Abstract
Retroviral and lentiviral vectors integrate their DNA into the host cell genome leading to stable transgene expression. Integration preferentially occurs in the proximity of active genes, and may in some case disturb their activity, with adverse toxic consequences. To efficiently analyze high numbers of lentiviral insertion sites in the DNA of transduced cells, we developed an improved high-throughput method called vector integration tag analysis (VITA). VITA is based on the identification of Genomic Tags associated to the insertion sites, which are used as signatures of the integration events. We use the capacity of MmeI to cleave DNA at a defined distance of its recognition site, in order to generate 21 bp long tags from libraries of junction fragments between vector and cellular DNA. The length of the tags is sufficient in most cases, to identify without ambiguity an unique position in the human genome. Concatenation, cloning and sequencing of the tags allow to obtain information about 20–25 insertion sites in a single sequencing reaction. As a validation of this method, we have characterized 1349 different lentiviral vector insertion sites in transduced HeLa cells, from only 487 sequencing reactions, with a background of <2% false positive tags.
INTRODUCTION
Retroviral elements integrate DNA copies of their genome into the host cell chromosome. This property has allowed them to spread into the genome of vertebrates and contribute to their evolution (1). It is also exploited in retrovirus-derived vectors that are commonly used for stable gene transfer in experimental and therapeutic settings. Successful gene therapy treatments using retroviral vectors have also revealed the genotoxicity of retroviral integration and called for a better understanding of the integration process (2,3). Retroviral DNA integration is mostly independent of the sequence context and has long been considered to occur at random locations in the host cell genome (4). Recently however, it has become possible to perform genome-wide analysis of retroviral integration events, and these ‘integrome’ studies have revealed strong biases in the distribution of integration sites (5). Lentiviruses (HIV-1 and SIV-1), MLV and ASLV, as well as gene transfer vectors derived from them, all display a more or less pronounced preference for transcription units (6,7). Different integration behaviours have been demonstrated among retroviruses, with lentiviruses integrating their proviral DNA all along the transcription units, while MLV (a gammaretrovirus) display a marked preference for the proximal region of active genes (8), ASLV shows only a weak tropism for transcription units without any preference for the integration position. Integrome profiles are also dependent on the host cell origin and physiological state (7,9). Further biases with other genomic features may exist and could be discovered through larger integrome analysis.
Yet, these studies require the characterization of hundreds of integration sites in order to be informative. Today the most popular technique is linear amplification mediated PCR (LAM-PCR) (10) which is derived from linker mediated PCR (LM-PCR) and involves the isolation and sequencing of individual junction sequences between proviral and host genomic DNA. Our goal here was to increase the throughput of current integration site analysis by allowing for the simultaneous analysis of multiple sites in a single sequencing reaction. We describe a method called vector integration tag analysis (VITA), based on the identification of genomic tags (11) close to the insertion sites which can be used as a signature of the integration event. As in the long-SAGE technique (12), we use the capacity of the tagging MmeI enzyme to cleave DNA at a defined distance of its recognition site, in order to generate 21 and 22 bp long tags. This length is sufficient, in most cases, to identify without ambiguity a unique position in the human genome. Concatenation, cloning and sequencing of tags allow to obtain information about 20–25 insertion sites in a single sequencing reaction.
We report the characterization using VITA, of a population of integration sites in the genome of human cells transduced with a lentiviral vector. We show that the method accurately and efficiently describes the integrome and its association with gene-enriched regions of the genome.
MATERIALS AND METHODS
Cells
Cells from HT1080, a human fibrosarcoma cell line, were transduced with a VSV-G pseudotyped Self-Inactivated lentiviral vector containing a GFP gene (pRRLSINcPPT_PGKGFP_WPRE) at a multiplicity of infection (MOI) of 1. A clone (HT3) was isolated by limiting dilutions and cell sorting on the basis of GFP expression level, using a MoFlo cell sorter. HeLa cells were transduced twice with the same HIV-1 based vector (1.1010 TU/ml) at a MOI of 5, and were harvested after an average of 28 doublings. A subpopulation representing 25% of the cells was isolated on the basis of its high GFP expression levels.
Insertion site fragments (ISF) library
Five microgram of purified DNA from transduced cells were digested with 10 U of NlaIII (Biolabs).
The NlaIII compatible adaptor (OL) was obtained with two synthetic oligonucleotides (OLS and OLL, Table 1) which were mixed, heated at 95°C for 10 min and cooled slowly to room temperature. A 5-fold molar excess of this adaptor (0.45 μg) was ligated with 100 ng of NlaIII digested DNA using 1 U of T4 DNA ligase (Invitrogen) in a 20 μl reaction volume. A first linear amplification of integration sites was done with 10 μl of this ligation and 1 pmol of biotinylated oligonucleotide (bOC) (Table 1) (six cycles: 30 s, 92°C; 30 s, 58°C; 1 min 30, 72°C) in 5 mM MgCl2, 200 nM dNTPs, and 1 U of Platinum® Taq DNA polymerase in its buffer (Invitrogen). This amplification provided biotin-labelled DNA fragments starting in the vector and finishing at the first NlaIII restriction site in the host genome beyond the vector 5′ end. Streptavidin-coated paramagnetic beads (0.5 mg Dynal Biotech) were washed in 50 μl of 5 mM Tris–HCl pH7.5, 0.5 mM EDTA, 1 mM NaCl and mixed with DNA fragments for 20 min at room temperature. DNA attached to the beads was amplified by 15 cycles of PCR: 15 s, 92°C; 30 s, 59°C; 1 min 30, 72°C. 1 U Platinum® Taq DNA polymerase, 5mM MgCl2, 200 nM dNTPs, 25 μmol of both PVE and PLE (Table 1). PCR products were diluted 1000-fold and submitted to a second amplification (30 cycles) using a nested set of primers (PVI and PLI, Table 1) under the same conditions except for a reduced annealing temperature (56°C). Products of this PCR constitute the ISF library. An aliquot was cloned into pCR®2.1-TOPO (Invitrogen).
Table 1.
Oligonucleotides used in VITA protocol
| Oligo | Sequence | Description |
|---|---|---|
| bOC | b-AAAAAAAAAATACTGACGCTCTCGCACCCATCTCT | Position 383–407 on HIV genome, linear amplification of junctions |
| PVE | TCTCGCACCCATCTCTCTCC | Position 379–398 on HIV genome, amplification of junctions fragments |
| PVI | TGGTTTCCCTTTCGCTTTCA | Position 255–274 on HIV genome, amplification of junctions fragments |
| OLS | TCGGAGAAGAGGATACGGATTGTAGGCAGGGGGACGCCTCAAGAATAAGGGCT-a | |
| OLL | CCCTTATTCTTGAGGCGTCCCCCTGCCTACAATCCGTATCCTCTTCTCCGACATG | |
| PLE | CCTTATTCTTGAGGCGTCCC | |
| PLI | GGCGTCCCCCTGCCTACAAT | |
| bPLN | b-CGACGAGACACTGCCCCTGA | |
| bPLI | b-GTCCCCCTGCCTACAATCCG | |
| LNS | AACATGTTTGAATCTGTTTCAGGGGCAGTGTCTCGTCGGGA-a | |
| LNL | GAGTCCCGACGAGACACTGCCCCTGAAACAGATTCAAACATGTTNN |
b = biotin, a = amine.
Tags extraction
After ethanol precipitation, the ISF library was digested with 4 U of the tagging enzyme (MmeI, 1 h at 37°C). This resulted in the release of the adaptor with the adjacent genomic DNA (59 bp fragment) which was purified through a 12% polyacrylamide gel (Spin-X®, Costar®). Purified DNA was linked to a second adaptor, LN, adding a new NlaIII site and a TT dinucleotide that introduces an asymmetry for orientation purposes. LN degenerate adaptor was prepared by annealing two synthetic oligonucleotides (LNL and LNS, see Table 1) as described above for OL. Ligation products were diluted 200-fold and used in 200 individual PCR under the following conditions; 92°C, 30 s; 58°C, 30 s; 72°C, 1 min 30 s; 30 cycles, 50 μl reaction in 5 mM MgCl2, 200 nM dNTPs, 500 nM biotinylated primers bPLN and bPLI (Table 1) and 1 U of Platinum® Taq DNA polymerase with its buffer. PCR products were pooled and ethanol precipitated. The resulting 97 bp DNA fragment was purified through a 12% polyacrylamide gel and digested with 2 × 100 U of NlaIII in a 400 μl-reaction (2 × 1 h at 37°C). After digestion, tags were purified away from biotinylated fragments using streptavidin coated magnetic beads. Unbound tags were recovered in the supernatant and precipitated before purification on a 12% polyacrylamide gel. The purified tags were then concatenated using 5 U of T4 DNA ligase in 10 μl as recommended by the supplier, for 3 h at 16°C. Concatemers (600–1000 bp long) were purified on a 1.5% agarose gel (Kit Qiaquick, Qiagen) and cloned into SphI-site of a pZero-1 plasmid (Invitrogen). Recombinant clones obtained after electroporation of TOP10 cells (Invitrogen) were selected on LB-low salt plates containing 50 μg/ml zeocin.
Sequencing and analysis
Sequencing reactions were performed using Big Dye terminator sequencing chemistry (Applied Biosystems) from the M13 forward or M13 reverse primers and run on a 377-XL Applied Biosystems automated sequencer. Sequences obtained from the ISF were matched on the human genome using the BLAT program (UCSC Human Genome Working Draft, May 2004 freeze). A sequence was considered to be a ‘bona fide’ vector-genome junction if: (i) it contains the sequence of the 5′ end of the vector and those of the adaptor OL; (ii) it matches with one genomic locus showing >95% identity. Integration was considered to have occurred in gene only if it was located within the boundaries of one of the RefSeq genes (UCSC Genome Browser on Human May 2004 Assembly).
Sequenced tags were extracted and analyzed using a home-made software (available upon request) that performs the following steps; (i) Locate the NlaIII sites (CATG) within the tags concatemers; (ii) Extract tags of length 21–30 bp which fall between these sites and orientate them (two consecutive T are used to orientate the adjacent tags). (iii) Exclude ambiguous tags (Tags with Ns, tags unable to be orientated) (iv) Count number of occurrences of each tag; (v) Remove repeated occurrences of tags.
The result of this process is a fastA file of tags which can be used as an entry to a similarity search of the genome (Goldenpath). The Blast software was used to map tags on the human genome with parameters which force perfect alignments (−W = 21; −e = 0.1). To test whether the number of integrations was correlated to the number of genes per chromosome, we used the Pearson's correlation test which measures the strength of the linear relationship between two variables.
RESULTS
VITA overview
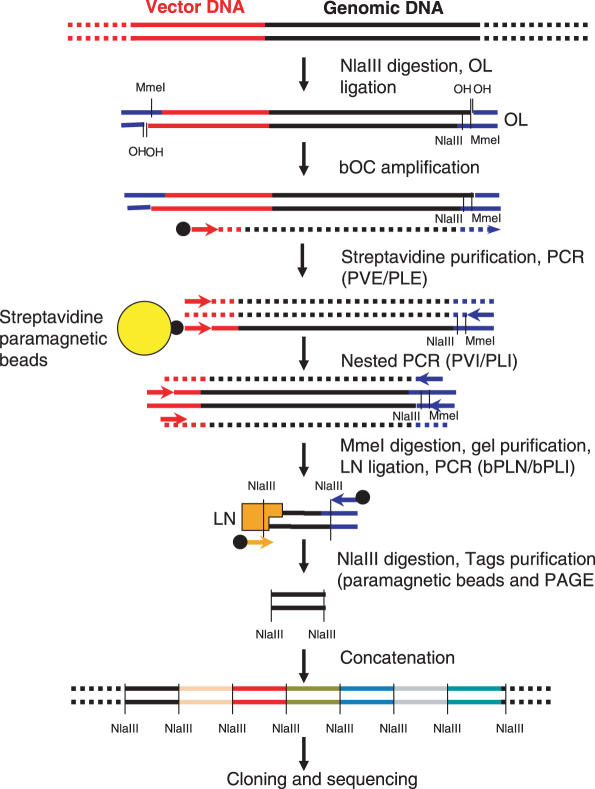
Twenty-one nucleotide long tags are enough to unambiguously define a genomic site in 70% of cases. The method described here is designed to extract and analyze such short sequences in the vicinity of retroviral integration sites. It is based in the ability of the MmeI restriction enzyme to cut DNA outside of its recognition site and includes two steps: the production of an ISF library and the extraction of a 21 bp tag from each fragment. Figure 1 gives a general outline of the method and the different oligonucleotides used are listed in Table 1.
Figure 1.
Overview of VITA method. All steps are detailed in ‘Materiel and Methods’. For oligonucleotides information, see Table 1.
ISF library
The genomic DNA from cells transduced with a lentiviral vector was digested with a high frequency cutting restriction enzyme [NlaIII (CATG), anchoring enzyme]. Restriction fragments were then ligated with a NlaIII compatible adaptor which contains a MmeI restriction site (OL, see Table 1). OL oligonucleotides were not phosphorylated, resulting in a ligation reaction in which only their 3′ protruding end was covalently bound to genomic DNA.
A linear amplification of insertion site fragments containing vector-genomic DNA junctions was then performed using a bOC (Table 1) that hybridizes with the vector sequence outside of the LTRs. In this reaction, the OL adaptor and its MmeI site were replicated at the end of the ISFs which were then purified with paramagnetic streptavidin-coated beads.
ISFs were then amplified by nested PCR with primer in the vector (PVE/PLE, Table 1) and in the OL adaptor (PVI/PLI, Table 1). This step was necessary, because it eliminated unwanted NlaIII fragments carrying the OL adaptor which could contaminate the ISF, but did not contain vector sequences.
Tag extraction
The ISFs library was digested by MmeI, yielding 21–22 bp DNA tags linked to the OL sequences. MmeI produces a 2 nt 3′ protruding end that cannot be filled by DNA polymerase. In order to preserve the information from this end, the MmeI products were ligated to an adaptor oligonucleotide with a random 2 nt 3′ protruding end (LN, Table 1).The LN adaptor also contains a NlaIII site for tag concatemer formation (see below). A TT dinucleotide was added at the 3′ end of LN. This ‘tag in the tag’ introduces an asymmetry that permits to orientate the tags. Ligation products were PCR amplified using two biotinylated primers hybridizing to OL and LN (bPLN/bPLI, Table 1) and cleaved by NlaIII. Biotinylated fragments were then eliminated with streptavidin-coated beads, 21 bp tags with NlaIII remaining in solution were gel-purified, ligated into concatemers and cloned in a bacterial plasmid.
Complex integrome analysis
In order to validate the VITA method, we analyzed the integration sites in a HeLa cell population transduced with a lentiviral vector (pRRLSINcPPT_PGKGFP_WPRE) (13) at a MOI of 5.
HeLa ISF library
An ISF library was obtained as described above and introduced into pCR®2.1-TOPO plasmid. 82 clones were sequenced in order to analyze the redundancy of the ISF population. Seventy-five clones had the expected structure, including 274 bp from the provirus, a variable length of genomic DNA and the OL adaptor sequence next to a NlaIII site. Of those, 66 (91%) were long enough to be analyzed by BLAT (14), 8 were in repeated regions and 58 could be assigned to a specific genomic site, identifying 54 different integration sites.
HeLa tag library
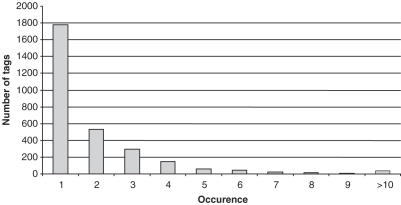
After tags purification, concatenation and cloning, we performed 487 sequencing reactions that yielded 7831 tags after extraction using a home-made software. Non-oriented tags starting with AA and finishing with TT were excluded from the analysis. 6872 tags had the expected size, did not contain sequence ambiguities and could be precisely oriented using the TT dinucleotide on the LN adaptor. Tags redundancy is shown in Figure 2.
Figure 2.
Redundancy of sequenced tags in a typical VITA experiment. VITA was performed onto a HeLa cell population transduced with a lentiviral vector.2480 21 or 22 bp long tags were generated and counted. Boxes represent the number of tags sequenced once, twice, until more than 10 times.
A total of 2480 different tags were obtained and mapped on the human goldenpath (hg17) by using BLAST with parameters that force perfect alignment (−W = 21, −e = 0.1). Among these tags, 1349 were localized at unique positions and designated ‘one hit’. Tags without match on the genome (n = 632) were called ‘no hit’ and tags with multiple hits (n = 499) were called ‘multi hit’.
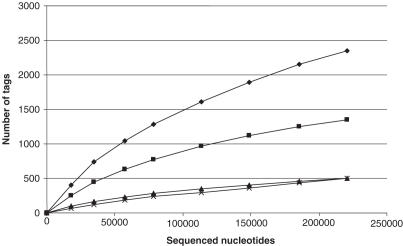
In order to estimate the complexity of the tag population we represented the number of different tag in each category (‘one hit’, ‘no hit’, ‘multi hit’) as a function of the cumulated number of sequenced nucleotides (Figure 3). All the curves are far from the asymptote, indicating that the tag population can still be extensively exploited.
Figure 3.
Evolution of the number of sequenced tags in function of cumulated number of sequenced nucleotides. The 2480 tags generated by running VITA on a HeLa cells were mapped on the human genome and classified in different categories. Curves represent ‘total different tags’ (diamond), ‘unique hits’ (square), ‘no hits’ (triangle) and ‘multiple hits’ (cross) (see Discussion).
Examination of the ‘no hit’ population indicated that 20% of them (132/632) contained sequences of proviral origin, due to insertions very close to a NlaIII site on the genome. The remaining ‘no hit’ tags could originate from insertions into non sequenced parts of genome, but most are probably mutant versions of real tags, that appeared during one of three PCR included in the protocol. Reciprocally, this raised the possibility that certain tags with hits in the genome could also be the result of error-prone PCR. In order to estimate the probability of wrongly identifying insertion sites, we selected a population of 303 ‘bona fide’ 21 bp tags deduced from sequenced ISF clones and created a virtual population of ∼50 000 mutated tags, by changing one position in each of them (base change, deletion or insertion). None of the virtual tags could be matched on the genome and therefore would not lead to erroneous insertion site identification.
Integrome analysis in a cell clone
The accuracy of the VITA method as well as the possibility to generate ‘false tags’ were further tested on a cellular clone containing a known set of integrated proviral genomes. The HT3 clone was obtained after transduction of HeLa cells as described above (see Materials and Methods), and the number of integration sites was determined to be at least seven by Southern blot analysis (data not shown).
HT3 ISF library
An ISF library was obtained from the HT3 clone DNA, as described above. We sequenced 83 clones, and 82 of them were found to represent actual junctions between vector and genomic DNA. These junctions corresponded to eight different integration sites. Each integration event was sequenced 4–13 times (average: 10.25, median: 11.5), which suggested that we had saturated our junctions library analysis and that integration sites had been identified. One of sequenced clones did not have the expected vector-genome DNA structure. A tag library was then constructed and analyzed, in order to evaluate the number of false positive generated by the method.
HT3 tag library
We obtained 913 tags by sequencing 70 concatemer clones. Of those 266 corresponded to one of the eight expected insertion sites. Among the remaining tags, 533 could be oriented and were analyzed by BLAST, 517 were ‘no hits’, 8 were ‘one-hit’ and 8 were ‘multi-hit’. The origin of ‘no-hit’ tags was further investigated. The 517 ‘no-hit’ tags were compared with the nine expected tags from the nine ISF library clones (eight true tags and one contaminant). A total of 508 out of 517 ‘no hit’ tags were very similar to one of the nine expected tags suggesting that the principal source of ‘no-hit’ tags were PCR associated mutations. These 517 ‘no hit’ would be discarded in a standard analysis (see above) and therefore, we estimate the background of false positive to be 8 + 8 = 16 tags out of 533 + 266 = 799, or ∼2%.
Vector insertion profile
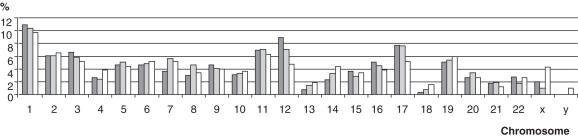
Lentiviral vectors are known to integrate preferentially into transcribed regions of the genome (5). As a further validation of our method, we checked whether a similar bias was observed in the ISF library and in the tag populations. Additional HeLa ISF library clones (n = 932) were sequenced and 607 insertion sites could be identified. As expected, the number of insertion sites found on each chromosome was correlated with the number of genes (Pearson's correlation rate = 0.79, P-value = 2.072e−6). The same correlation was observed with the 1349 ‘one-hit’ tags described in HeLa tag library (Pearson's correlation rate = 0.86, P-value = 2.84e−8). These results suggest that the tag population properly reflects the actual integrome (Figure 4).
Figure 4.
Correlation between ISF clones/tags and genes per chromosome. ISFs (607 ISFs) and one-hit tags (1349 tags) obtained by VITA technique on HeLa cells were mapped into human genome. Dark grey boxes and clear grey boxes represent the percentage of ISF and tags identified per chromosome respectively. White boxes represent the percentage of genes per chromosome.
DISCUSSION
LAM-PCR is the reference method for analyzing retroviral integration sites. In this method, a library of ‘insertions site fragments’ is cloned and sequenced. Each clone gives information about one insertion site. Clone sequencing is the limiting step of this technique. In order to accelerate the discovery of insertion sites, we propose a new method called VITA based on LAM-PCR with a supplementary step to obtain genomic tags of each clones. Tags are obtained and concatenated as in the ‘long-SAGE’ method of transcriptome analysis (12) resulting in a 20–25-fold increase of data output.
The first part of VITA is similar to the LAM-PCR protocol published by Schmidt et al. (10) with some differences: the first step in LAM-PCR is a 100 cycles of linear amplification followed by a double strand synthesis with random oligonucleotides and anchoring enzyme digestion. Compatible adaptors are then added to perform a nested PCR of ISF fragments. In our method the first step is the anchoring enzyme digestion, followed by compatible adaptor ligation and six cycles of linear amplification. During this step, the adaptor strand which will be targeted by the nested PCR is amplified together with the ISF fragment. No double strand synthesis is included in our method. In our hands as much as 90% of obtained clones have the expected structure compared to 60% published by Burgess (8) who used LM-PCR.
Genomic tags are extracted using the type IIS restriction enzyme MmeI which cuts 20–22 nt apart its recognition site. The length of the tags generated by the method (21 bp)is a compromise between the informational content which allows to unambiguously map them on the genome and the capacity to concatenate a lot of them to be sequenced in a single run. There are 54 586 284 predicted 21 nt long NlaIII tags in the human genome reference sequence (hg 17, International Human Genome Sequencing Consortium), and 73% of them are unique. If we consider that the integration pattern is not correlated to the distribution of genomic NlaIII sites, then 73% of identifiable tags generated by VITA should point at unique genomic positions. Consistently, we obtain 68% ‘one hit’ tags when the predicted tags from 443 ISF clones are searched against the human genome.
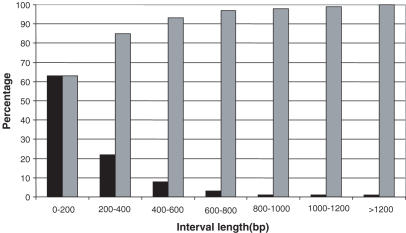
In contrast to ISF cloning and sequencing which localize the exact point of vector integration site, VITA indicates the coordinates of the NlaIII restriction fragment containing the vector insertion. The accuracy of VITA thus depends on the average length of NlaIII genomic fragments. An in silico NlaIII restriction analysis of the human genome indicates that in 85% of cases, VITA will have preciseness better than 400 nt and in 99% of cases better than 1200 nt (Figure 5). Since the actual junction sequences are not determined by the method, it is critical to make sure that the proportion of ‘false tags’ which map on the genome but do not correspond to real insertion sites is kept minimal. For this reason it is paramount to perform a careful quality control of the ISF library. Using the protocol described here, we routinely obtain >90% of ISF clones that contain true insertions. The other would not contribute to the tag population because they lack the MmeI site or do not match the genome. In addition, tag population show the same bias for gene rich regions as the ISF population, suggesting that tag libraries properly reflect the composition of ISF libraries.
Figure 5.
Distribution of NlaIII fragment size in the human genome. Human genome was digested in silico by NlaIII. The proportion of NlaIII fragments of a defined length is represented in black boxes. Cumulated percents are represented in grey boxes.
In this study we used NlaIII as anchoring enzyme with the HIV1-SIN vector. VITA oligonucleotides can be adapted to other restriction enzymes or vectors, with the following rules: (i) bOC, PVE and PVI should not be chosen in the LTR to avoid a vector internal amplification fragment. (ii) the anchoring enzyme should not cut between bOC and the end of the vector. A few base recognition site is better because ISFs will be shorter and the nested PCR less biased. NspI which generates compatible ends with NlaIII can be alternatively used without any change in OL adaptor oligonucleotides (data not shown).
As a PCR-based technique, VITA can present amplification biases. We tried to minimize this problem by choosing a frequently cutting anchoring enzyme to reduce the length of ISF fragments. The tag recovery step uses nested PCR over a constant length tag population with common primers and should not introduce important amplification biases. SAGE, which features a similar step is considered a ‘quantitative’ method. In conclusion, VITA biases should not be more pronounced than in LM or LAM-PCR.
VITA can be further improved. The LN adaptor introduces a TT dinucleotide in the tags for orientation purpose, but tags starting with AA cannot be properly oriented. The proportion of ‘unorientable’ tags can be strongly reduced by adding two additional nucleotides. We have used a TTTT sequence in LN which generates 25 nt tags (instead of 23) without loss of efficiency (data not shown). Alternatively, more specific anchoring enzyme (5 or 6 nt recognition site) can be used with a more processive DNA polymerase to compensate the increment of ISF size. This should allow adapting VITA to other vectors carrying NlaIII site between bOC and the end of the LTR. Other commercially available polymerases with editing activity could also be used to decrease the ‘mutated tag’ rate.
Acknowledgments
We thank Genomining SA for their help in bioinformatics analysis. This work was supported by Association Française contre les Myopathies and CONSERT European project (CONserted Safety and Efficiency evaluation of Retroviral Transgenesis, LSHB CT-2004-005242). Funding to pay the Open Access publication charges for this article was provided by Genethon.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E., Radford I., Villeval J.L., Fraser C.C., Cavazzana-Calvo M., et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 3.Ott M.G., Schmidt M., Schwarzwaelder K., Stein S., Siler U., Koehl U., Glimm H., Kuhlcke K., Schilz A., Kunkel H., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nature Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 4.Bushman F., Lewinski M., Ciuffi A., Barr S., Leipzig J., Hannenhalli S., Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nature Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 5.Schroder A.R., Shinn P., Chen H., Berry C., Ecker J.R., Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 6.Hematti P., Hong B.K., Ferguson C., Adler R., Hanawa H., Sellers S., Holt I.E., Eckfeldt C.E., Sharma Y., Schmidt M., et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell R.S., Beitzel B.F., Schroder A.R., Shinn P., Chen H., Berry C.C., Ecker J.R., Bushman F.D. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 9.Ciuffi A., Mitchell R.S., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F.D. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol. Ther. 2005;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt M., Carbonaro D.A., Speckmann C., Wissler M., Bohnsack J., Elder M., Aronow B.J., Nolta J.A., Kohn D.B., von Kalle C. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. Nature Med. 2003;9:463–468. doi: 10.1038/nm844. [DOI] [PubMed] [Google Scholar]
- 11.Dunn J.J., McCorkle S.R., Praissman L.A., Hind G., Van Der Lelie D., Bahou W.F., Gnatenko D.V., Krause M.K. Genomic signature tags (\s): a system for profiling genomic DNA. Genome Res. 2002;12:1756–1765. doi: 10.1101/gr.306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha S., Sparks A.B., Rago C., Akmaev V., Wang C.J., Vogelstein B., Kinzler K.W., Velculescu V.E. Using the transcriptome to annotate the genome. Nat. Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- 13.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]