., Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18:S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 13.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiffin N., Adie E., Turner F., Brunner H.G., van Driel M.A., Oti M., Lopez-Bigas N., Ouzounis C., Perez-Iratxeta C., Andrade-Navarro M.A., et al. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badano J.L., Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature Rev. Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 16.Gandhi T.K., Zhong J., Mathivanan S., Karthick L., Chandrika K.N., Mohan S.S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- 17.Oti M., Snel B., Huynen M.A., Brunner H.G. Predicting disease genes using protein-protein interactions. J. Med. Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke L., Bakel H., Fokkens L., de Jong E.D., Egmont-Petersen M., Wijmenga C. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez-Sanchez G., Childs B., Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- 20.George R.A., Spriggs R.V., Bartlett G.J., Gutteridge A., MacArthur M.W., Porter C.T., Al-Lazikani B., Thornton J.M., Swindells M.B. Effective function annotation through catalytic residue conservation. Proc. Natl Acad. Sci. USA. 2005;102:12299–122304. doi: 10.1073/pnas.0504833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George R.A., Heringa J. Protein domain identification and improved sequence similarity searching using PSI-BLAST. Proteins Struc. Func. Genet. 2002;48:672–681. doi: 10.1002/prot.10175. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader G.D., Cary M.P., Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown K.R., Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 29.Bader G.D., Betel D., Hogue C.W. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 31.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones R.B., Gordus A., Krall J.A., MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 33.Ingham R.J., Colwill K., Howard C., Dettwiler S., Lim C.S., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 38.Ramani A.K., Bunescu R.C., Mooney R.J., Marcotte E.M. Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The Ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy M.I., Smedley D., Hide W. New methods for finding disease-susceptibility genes: impact and potential. Genome Biol. 2003;4:119. doi: 10.1186/gb-2003-4-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 42.Bandyopadhyay S., Sharan R., Ideker T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 2006;16:428–435. doi: 10.1101/gr.4526006. [DOI] [PMC free article] [PubMed] [Google Scholar]