Abstract
The Drosophila non-long terminal repeat (non-LTR) retrotransposons TART and HeT-A specifically retrotranspose to chromosome ends to maintain Drosophila telomeric DNA. Relatively little is known, though, about the regulation of their expression and their retrotransposition to telomeres. We have used rapid amplification of cDNA ends (RACE) to identify multiple transcription initiation and polyadenylation sites for sense and antisense transcripts of three subfamilies of TART elements in Drosophila melanogaster. These results are consistent with the production of an array of TART transcripts. In contrast to other Drosophila non-LTR elements, a major initiation site for sense transcripts was mapped near the 3′ end of the TART 5′-untranslated region (5′-UTR), rather than at the start of the 5′-UTR. A sequence overlapping this sense start site contains a good match to an initiator consensus for the transcription start sites of Drosophila LTR retrotransposons. Interestingly, analysis of 5′ RACE products for antisense transcripts and the GenBank EST database revealed that TART antisense transcripts contain multiple introns. Our results highlight differences between transcription of TART and of other Drosophila non-LTR elements and they provide a foundation for testing the relationship between exceptional aspects of TART transcription and TART's specialized role at telomeres.
INTRODUCTION
The TART and HeT-A families of Drosophila non-long terminal repeat (non-LTR) retrotransposons transpose specifically to the chromosome termini, presumably by using the 3′ hydroxyl at the chromosome terminus as a primer for target-primed reverse transcription (TPRT) (1–3). They are thought to provide an essential role in maintaining telomeric DNA and are the only retrotransposons that have been identified as having a beneficial role for the cells in which they reside. As a result of successive retrotranspositions, the termini of most Drosophila chromosomes are composed of tandem head-to-tail arrays of HeT-A and/or TART elements with the 5′ end of each element oriented towards the terminus (1,2). In contrast, short DNA repeats maintained by telomerase are present at telomeres in most other eukaryotes (4). Extension of telomeres by both telomerase and retrotransposition involves reverse transcription (RT) of an RNA template, but these RNA templates are very different in length. A recent detailed analysis of six telomeres from one Drosophila melanogaster strain identified a third family of telomeric non-LTR retrotransposon, TAHRE, for which only a single complete copy was described (5).
The longest known copies of TART and TAHRE have two open reading frames (ORFs), ORF1 and ORF2, separated by a short spacer (5,6). The predicted TART ORF2 protein contains putative reverse transcriptase and endonuclease domains (3,6,7), and the predicted TART ORF1 protein has a cluster of three CCHC-type zinc knuckles (6), as do proteins encoded by many other non-LTR retrotransposons. HeT-A elements do not encode reverse transcriptase and their single ORF codes for a protein with zinc-knuckle motifs that can associate with telomeres when overexpressed in cultured Drosophila cells (2). All three families of telomeric retrotransposons have unusually long 3′-untranslated regions (3′-UTRs) (2,3,5,6) and TART contains a direct repeat of the 5′-UTR and beginning of ORF1 at a subterminal location in the 3′-UTR (6). Three subfamilies of TART have been described in D.melanogaster, called TART-A, TART-B and TART-C [(6); F.Sheen, L.Tolar, J.Stolk and R.Levis, unpublished data]. The nucleotide sequences of both ORFs are >90% identical in pairwise alignments of elements from any of the three subfamilies, but the lengths and sequences of their 3′-UTRs have diverged.
Most non-LTR retrotransposons that have been examined are transcribed from internal promoters that are positioned near the very 5′ ends of the elements (8–17). Transcripts initiating at the 5′ ends serve as templates for RT of full-length copies of these elements. The presence of TART, HeT-A and TAHRE at telomeres may present special challenges for their expression. DNA is lost from the termini of Drosophila chromosomes at a rate of ∼75 bp per fly generation, presumably due to incomplete replication, regardless of whether there are TART or HeT-A elements at the termini (1–3). Because of the invariant orientation of these transposons at the telomeres, incomplete replication of telomeric DNA leads to the progressive loss of DNA from the 5′ ends of TART, HeT-A or TAHRE elements exposed at telomeres. If the only promoter was located near the 5′ end, the expression of the element and its ability to spawn new copies would be eliminated soon after it transposed to the terminus. For HeT-A, transcription initiates in the 3′ end of one element and reads through an adjacent downstream element to generate an RNA template for RT (18). The unusual location of this promoter could be advantageous by protecting it from frequent loss. Northern analyses of TART have shown that TART produces multiple sense and antisense transcripts, including transcripts approximately the same size as genomic copies of TART (19). In the same study, it was reported that, in unpublished preliminary experiments, a 900 bp fragment from the 3′ end of either TART-A or TART-B had antisense, but not sense promoter activity, in an assay in which lacZ reporter constructs were transiently transfected into cultured Drosophila cells (19).
We mapped transcription initiation and polyadenylation sites for the three TART subfamilies in D.melanogaster. Initiation of sense and antisense transcription occurs at different sites in the 3′ half of the direct repeats in TART and we identified initiation and polyadenylation sites for putative subgenomic RNAs. As a result of our mapping, we also found that TART antisense transcripts contain multiple introns.
MATERIALS AND METHODS
Fly strains and cultured cells
D.melanogaster wild-type strains Oregon-R and Mk-G(II)12 were used in this study. Mk-G(II)12 was originally designated as Chepachet 74i, since the initial population was collected by Margaret Kidwell in Chepachet, Rhode Island in 1974 and was obtained from William Engels (University of Wisconsin, Madison). Flies were raised at room temperature on standard cornmeal medium supplemented with live yeast.
Schneider2 (S2) cells were obtained from Invitrogen and were grown in Ultimate Insect Serum-Free Medium (Invitrogen) at 25°C.
Rapid amplification of cDNA ends (RACE) analysis
Total RNA samples were prepared from S2 cells, wandering third instar larvae and adults (one to several days after eclosion) using the RNAqueous kit (Ambion). Approximately 5 × 106 S2 cells were pelleted, washed in 1 ml of phosphate-buffered saline (PBS) and vortexed in 300 μl of lysis/binding solution (RNAqueous kit, Ambion) to disrupt cells. Larvae and adults were ground by hand in a ground glass homogenizer in lysis/binding solution (30–40 larvae or adults in 300 μl or 80–90 larvae or adults in 600 μl) and homogenates were transferred to 1.5 ml polypropylene microcentrifuge tubes. After a clarifying spin for 2 min at maximum speed in a microcentrifuge, homogenates were carried through the RNAqueous protocol. Eluted RNA was treated with RNAsecure (Ambion) at 60°C for 10–20 min to inactivate any RNases. Samples were then heated at 95–96°C for 6 min (to denature any DNA:RNA hybrids) and digested with 2 U of DNase I (DNA-free kit, Ambion) at 37°C for 1 h. DNase I was removed using DNase I inactivation reagent (DNA-free kit, Ambion) following the manufacturer's recommendations.
The 5′ and 3′ RACE analyses were performed using reagents in the First Choice™ RLM-RACE kit (Ambion), following the general guidelines provided with the kit. Briefly, for 5′ RACE, total RNA (10 μg) was treated with calf intestinal phosphatase at 37°C for 1 h, followed by phenol extraction and ethanol precipitation. Samples were then treated with tobacco acid pyrophosphatase (TAP) at 37°C for 1 h to decap mRNA. An RNA adapter was ligated to the exposed 5′ phosphates using 5 U of T4 RNA ligase at 37°C for 1 h and samples were reverse transcribed using random decamers and MMLV–RT at 52°C for 1 h. Control samples that received mock treatment with TAP (incubation in 1× TAP buffer without TAP enzyme) were processed in parallel with test samples. For 3′ RACE, up to 1.5 μg of total RNA was reverse transcribed with the provided oligo(dT) adapter and MMLV–RT at 50°C for 1 h. Control reactions for 3′ RACE were mock reverse transcribed in parallel with test samples.
One microliter of each 20 μl RT reaction was used as a template for nested PCR with nested TART-specific and adapter-specific primer pairs (in a few cases only a single round of PCR was performed). See Table 1 for a list of TART primers. PCR was performed using HotStarTaq (QIAGEN) according to the manufacturer's guidelines. Control and test RT reactions for each RNA source were amplified in duplicate with each primer pair. Reproducible products specific to the test samples were gel-purified, cloned into pCR2.1 (Invitrogen) or pGEM-T Easy Vector (Promega) and sequenced (BioResource Center, Cornell University, Ithaca, NY).
Table 1.
Sequences of primers
| Primer | Sequence (5′–3′) |
|---|---|
| ADR1a | GAGAATGGTCTCACTTGTACCAAGC |
| ADR2a | TTGTAACGTCGCTGCTGCGGAGAT |
| BDR1b | TCCGTTAGCCGGTATTACTGGTCA |
| BDR2b | TTCAGCGTCGCTGCACATCTCG |
| BFRb | AGCGGCAGTTGTGCGCGCTTTAGTTAGAG |
| HR1d | AGGTTGTCGGACATGGACATG |
| HR2d | CGTTTTGTTATGGCGGCAGCTTGTC |
| T31 | ATCCCTACCCAAAACCCACCCAC |
| TA3 | CTTGAACAATGGCGCGCAAAACTG |
| TA31 | CGGTTTTTGATGCTTTGGTTGCA |
| TA51 | CTAGAAAGAAGCAGGCAGAGGTAC |
| TA52 | AACTCCCCACCCTGCTGAGACTAG |
| TA53 | ACAAGCCCGCCGACAACTAGCA |
| TA54 | CCAACACCCATCTCACTCATTTC |
| TA55 | GTTGTACCCAAGGCTGACGGCA |
| TA56 | GGAAACGGGTATCGAAGGATC |
| TA57 | ATGCTCTCAACATACGCAGACGACA |
| TAA1a | GTAACATCAGTCAGACTGTACTCAC |
| TAA2a | CTCCAACACATGTTTTCCTCCA |
| TAB1b | CTCCGACTCCTTACTTGACCACTG |
| TAB2b | GAGATGTGCAGCGACGCTGAAAGC |
| TAB3b | TCCTGGCGAGATTGTTAAGGCATC |
| TAB4b | ATCTGCTACTGCGCCTTGCGTC |
| TAC3c | GAGCAACGACACTGAAAGCAAC |
| TAC4c | ATGCTAGAACATCGAGTACCAGAC |
| TAD1a | TCGTCGTCATCTCCGTCGCACA |
| TAD2a | TGGCTGCTGCTGGTGAGTGTGT |
| TAR1a | AGGTATAGTTGAGCGATGACTTG |
| TAR2a | TTGCGCGCCATTGTTCAAGTGTAG |
| TBD1b | CAGGATCATCGTCGTCGTACAG |
| TBD2b | CTGATGTTGCTGCTATCCTAGCTG |
| TBIN1b | CAACTGCCGCTGATCCGAAATTG |
| TBR1b | TTCGGATCAGCGGCAGTTGTGC |
| TBR2b | TGGCGCAGGAATCCAGCGATGA |
| TC3c | ACTATGGCGCGCAAAATGACTAC |
| TCR1c | GGTTAACTTGGCTGCGGTTATGC |
| TCR2c | GTAGTCATTTTGCGCGCCATAG |
| TR1 | ATGTGTTAGTGGCGAGGAAGTGCT |
| TR1R | GGTGGTTTTCTGCTAGCTGGAACAG |
| TR2 | CATTGGCTTCTGTGGTATTAGCAGT |
| TR2F | CCAATATGGCAGTATGCCATAGCAGC |
| TR3 | GTTGGCGTGGTGTCCAGCGATGA |
| TR4 | GAGTGTCTTGGCTGCTATGCTTGT |
| TR5 | GCGGGCTGTGTTGGCTTGGTTTC |
| TR6 | GCTGCGTCTGTTGTTTTCAGGCA |
| TR7 | TTGGGCTAAGTATGTGAAATATCCTG |
| TR8 | TGTCGTCTGCGTATGTTGAGAGCAT |
| TR9 | GGTGACCCTGGCTATTTGCTGCTC |
| TR10 | GCTTCCCAGGAGTTTTAGGAGA |
| TX1a | GACCGTCATCACAAACGCTCTCA |
| TY1b | AGCAGCTACCCAAAGTGAACAC |
aTART-A specific.
bTART-B specific.
cTART-C specific.
dThese primers correspond to HeT-A sequences and are the only primers that do not correspond to TART sequences.
To determine whether 3′ RACE products resulted from internal priming at A-rich sequences, we examined the sequence from +10 to −10 relative to the putative polyadenylation site. A product was considered to have resulted from internal priming if at least six consecutive adenines or seven adenines in a 10 nt window were present, which is similar to the criteria used by other groups (20,21). Sites lacking A-rich sequences were only considered to represent true polyadenylation sites if the first position of the hexamer AAUAAA or a sequence varying from the hexamer by only 1 nt was found 10–30 nt upstream of the sites. Polyadenylation signal sequences in Drosophila have not been fully characterized, but an analysis of a large set of expressed sequence tag (EST) sequences demonstrated that ∼40% of the ESTs lacked an AAUAAA sequence and many variant hexamers with single substitutions were identified (22). Additionally, there are multiple examples of experimentally verified polyadenylation sites in Drosophila that map downstream a short distance from a hexameric sequence that differs from AAUAAA by a single nucleotide (23–25).
EST database searches and EST clone accession numbers
The public EST database available from GenBank during July 2006 was searched using default settings for standard nucleotide–nucleotide BLAST (26) and complete sequences of TART-A1, TART-B1 and TART-C1. EST clones obtained from these searches were included in our analysis of TART transcripts only if they produced a BLAST score above 200 bits. The absence of sequences corresponding to introns in EST clones corresponding to antisense TART transcripts and the exact boundaries of these introns were determined from alignments by visual inspection and manual editing of the alignments. Since a large number of clones were obtained from these searches, only a partial list of accession number for clones is provided. When appropriate, any introns supported by the sequence present in a given clone are indicated in parentheses after the accession number (also see Results and Table 4). Sequences corresponding to sense and antisense transcripts are listed separately.
TART-A1—Sense: AW940620.1, BG632572.1, BG632770.1, BG632805.1, BG639264.1, BI565296.1, BI573240.1, BI586393.1, BI591592.1, BI616292.1; Antisense: AI107163.1 (1st), AI238808.1 (1st + 2nd), AI516867.1 (1st + 2nd), BI227572.1 (1st + 2nd), AI258367.2 (2nd + 3rd), AI062230.1 (3rd), AI404467.1 (4th + 5th), AI238203.1 (4th), AI107444.1 (5th), AI238878.1 (6th), AA697502.2 (7th), AA439229.1 (8th), BE976304.1, BI606888.1, BI634958.1, BI564924.1, AI108616.2, AI135720.1, AI293246.2.
TART-B1—Sense: BE978694.1, BG639637.1, BI351764.1, CO318210.1, CO318322.1; Antisense: BI636535.1 (1st + 2nd), BI632694.1 (1st + 2nd), BG637047.1 (1st + 2nd), AI531805.1 (1st + 2nd), AA696087.1 (1st + 2nd), BI636602.1 (2nd), AI533065.1 (3rd), BI641081.1 (3rd), BG641352.1 (4th), BG635806.1 (4th), BI640910.1 (4th).
TART-C1—Sense: CK604564.1, CO316146.1, CO339374.1; Antisense: BF493305.2 (1st + 2nd + 3rd), BI369114.1 (1st + 2nd + 3rd), BG640555.1 (1st + 2nd + 3rd), AI531204.1 (1st + 2nd), BI639831.1 (2nd + 3rd), BI637557.1 (2nd + 3rd), CK604348.1 (2nd + 3rd + 4th), CO337976.1 (5th), BI638964.1 (6th).
TART sequences
We used the cloned TART-A1, TART-B1 and TART-C1 elements as prototypes for the three subfamilies of TART. The GenBank accession numbers for their sequences are AY561850, U14101 and AY600955, respectively. The nucleotide positions for sites that we refer to in the Results are relative to these sequences. The TART 3′-UTR sequences of TART-A1, TART-B1 and TART-C1 are 5.5, 3.3 and 4.5 kb, respectively and are divergent enough in sequence that their sequences cannot be reliably aligned across their entire lengths. There is 3′-UTR sequence heterogeneity even among TART elements classified within a single subfamily. For example, the 3′-UTR of the TART-A441 element is 330 bp shorter than the TART-A1 element.
RESULTS
General RACE strategy
RACE analysis was used to obtain sequence information for the 5′ and 3′ ends of TART transcripts using total RNA from five sources: third instar larvae and adults of two wild-type D.melanogaster strains [Oregon-R and Mk-G(II)12] and the D.melanogaster S2 cultured cell line. Each reaction mapped the 5′ or 3′ ends within a specific segment of TART. We were able to design primers that corresponded to TART ORF sequences that would anneal to the transcripts of all three subfamilies. However, we designed subfamily-specific primers that corresponded to TART UTR sequences (see Table 1).
5′ and 3′ RACE products from multiple regions of TART
We assigned arbitrary designations to each of the five 5′ and five 3′ ends that we mapped (5a–5e and 3a–3e, Figures 1 and 2). All 5′ and 3′ ends were mapped using RNA from all five sources with the following three exceptions: ends 5b, 5e and 3a were only mapped using S2 cell RNA, Mk-G(II)12 adult RNA and Mk-G(II)12 larval and adult RNA, respectively. Transcripts for all three TART subfamilies were mapped for Mk-G(II)12 and Oregon-R RNA samples, but we were unable to map any ends for TART-A in S2 cells.
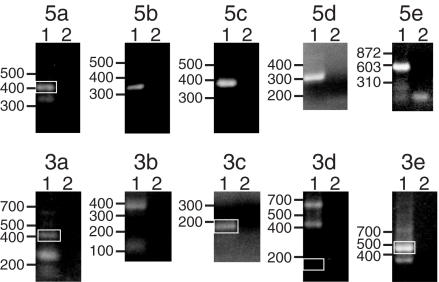
Figure 1.
Representative 5′ and 3′ RACE reactions. Each panel is an ethidium bromide stained agarose gel of RACE reactions. Above each panel is a 5′ end or 3′ end designation (5a–5e or 3a–3e, respectively) used for discussion purposes. Lanes 1 and 2 are the experimental and control lanes, respectively. The sources of RNA for the reactions shown were Oregon-R adults (5a, 5c, 5d, 3c, 3d and 3e), S2 cells (5b and 3b), Mk-G(II)12 adults (5e) or Mk-G(II)12 third instar larvae (3a). The TART primers used for the reactions shown were as follows (both outer and inner primers are listed for reactions in which two rounds of PCR with nested primers were used): 5a: TR1 + TR2; 5b: TR6 + TR7; 5c: TCR1 + TCR2; 5d: TAB1; 5e: ADR1 + ADR2; 3a: TA53 + TA54; 3b: TA51 + TA52; 3c: TA3; 3d: TR8 + TR9; and 3e: TA31. White boxes indicate products (which in some cases are very faint) that either corresponded to the major 5′ end used for sequence comparisons (5a) or that met our criteria for representing polyadenylation sites (3a, 3c, 3d and 3e), as determined by sequencing of cloned products (see Materials and Methods). The migration of DNA standards (in bp) is indicated to the left of each panel.
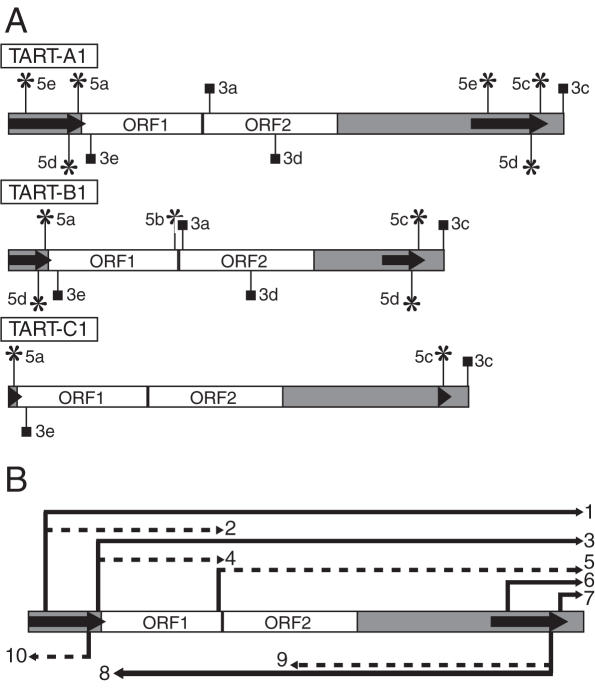
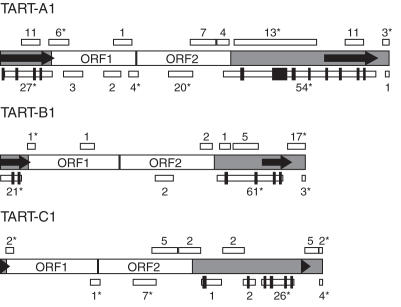
Figure 2.
Relative positions of the 5′ and 3′ ends suggested by RACE analysis and a putative transcript array for TART. Schematic representations of the positions of 5′ and 3′ ends and a putative array of TART transcripts are shown. TART ORFs are indicated by white boxes, the UTRs are indicated by gray regions and the direct repeats are indicated by arrows. The relative lengths of the UTRs correspond to those of TART-A1, -B1 and -C1, but the lengths of UTR sequences vary for other members of each subfamily. (A) Vertical lines ending in asterisks or squares indicate 5′ and 3′ ends, respectively. Each end is distinguished using designations introduced in Figure 1 that appear to the left or right of each vertical line. Those ends drawn above each element correspond to sense transcripts and those drawn below to antisense transcripts. Note that in a few cases a particular end was only identified for one of the three subfamilies (see text). (B) A generic TART element is diagrammed. Arrows above and below the element correspond to potential sense and antisense transcripts, respectively, that could be produced if all the ends shown in (A) are considered. Solid lines represent transcripts supported by previous northern blots of TART transcripts (19) and dotted lines represent transcripts not clearly supported by those same northern blots. Each transcript is numbered at its 3′ end for discussion purposes (see Discussion).
Typically, 5′ RACE reactions yielded a single product, but 3′ RACE reactions generally yielded multiple products (Figure 1). Some 3′ RACE products may result from annealing of the oligo(dT) primer to A-rich sequences at internal sites in the RNA, rather than the poly(A) tail. We therefore analyzed the sequences of multiple gel-purified products for most 3′ RACE reactions to try to distinguish sites of internal priming from polyadenylation sites (see Materials and Methods).
Sense and antisense transcription initiation occurs in the direct repeats of TART
Three of the four sense strand 5′ ends and the single antisense 5′ end identified were mapped to sites within the direct repeats of TART (Figure 2A and Table 2), suggesting an important role for the direct repeats in TART transcription. Ends 5a and 5c correspond to the same start site in the direct repeats, but were mapped to either the 5′- or 3′-UTR, respectively by using primers to unique downstream sequences. Since 5d and 5e were mapped using primers to the direct repeats, they could reflect transcription initiating in either one or both copies of the direct repeats (Figure 2A and Table 2). In contrast, 5b mapped upstream of ORF2.
Table 2.
Positions of transcription initiation identified by 5′ RACE
| TART Subfamily | Positions of 5′ endsa | ||||
|---|---|---|---|---|---|
| 5a | 5b | 5c | 5db | 5e | |
| A | 1683 (4) | No clonec | 12 861 (3) | 12 636 (4) | 413 (1) |
| 1613 (1) | 12 672 (1) | ||||
| 1685 (1) | 12 664 (1) | ||||
| 12 653 (2) | |||||
| 12 645 (2) | |||||
| 12 629 (1) | |||||
| 12 621 (1) | |||||
| B | 886 (12) | 4021 (2) | 9916 (4) | 9749 (11) | No clone |
| 885 (1) | 4140 (1) | 9768 (1) | |||
| 889 (1) | 9767 (1) | ||||
| 9746 (1) | |||||
| 9736 (1) | |||||
| C | 131 (3) | No clone | 10 513 (3) | No clone | Not testedd |
| 10 516 (1) | |||||
aPositions listed are on the sense strand and ends correspond to those diagrammed in Figure 2A. The number in parentheses indicates the number of cloned RACE products obtained corresponding to the given site. Positions in bold are those most commonly found for a given 5′ end and remaining positions are listed starting with those that are furthest upstream.
bPositions listed for 5d are from the 3′ direct repeat. The corresponding positions in the 5′ direct repeat for TART-A are 1458, 1494, 1486, 1475, 1467, 1451, 1443 and for TART-B are 719, 738, 737, 716 and 706 (the order corresponds to the order in the table).
cIndicates that no specific or reproducible 5′ RACE product was obtained.
dPrimers were not designed to test for this end in TART-C.
Three of the 5′ ends (5b, 5d and 5e) were identified only for one or two of the three TART subfamilies (Table 2 and Figure 2A). However, sequences present in the GenBank EST database suggest that antisense transcripts do initiate at a site corresponding to 5d in TART-C elements. The likely reason that we were unable to identify end 5d for TART-C is that one of the TART-C primers used to test for 5d overlaps the splice junction for an intron in TART-C antisense transcripts (see below for a discussion of TART antisense introns). The presence of introns in the antisense transcripts was not known at the time when these 5′ RACE reactions were performed. End 5e was only amplified using TART-A primers and sequences of cloned products corresponded to another cloned copy from the TART-A subfamily, TART-A441 [(3); GenBank accession no. U02279], rather than TART-A1. 5e mapped to position 6586 of TART-A441, which corresponds to positions 413 and 11 591 in the 5′ and 3′ direct repeats of TART-A1 (Table 2).
TART transcripts may be polyadenylated at multiple sites
All polyadenylation sites identified mapped to unique sequences in the ORFs or in the 3′-UTR downstream of the end of the 3′ direct repeat (Figure 2A and Table 3). Two of these ends, 3a and 3d, were only mapped for the TART-A and TART-B subfamilies (Figure 2A and Table 3). Site 3a was mapped to position 4210 of TART-B1, just downstream of an AAUAAA sequence at positions 4194–4199 and to position 4862 of TART-A1, just downstream of the sequence AACAAA at positions 4851–4856. None of the sites mapped by cloned products for 3b met our criteria for true polyadenylation sites because of A-rich sequences present in this region of TART (see Materials and Methods). Site 3c mapped to the expected polyadenylation site at the 3′ end of each subfamily. However, it should be noted that the sequence at the very 3′ end of each TART subfamily is an oligo(A) tract and that the distance from the start of the nearest polyadenylation signal sequence to site 3c was 33–36 nt. Since the oligo(A) tract at the 3′ end of TART presumably is the result of RT of TART mRNA polyadenylated at this site, we made exceptions for site 3c from our criteria for true polyadenylation sites (see Materials and Methods). Site 3d was mapped for TART-B1 at locations 14–21 nt downstream of an AAUAAA sequence (on the antisense strand) and for TART-A1 to a position 8 nt downstream of the sequence AAUAGA (on the antisense strand). Mulitple cloned products mapped 3e to a site ∼15–20 nt downstream of the sequence AAUAAA (on the antisense strand).
Table 3.
Positions of polyadenylation sites identified by 3′ RACE
| TART Subfamily | Positions of 3′ endsa | ||||
|---|---|---|---|---|---|
| 3a | 3b | 3c | 3d | 3e | |
| A | 4862 (2) | No cloneb | 13 411 (6) | 6450 (1) | 1985 (2) |
| 13 409 (1) | 1987 (1) | ||||
| 13 410 (5) | 1986 (1) | ||||
| B | 4210 (1) | No clone | 10 515 (9) | 5858 (2) | 1189 (3) |
| 10 513 (5) | 5865 (1) | 1195 (1) | |||
| 10 514 (1) | 5860 (1) | 1187 (2) | |||
| C | No clone | No clone | 11 102 (5) | No clone | 430 (2) |
| 11 001 (1) | 434 (1) | ||||
| 11 101 (1) | 432 (1) | ||||
aPositions listed are on the sense strand and ends correspond to those diagrammed in Figure 2A. The number in parentheses indicates the number of cloned RACE products obtained corresponding to the given site. Positions in bold are those most commonly found for a given 3′ end and remaining positions are listed starting with those that were furthest upstream.
bIndicates that all cloned 3′ RACE products for a given subfamily and 3′ end corresponded to sites that did not meet our criteria for potential polyadenylation sites (see Materials and Methods).
Additional 3′ end-processing signals for animals include the presence of a CA or UA dinucleotide at the cleavage and subsequent polyadenylation site, as well as U-rich sequences downstream of the poyladenylation site (22). At least one position listed for each polyadenylation site except TART-B1 3a corresponds to a CA or UA dinculeotide (Table 3). Also, U-rich sequences containing four or five uracils in a 5 nt window were present <30 nt downstream of each polyadenylation site, except for TART-B1 site 3a (data not shown).
Confirming the reliability of the 5′ RACE data
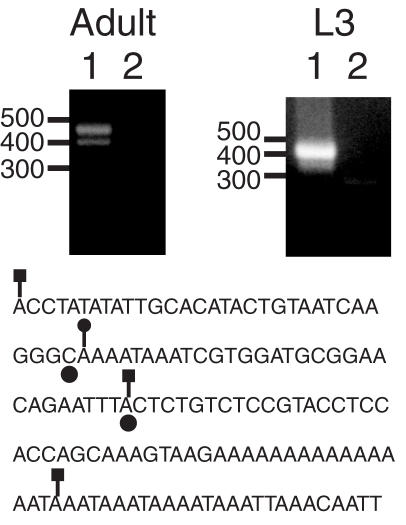
We initially expected to map a TART 5′ end within the first few nucleotides of TART sequences or in previously unidentified upstream 5′-UTR sequences, so we took steps to confirm the reliability of the 5′ RACE mapping. The results of RT–PCR experiments using different combinations of primers upstream and downstream of the site of 5a were consistent with most TART transcripts initiating at site 5a, rather than at sites further upstream in the 5′-UTR (data not shown). In addition, we used our samples and RACE method to map HeT-A transcription initiation sites. HeT-A transcription start sites were previously mapped to HeT-A 3′-UTR sequences from a second element upstream of the element being transcribed, at positions −62 and −31 [(18), the C and A denoted with circles in Figure 3]. We mapped HeT-A 5′ ends to positions −93 and −31 in the 3′-UTR using adult RNA (Figure 3, lines with squares) and to position −61 using larval RNA (Figure 3, line with a circle). The −93 site we mapped could be a site that is used in flies but not in S2 cells (the −62 and −31 sites were mapped in S2 cells) or it could be that the RACE method we used was able to map a longer 5′ end than the primer extension method used previously (18). Our ability to confirm the transcription start sites of HeT-A suggests that our 5′ RACE mapping of TART was reliable.
Figure 3.
5′ RACE confirms the location of transcription intiation in HeT-A. The two panels show ethidium bromide stained agarose gels of 5′ RACE reactions using the HeT-A outer and inner primers HR1 and HR2. The sources of RNA were Oregon-R adults or third instar larvae, as indicated. Lanes 1 and 2 correspond to the experimental and control lanes, respectively. The migration of DNA standards (in bp) is indicated to the left of each panel. Below the panels is a sequence corresponding to the largest cloned RACE product obtained from adult RNA. Positions 1–92 and 111–137 are 91/92 and 27/27 matches, respectively, to positions 6999–7090 and 7136–7162 in HeT-A 23Zn (accession no. U06920). Filled circles below two bases indicate the previously identified transcription initiation sites (18). Lines ending in squares indicate the sites corresponding to three RACE products obtained from adult RNA and a line ending in a circle indicates the site corresponding to the major RACE product from L3 RNA.
Promoter sequence motifs near putative TART transcription initiation sites
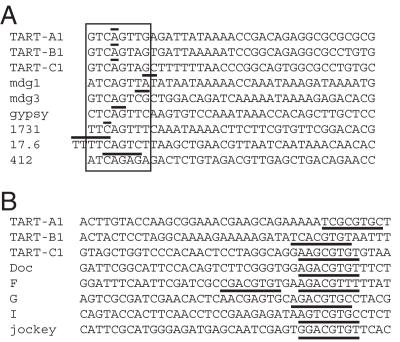
We identified good matches to an initiator consensus at end 5a (and 5c, since these are the same sites in the direct repeats). Boxed sequences for TART-A, -B and -C in Figure 4A match seven or eight positions in a consensus sequence for a Drosophila initiator, A/T/G-TCA-G/T-T-C/T-G (27). Similar sequences overlap the transcription start sites of five D.melanogaster LTR retrotransposons and the D.melanogaster retrovirus gypsy (Figure 4A).
Figure 4.
Alignment of TART transcription initiation sites with initiation sites of other Drosophila retrotransposons. (A) The 5a end of each TART subfamily is aligned with the 5′ start sites of several D.melanogaster LTR retrotransposons. An 8 nt sequence that is a close match to a consensus D.melanogaster initiator sequence, A/G/T-T-C-A-G/T-T-C/T-G (27), is boxed in all of the sequences. Experimentally determined transcription initiation sites are indicated with horizontal lines above one or more bases; for the TART sequences only the most common start site is indicated (bold positions in Table 2). Each sequence begins with the initiator consensus or the transcription initiation site. References for these sites are: mdg1, mdg3 and gyspy (40), 1731 (41), 17.6 (42) and 412 (43). (B) The 5d end of each TART subfamily is aligned with the 5′ start sites of several D.melanogaster non-LTR retrotransposons. Each alignment begins with the transcription initiation site. The corresponding sites for TART-A1, -B1 and -C1 are 12 645, 9749 and 10 133, respectively and the sequences shown are the antisense strand. As described in the text, the start site for TART-C1 end 5d was taken from an EST sequence in the GenBank database. Underlined sequences match an extended version of a downstream promoter element (13,17,44). References for the other non-LTRs are: Doc (13), F (10), G (17,45), I (11) and jockey (8).
Sequences just downstream of antisense end 5d for TART-A1, -B1 and -C1 match five or six of eight positions in a consensus downstream promoter element found near the start sites for sense strand transcription of several D.melanogaster non-LTR retrotransposons (Figure 4B, underlined sequences). The consensus is A/G-G-A/T-C-G-T-G/T-C/T (13) and it is found around positions +29 to +36 of Doc, F, G, I and jockey elements, though F elements have an additional copy closer to the transcription start site (8,10,11,13,17). The sequence shown for TART-C1 is from a clone in the GenBank EST database (accession no. AI531204.1) that starts at position 10 133 of TART-C1, since our RACE mapping did not identify end 5d for TART-C. Also, the TART-A1 sequence shown begins at position 12 645, rather than 12 636 (Table 2), since this produced a better alignment of the downstream promoter element with the other sequences (Figure 4B). Additionally, sequences at positions 12 652–12 642 of TART-A1 and 9756–9746 of TART-B1 match 9 of 11 positions in the consensus C/T-G-G-T-C-A-C-A-C-T-A/G, a common motif identified through a large-scale analysis of Drosophila promoters (28). This motif is preferentially found at positions −15 to +5 relative to the transcription start sites (28) and these sequences overlap the 5d start sites (Table 2).
No obvious matches to motifs associated with transcription start sites were seen near end 5b and only weak matches to two motifs were found near end 5e.
Analysis of ESTs containing TART sequences
We analyzed EST clones containing TART sequences that we obtained by using the full sequences of TART-A1, -B1 and -C1 to search the GenBank EST database (Figure 5). Of the 262 ESTs analyzed, 213 were annotated as either 5′ or 3′ ESTs. Of those 213 ESTs, 189 were 5′ ESTs and only 24 were 3′ ESTs. Most of the 3′ ESTs (20/24) contained TART sense strand sequences from various regions along the length of TART, even though most of the ESTs that we analyzed (179/262) contained TART antisense strand sequences. We noted two additional aspects of the set of EST sequences we obtained. First, the sites of RACE ends 5c and 5d were supported by multiple EST clones. Four TART-B ESTs (CO318210.1, CO318322.1, EC204298.1 and EC089660.1) and three TART-C ESTs (CO316146.1, EC251001.1 and EC202860.1) containing 3′-UTR sequences started precisely at sites corresponding to 5c. Twelve antisense TART-A ESTs and 36 antisense TART-B ESTs started at positions 12 619–12 662 and 9720–9753, respectively, near to the site of end 5d for each of these subfamilies (Table 2).
Figure 5.
Regions of TART sequence represented in the GenBank EST database and positions of antisense introns. TART elements are drawn as in previous figures and each drawing is to scale. White boxes above and below each drawing indicate regions of TART sense and antisense sequences, respectively represented in the EST database (note that some boxes are so small that they appear as vertical lines). A number near each box is the number of ESTs with sequences corresponding to the given region of TART. ESTs containing only direct repeat sequences were included in the totals for both the 5′ and 3′ direct repeat regions. Asterisks indicate that the number of ESTs listed includes one or more ESTs that contained sequences from non-contiguous segments of TART and that were counted as representing each of those non-contiguous segments. Black boxes (some of which look like vertical lines) that are taller than the white boxes mark the positions of intron sequences not present in many of the ESTs corresponding to antisense transcripts.
Second, the sequences present in several ESTs suggest that read-through transcription occurs from one TART element into a second TART element or from a HeT-A element into a TART element (accession nos BG632401.1, BG632770.1, BG632805.1, BI636602.1, BI565296.1, BG632827.1, BG639264.1 and BI351764.1). In the latter case, the EST sequences begin with HeT-A sequences that are within 15 nt of HeT-A transcription initiation sites [Figure 3 and (18)] and end with TART sense strand sequences.
Introns are removed from TART antisense transcripts
Sequence analysis of 14 of 16 TART-B products and three of 13 TART-A products for end 5d (Figure 2A) identified two putative TART-B introns and one putative TART-A intron in TART antisense transcripts (Table 4). We identified eight TART-A, four TART-B and six TART-C introns in antisense transcripts through our analysis of TART sequences present in EST clones (Figure 5 and Table 4). If introns in the direct repeats are counted twice for being in both the 5′- and 3′-UTRs, this number increases to 12 and 6 for TART-A and -B, respectively (Figure 5 and Table 4). Good matches to splice site consensus sequences are present at the exon/intron boundaries for all the TART antisense introns (data not shown).
Table 4.
TART antisense introns
| TART Subfamily | Putative introns | ||
|---|---|---|---|
| Total | Corresponding positionsa | Intron sizeb | |
| A | 8 | 12 595–12543c, 12 382–12 319, 11 772–11 707, 11 296–11 237, 10 696–10 643, 10 178–10 075, 9886–9398, 8394–8304 (1417–1365, 1204–1141, 594–529, 118–59)d | 53, 64, 66, 60, 54, 104, 489, 91, (53, 64, 66, 60) |
| B | 4 | 9701–9638c, 9491–9425c, 8862–8788, 7841–7756, (671–608, 461–395)d | 64, 67, 75, 86, (64, 67) |
| C | 6 | 10 088–10 028, 9875–9816, 9401–9320, 9164–9106, 8588–8528, 7097–6996 | 61, 60, 82, 59, 61, 102 |
aThe positions are written using the corresponding sense strand positions, but in reverse numerical order to reflect that the introns are removed from the antisense strand.
bSizes listed in each row correspond to the introns listed in the same row of the column indicating intron positions.
cThese introns were confirmed by RACE analysis presented in this work.
dThe first four and two introns listed for TART-A and TART-B, respectively, are in the 3′ direct repeats and the corresponding positions of these introns in the 5′ direct repeats are indicated in parentheses.
The similar sizes and spacing of introns seen in all three TART subfamilies is intriguing, since the UTRs of each subfamily are very diverse. However, we were unable to find significantly long predicted ORFs in spliced versions of TART antisense sequences, though we did not consider all possible alternative splice variants for each TART subfamily (data not shown).
DISCUSSION
All three subfamilies of TART contain a direct repeat of the 5′-UTR and the 5′ end of ORF1 at a subterminal position in the 3′-UTR, unlike the non-LTR retrotransposons to which TART is most closely related by phylogenetic analysis (7). Most 5′ ends we identified mapped to TART's direct repeats, so these repeats may play an important role in TART transcription. The presence of transcription initiation sites in sequences repeated in the 5′- and 3′-UTRs is reminiscent of transcription initiation within the LTRs of LTR retrotransposons (29). Also, an initiator motif identified at the site of end 5a was found to be more similar to sequences present at the transcription initiation sites of Drosophila LTR retrotransposons than Drosophila non-LTR retrotransposons (Figure 4A) (30). HeT-A transcription can initiate in the 3′ end of one element and read-through to the very 3′ end of a downstream element, producing a terminally redundant transcript similar to the terminally redundant transcripts of LTR retrotransposons (18). The parallels between transcription of Drosophila non-LTR telomeric retrotransposons and LTR retrotransposons are intriguing and may reflect the adaptations that telomeric retrotransposons have evolved to cope with the loss of DNA from their 5′ ends as a result of incomplete replication.
The RNA transcripts of retrotransposons have at least two essential roles in transposition: they are reverse transcribed into DNA copies and they are translated into proteins. Neither the relative nor the absolute levels of TART transcripts required for either of these roles is known. It has been proposed that TART and HeT-A may cooperate in their roles at telomeres and that TART may reverse transcribe HeT-A RNA (2). Therefore, it is conceivable that the primary importance of many TART transcripts is to serve as templates for translation of TART proteins, rather than to serve as templates for RT. The transcription initiation site 5a that we identified just upstream of ORF1 would be well suited to produce transcripts for expression of TART proteins. Genomic copies of TART that have 5′-UTR sequences upstream of site 5a could be produced from rare transcripts that initiate further upstream of site 5a or that are produced by read-through transcription from the 3′ end of a HeT-A or TART element into a downstream TART element. An alternative possibility is that when the TART reverse transcriptase reaches the 5′ direct repeat, it makes a template jump to the 3′ direct repeat and continues RT to generate additional 5′-UTR sequences upstream of site 5a.
TART produces an array of transcripts
Our identification of multiple 5′ and 3′ ends for TART transcripts is consistent with the production of a heterogeneous array of TART transcripts (Figure 2B). The conservation of the locations of many of the 5′ and 3′ ends among all three subfamilies is consistent with them being functionally important. Figure 2B shows a comparison between our data and data obtained from previously published northern blots of TART transcripts (19). We estimate from inspection of those published northern blots that the primary sense transcripts that were detected were ∼8, 9–9.5, 9.5–10, 10.5–11 and 11.5–13 kb, depending on the probe and RNA source. Transcript 1 would be 13 kb long for TART-A1 and transcript 3 would be 11.7, 9.6 and 11.0 kb for TART-A1, -B1 and -C1, respectively (Figure 2B). The predicted sizes of these transcripts overlap the sizes of most of the sense transcripts detected on northern blots. TART 3′-UTR probes also detected small sense transcripts that were ∼1 kb (19) that could correspond to transcripts 6 and 7, which would be ∼1.8 and 0.5–0.6 kb, respectively. We identified site 5b only from S2 cell RNA, raising the possibility that it corresponds to a previously reported 5.5 kb sense transcript specific to S2 cell RNA that hybridized to an ORF2 probe (19). However, transcript 5 would be 6.5 kb for TART-B1 (Figure 2B), so it is unclear whether it corresponds to that transcript.
TART antisense transcripts were typically in the ranges of 8–8.5, 9–9.5, 10–11 and ∼13 kb (19). Transcript 8 (Figure 2B) would be 10.7, 8.6 or 9.7 kb for TART-A1, -B1 or -C1, respectively (using the start site in GenBank EST clone AI531204.1 for TART-C1 site 5d). Splicing of introns in transcript 8 would reduce the sizes to 9.7, 8.3 or 9.3 kb, respectively. These predicted sizes overlap most of the sizes of antisense transcripts detected on northern blots (19). Additional signals for antisense transcripts varying in size from ∼6–8 kb were present on some northern blots (19). Transcript 9 for TART-A1 would be 6.2 or 5.2 kb if it were spliced or unspliced, respectively, but the northern data do not clearly support transcript 9.
Overall, transcripts 2, 4, 5, 9 and 10 (Figure 2B) are not clearly supported by the data from northern blots of TART transcripts (19). This may be due to the greater sensitivity of the RACE method, but further work will be needed to resolve this difference. Transcripts lacking one or both TART ORFs or most TART UTR sequences could be differentially localized, differentially utilized as RNA templates for TPRT, used to produce different relative levels of ORF1 and ORF2 proteins or be used to regulate TART expression. It has been proposed that partial-length (subgenomic) transcripts of some elements regulate the relative expression levels of element-encoded proteins (31,32) or represent a means of inhibiting expression of an element through premature polyadenylation (33,34).
Repeat-associated small interfering RNAs corresponding to TART transcripts have been identified (35) and mutations in genes involved in RNA interference eliminate these short RNAs in ovaries and lead to increased TART retrotransposition (36). These observations indicate that TART antisense RNA most likely anneals to TART sense RNA and activates an RNA interference pathway that inhibits TART retrotransposition. Our mapping data are consistent with this negative role for TART antisense transcripts. The antisense transcripts we mapped do not contain sequences absent from sense transcripts and do not appear to encode proteins, even though they contain introns, so they are unlikely to contribute sequence information necessary for retrotransposition.
We unexpectedly identified introns in TART antisense transcripts; however, the presence of introns is not unique to TART elements. Introns have been identified in transcripts of L1 (37), Tad (14), kangaroo-1 (38), Penelope and Athena elements (39), but their potential importance remains to be established. The TART introns are present in sequences that are very divergent between the three TART subfamilies, yet there are similarities in the sizes and spacing of the introns for all three subfamilies. This may reflect some conserved structural elements in TART RNA. Alternatively, the introns themselves could have some function. For example, the small intron RNAs could play some role in regulating TART expression, either instead of or in addition to any regulation mediated by longer antisense transcripts.
Overall, our results provide an important foundation for investigations of TART expression and mobility. Our analysis of transcription initiation sites and sequence motifs that may be important for TART transcription will enable analyses of cis- and trans-acting factors important for TART expression.
Acknowledgments
The authors thank A. Dingwall, S. Erdman and D. Sullivan for helpful discussions. This research was supported in part by a grant from the National Institute of General Medical Sciences (R01 GM38259 to R.W.L.). J.M.B. is supported by grant MCB-0416647 from the National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Biessmann H., Mason J.M. The unusual telomeres of Drosophila. Trends Genet. 1995;11:58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- 2.Pardue M.L., DeBaryshe P.G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 3.Levis R.W., Ganesan R., Houtchens K., Tolar L.A., Sheen F.M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 4.McEachern M.J., Krauskopf A., Blackburn E.H. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 5.Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 6.Sheen F.M., Levis R.W. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc. Natl Acad. Sci. USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik H.S., Burke W.D., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 8.Mizrokhi L.J., Georgieva S.G., Ilyin Y.V. Jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- 9.Swergold G.D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minchiotti G., Di Nocera P.P. Convergent transcription initiates from oppositely oriented promoters within the 5′ end regions of Drosophila melanogaster F elements. Mol. Cell. Biol. 1991;11:5171–5180. doi: 10.1128/mcb.11.10.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLean C., Bucheton A., Finnegan D.J. The 5′ untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol. Cell. Biol. 1993;13:1042–1050. doi: 10.1128/mcb.13.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumann G., Zundorf I., Hofmann J., Marschalek R., Dingermann T. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium repetitive element, from Dictyostelium discoideum. Mol. Cell. Biol. 1994;14:3074–3084. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Contursi C., Minchiotti G., Di Nocera P.P. Identification of sequences which regulate the expression of Drosophila melanogaster Doc elements. J. Biol. Chem. 1995;270:26570–26576. doi: 10.1074/jbc.270.44.26570. [DOI] [PubMed] [Google Scholar]
- 14.Sewell E., Kinsey J.A. Tad, a Neurospora LINE-like retrotransposon exhibits a complex pattern of transcription. Mol. Gen. Genet. 1996;252:137–145. [PubMed] [Google Scholar]
- 15.DeBerardinis R.J., Kazazian H.H., Jr Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics. 1999;56:317–323. doi: 10.1006/geno.1998.5729. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi H., Fujiwara H. Transcription analysis of the telomeric repeat-specific retrotransposons TRAS1 and SART1 of the silkworm Bombyx mori. Nucleic Acids Res. 1999;27:2015–2021. doi: 10.1093/nar/27.9.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutach A.K., Kadonaga J.T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danilevskaya O.N., Arkhipova I.R., Traverse K.L., Pardue M.L. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 19.Danilevskaya O.N., Traverse K.L., Hogan N.C., DeBaryshe P.G., Pardue M.L. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol. Cell. Biol. 1999;19:873–881. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA poyladenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Currie P.D., Sullivan D.T. Structure and expression of the gene encoding phosphofructokinase (PFK) in Drosophila melanogaster. J. Biol. Chem. 1994;269:24679–24687. [PubMed] [Google Scholar]
- 24.Benoit B., Nemeth A., Aulner N., Kuhn U., Simonelig M., Wahle E., Bourbon H.M. The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res. 1999;27:3771–3778. doi: 10.1093/nar/27.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talamillo A., Fernandez-Moreno M.A., Martinez-Azorin F., Bornstein B., Ochoa P., Garesse R. Expression of the Drosophila melanogaster ATP synthase α subunit gene is regulated by a transcriptional element containing GAF and Adf-1 binding sites. Eur. J. Biochem. 2004;271:4003–4013. doi: 10.1111/j.1432-1033.2004.04336.x. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arkhipova I.R. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–1369. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohler U., Liao G.C., Niemann H., Rubin G.M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0087. research 0087.1–87.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeke J.D., Stoye J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 30.Arkhipova I.R., Ilyin Y.V. Properties of promoter regions of mdg1 Drosophila retrotransposon indicate that it belongs to a specific class of promoters. EMBO J. 1991;10:1169–1177. doi: 10.1002/j.1460-2075.1991.tb08057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brierley C., Flavell A.J. The retrotransposon copia controls the relative levels of its gene products post-transcriptionally by differential expression from its two major mRNAs. Nucleic Acids Res. 1990;18:2947–2951. doi: 10.1093/nar/18.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelisson A., Song S.U., Prud'homme N., Smith P.A., Bucheton A., Corces V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perepelitsa-Belancio V., Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nature Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 34.Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 35.Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 36.Savitsky M., Dmitry K., Georgiev P., Kalmykova A., Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nigumann P., Redik K., Matlik K., Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 38.Duncan L., Bouckaert K., Yeh F., Kirk D.L. Kangaroo, a mobile element from Volvox carteri, is a member of a newly recognized third class of retrotransposons. Genetics. 2002;162:1617–1630. doi: 10.1093/genetics/162.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arkhipova I.R., Pyatkov K.I., Meselson M., Evgen'ev M.B. Retroelements containing introns in diverse invertebrate taxa. Nature Genet. 2003;33:123–124. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 40.Arkhipova I.R., Mazo A.M., Cherkasova V.A., Gorelova T.V., Schuppe N.G., Ilyin Y.V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986;44:555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- 41.Ziarczyk P., Fourcade-Peronnet F., Simonart S., Maisonhaute C., Best-Belpomme M. Functional analysis of the long terminal repeats of Drosophila 1731 retrotransposon: promoter function and steroid regulation. Nucleic Acids Res. 1989;17:8631–8644. doi: 10.1093/nar/17.21.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inouye S., Hattori K., Yuki S., Saigo K. Structural variations in the Drosophila retrotransposon, 17.6. Nucleic Acids Res. 1986;14:4765–4778. doi: 10.1093/nar/14.12.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuki S., Inouye S., Ishimaru S., Saigo K. Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur. J. Biochem. 1986;158:403–410. doi: 10.1111/j.1432-1033.1986.tb09767.x. [DOI] [PubMed] [Google Scholar]
- 44.Burke T.W., Kadonaga J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 45.Di Nocera P.P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988;16:4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]