Abstract
We explore the interactions of CYT-19, a DExD/H-box protein that functions in folding of group I RNAs, with a well characterized misfolded species of the Tetrahymena ribozyme. Consistent with its function, CYT-19 accelerates refolding of the misfolded RNA to its native state. Unexpectedly, CYT-19 performs another reaction much more efficiently; it unwinds the 6-bp P1 duplex formed between the ribozyme and its oligonucleotide substrate. Furthermore, CYT-19 performs this reaction 50-fold more efficiently than it unwinds the same duplex free in solution, suggesting that it forms additional interactions with the ribozyme, most likely using a distinct RNA binding site from the one responsible for unwinding. This site can apparently bind double-stranded RNA, as attachment of a simple duplex adjacent to P1 recapitulates much of the activation provided by the ribozyme. Unwinding the native P1 duplex does not accelerate refolding of the misfolded ribozyme, implying that CYT-19 can disrupt multiple contacts on the RNA, consistent with its function in folding of multiple RNAs. Further experiments showed that the P1 duplex unwinding activity is virtually the same whether the ribozyme is misfolded or native but is abrogated by formation of tertiary contacts between the P1 duplex and the body of the ribozyme. Together these results suggest a mechanism for CYT-19 and other general DExD/H-box RNA chaperones in which the proteins bind to structured RNAs and efficiently unwind loosely associated duplexes, which biases the proteins to disrupt nonnative base pairs and gives the liberated strands an opportunity to refold.
Keywords: group I RNA, RNA folding, RNA unwinding, Tetrahymena ribozyme
Essentially all cellular processes that are mediated by structured RNAs also require one or more DExD/H-box proteins (1). These proteins use the energy from ATP binding and hydrolysis to accelerate RNA structural transitions, which can represent folding steps toward the native state or conformational switches between functional forms. The requirement for proteins presumably arises because RNA base pairs and other local structure can be highly stable even in the absence of enforcing structure, such that folding steps or rearrangements that require significant unfolding require assistance to proceed efficiently (2–4).
Despite their ubiquitous presence, key questions about the functions of DExD/H-box proteins remain largely unanswered. First, what interactions direct different DExD/H-box proteins to their physiological substrates? All of these proteins share a core “helicase” domain containing a set of conserved motifs, and most have additional domains, a few of which have been shown to recognize substrate RNAs or RNA–protein complexes (reviewed in ref. 5). On this basis, targeting has been proposed as a general role for these domains, and the specific interactions that target one DExD/H-box protein have been delineated (6–9). Nevertheless, in general, the interactions that direct DExD/H box proteins to their substrates remain to be identified.
Second, how do DExD/H-box proteins act to mediate RNA conformational transitions? Although they are homologous to DNA helicases, and several are known to possess helicase activity in vitro (10–17), rearrangements of structured RNAs may also involve disruptions of tertiary structure or even displacement of bound proteins (18–20). Studying the mechanisms of DExD/H-box proteins on their physiological substrates is difficult because the “starting state” of the RNA is typically poorly defined, as discrete misfolded species are difficult to populate and trap, and because most of the substrate RNAs do not have simple activities that can be used to distinguish the native state from structurally related misfolded species.
An attractive candidate for a detailed study of DExD/H-box protein targeting and function is the CYT-19 protein of Neurospora crassa (21, 22). CYT-19 is required for proper folding of several mitochondrial group I introns in vivo (21). These introns represent different subgroups and vary widely in their global architectures, suggesting that CYT-19 recognizes general features of RNA structure rather than a specific feature of a single RNA. The idea of general RNA recognition is extended by findings that, when heterologously expressed in yeast, CYT-19 can function in splicing of all 10 group I introns and even group II introns (23) and that it chaperones folding of group II introns in vitro (24).
Here we explore the action of CYT-19 on a long-lived misfolded species of the ribozyme derived from the self-splicing group I intron of Tetrahymena thermophila. The RNA folds through a progression of intermediates with increasing structure (25, 26), including a very long-lived misfolded species (27–32). This misfolded species can be populated by up to 90% of the RNA, and it behaves as a single species in refolding to the native state (31). Thus, it provides a well defined substrate RNA to probe the activity of a DExD/H-box protein. Indeed, a variant of this RNA was used previously to provide a physical signature for chaperone activity of CYT-19 (21). We find that CYT-19 accelerates refolding of the misfolded Tetrahymena ribozyme to its native state and that it performs another reaction much more efficiently; it unwinds the P1 duplex formed between the ribozyme and its oligonucleotide substrate. This unwinding is strongly enhanced by attachment of the P1 duplex to the ribozyme but is inhibited by the formation of tertiary contacts between P1 and the ribozyme body. The P1 unwinding activity is not specific to the Tetrahymena ribozyme, because it is also observed with one of the group I RNAs with which CYT-19 interacts in vivo. Together, these results suggest a model in which CYT-19 acts as a general chaperone by binding nonspecifically to structured RNAs and unwinding duplexes that are loosely associated with these RNAs. DED1, another DExD/H box protein that interacts functionally with multiple RNAs, displays analogous activity, suggesting that this mode of action may be common for DExD/H-box proteins that function as general RNA chaperones.
Results
Acceleration of Native Ribozyme Formation by CYT-19.
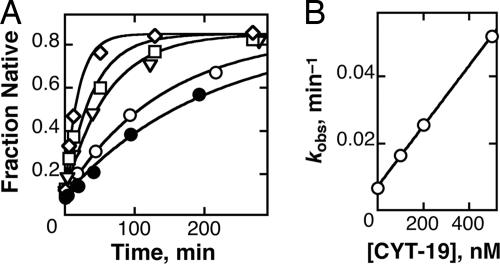
To investigate the interactions of CYT-19 with a misfolded group I RNA, we first determined whether it accelerates refolding of the misfolded Tetrahymena ribozyme to its native state by using the oligonucleotide cleavage activity of the native ribozyme as a probe for its formation (31). We found that CYT-19 accelerates formation of the native ribozyme and that the acceleration requires ATP (Fig. 1A). The observed rate constant for refolding increases linearly with CYT-19 concentration to at least 500 nM, giving a kcat/KM value of 9 × 104 M−1 min−1 (Fig. 1B). The lack of saturation indicates a lower limit of 500 nM on the Kd for functional binding by CYT-19,‡ consistent with previous results indicating relatively nonspecific binding to group I RNAs (21). In our refolding reactions, CYT-19 was inactivated before determination of the fraction of native ribozyme (see Materials and Methods and Fig. 7, which is published as supporting information on the PNAS web site), indicating that CYT-19 chaperones folding of the ribozyme to the native state, rather than giving formation of an RNA species that requires its continued presence for activity (24).
Fig. 1.
Acceleration of ribozyme refolding to the native state by CYT-19. (A) Misfolded ribozyme was incubated without CYT-19 (○), with 500 nM CYT-19 in the absence of ATP (●), or with 100 nM (▿), 200 nM (□), or 500 nM CYT-19 (◇) in the presence of 2 mM ATP-Mg2+. Rate constants for native state formation were 0.0071 min−1 (○), 0.0048 min−1 (●), 0.017 min−1 (▿), 0.026 min−1 (□), and 0.053 min−1 (◇). The faster reactions gave well defined end points of ≈0.85, and end points for the slower reactions were forced to this value. This end point represents essentially complete refolding of the ribozyme to the native state, because the remainder is inactive and presumably damaged (59). (B) Observed rate constants from A plotted against CYT-19 concentration. Three independent determinations gave a kcat/KM value of (8.8 ± 1.4) × 104 M−1 min−1 in the presence of 5 mM Mg2+. Analogous experiments at 10 mM Mg2+ gave an upper limit of 2 × 103 M−1 min−1 (Fig. 6, which is published as supporting information on the PNAS web site).
CYT-19 Efficiently Unwinds a Double-Stranded Region of the Ribozyme.
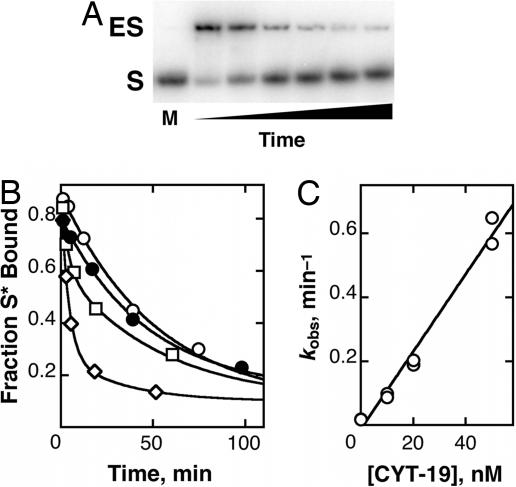
Surprisingly, we found that, in reactions analogous to those above, but with the oligonucleotide substrate CCCUCUA5 (S) initially bound to the misfolded ribozyme rather than added only after the refolding reaction, even low concentrations of CYT-19 resulted in rapid cleavage of S. This result suggested either that CYT-19 was much more efficient at refolding the ribozyme with bound S or that CYT-19 was giving efficient dissociation of S from the misfolded ribozyme by unwinding the P1 duplex formed between S and the ribozyme, allowing S to be bound and cleaved by the small fraction of native ribozyme present. To determine directly whether CYT-19 accelerates unwinding of the P1 duplex, releasing S to solution, we used a native gel mobility shift assay. Indeed, even at concentrations as low as 10 nM, CYT-19 substantially accelerated dissociation of S (Fig. 2). This result demonstrates that CYT-19 possesses RNA unwinding activity, as expected by analogy with other DExD/H-box proteins (10–17).§ An independent assay using the oligonucleotide cleavage activity of the ribozyme gave the same results within error (Fig. 8, which is published as supporting information on the PNAS web site), confirming that the gel shift assay was faithfully monitoring substrate dissociation. Similar to the ribozyme refolding reaction described above, acceleration of P1 duplex unwinding requires ATP (Fig. 2B), and the rate increases linearly with CYT-19 concentration. The dependence on CYT-19 concentration gave a kcat/KM value of 107 M−1 min−1, >1,000-fold larger than that for refolding the ribozyme to its native state under the same conditions.¶ Additional experiments using excess ribozyme demonstrated that CYT-19 is capable of performing multiple turnovers of P1 duplex unwinding (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 2.
Acceleration of substrate dissociation by CYT-19. (A) Native gel mobility shift assay to separate free substrate (S) from substrate bound to ribozyme (ES). In the marker lane at left (M), S was run alone. From left to right, the other lanes represent reaction in the presence of 30 nM CYT-19 for 1, 3, 4, 14, 30, and 45 min. The two bands shown in each lane accounted for >95% of labeled material. (B) Time courses of substrate dissociation from misfolded ribozyme. CYT-19 was absent (○, koffs = 0.019 min−1), present at 50 nM in the absence of ATP (●, koffs = 0.021 min−1), or present at 10 nM (□, koffs = 0.11 min−1) or 20 nM (◇, koffs = 0.19 min−1) in the presence of 2 mM ATP-Mg2+. In reactions with CYT-19 at concentrations of ≤20 nM, slower phases with rate constants of ≈0.02 min−1 were observed, presumably reflecting time-dependent inactivation of CYT-19 followed by substrate dissociation at its intrinsic rate. For these reactions, rate constants were calculated by multiplying the observed rate constant for the faster phase by the amplitude of the fast phase divided by the total amplitude. (C) Dependence of substrate dissociation on CYT-19 concentration. A linear fit gave a (kcat/KM) value of (1.1 ± 0.1) × 107 M−1 min−1.
Specificity for P1 Duplex Unwinding Contributed by the Ribozyme Structure.
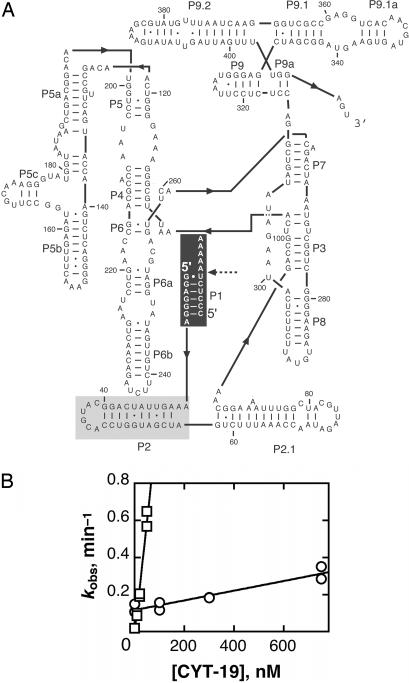
Unwinding the P1 duplex, which contains native base pairs, is not expected to be a physiologically productive activity of CYT-19, and we confirmed that the absence of P1 does not accelerate refolding of the misfolded ribozyme in vitro under near-physiological conditions (Fig. 10, which is published as supporting information on the PNAS web site). Nevertheless, we were interested in studying the P1 unwinding reaction as a means of gaining insight into the mechanism and specificity determinants for CYT-19 action. Thus, we explored whether this unwinding is purely nonspecific activity by CYT-19 or whether the protein is targeted by the presence of RNA structure adjacent to the duplex. We measured CYT-19-mediated unwinding of a duplex formed between S and the ribozyme strand that binds it, the internal guide sequence, under identical solution conditions and with the same molar concentration of the RNAs (Fig. 3). Strikingly, CYT-19 unwinds this duplex 50-fold less efficiently than when P1 is covalently attached to the ribozyme (Fig. 3B), suggesting that CYT-19 is recruited by forming additional contacts with the ribozyme. This interaction is most likely made with a different site on CYT-19 from the one responsible for RNA unwinding, because an interaction of the CYT-19 unwinding site with a site on the ribozyme away from P1 would most likely need to be broken to allow binding and unwinding of P1, such that the initial binding would be “nonproductive” and would therefore not increase the kcat/KM value (33).
Fig. 3.
CYT-19 unwinds the P1 duplex more efficiently when P1 is attached to the ribozyme than when it is free in solution. (A) Secondary structure of the ribozyme and P1 duplex. The black shaded region shows the nucleotides included in the model P1 duplex. The gray shaded region shows the additional nucleotides in the model duplex that includes P2 (see Results). (B) CYT-19-mediated unwinding of the isolated P1 duplex (○). Data from two independent determinations gave a second-order rate constant of (2.6 ± 0.3) × 105 M−1 min−1. Rate constants for CYT-19-mediated unwinding of the P1 duplex attached to the ribozyme are duplicated from Fig. 2C for comparison (□).
To further explore the possibility of a second RNA binding site on CYT-19, we compared the Mg2+ dependences for P1 duplex unwinding with and without the attached ribozyme. Although both reactions were less efficient at higher Mg2+ concentrations, the inhibition was stronger in the context of the intact ribozyme (Fig. 11, which is published as supporting information on the PNAS web site). These results provide further evidence for a second binding site, and they suggest that electrostatic interactions contribute substantially to RNA binding by this site or that the increased stability of the ribozyme at higher Mg2+ concentrations weakens CYT-19 binding.
CYT-19-Mediated Unwinding of the P1 Duplex Is Inhibited by Tertiary Docking of P1.
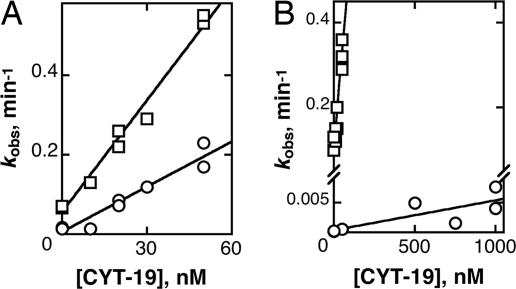
Initially to determine whether CYT-19 was more efficiently targeted to the misfolded ribozyme than the native ribozyme, we compared P1 duplex unwinding from the native and misfolded ribozyme species. CYT-19 efficiently unwound the P1 duplex from both the native and misfolded ribozymes, but the reaction was 2- to 3-fold less efficient from the native ribozyme (Fig. 4A). This result raised the possibility of a modest enhancement from features specific to the misfolded structure, but it was also possible that unwinding of the P1 duplex from the native ribozyme was inhibited by docking of P1 into stable tertiary contacts with the ribozyme core (34, 35). Because the small decrease in efficiency is approximately equal to the equilibrium constant for P1 docking to the native ribozyme under these conditions, it was possible that the inhibition from tertiary contact formation was very large but gave only a small effect because CYT-19 was able to unwind P1 efficiently during the time it spent undocked (see Calculation of Docking Equilibrium Constants in Supporting Text, which is published as supporting information on the PNAS web site).
Fig. 4.
Unwinding of the P1 duplex is inhibited by docking of P1 into the ribozyme core. (A) CYT-19-mediated unwinding of the P1 duplex from native (○) or misfolded (□) ribozyme. To prevent substrate cleavage by the native ribozyme, we used a modified substrate, −1d,rSA5, which has deoxyribose at the cleavage site and reacts poorly (61). The data gave second-order rate constants of (3.9 ± 0.2) × 106 M−1 min−1 and (9.3 ± 0.5) × 106 M−1 min−1 for P1 unwinding from native and misfolded ribozyme, respectively. (B) CYT-19-mediated dissociation of oligonucleotide product (CCCUCU) from native (○) or misfolded (□) ribozyme. These data gave second-order rate constants of (5.0 ± 1.7) × 103 M−1 min−1 and (4.5 ± 0.4) × 106 M−1 min−1 from native and misfolded ribozyme, respectively.
Therefore, we measured dissociation of a substrate that includes a 2′-methoxy substitution and does not form stable tertiary interactions with the native ribozyme (36) (see Table 1, which is published as supporting information on the PNAS web site, for a list of substrates). The preference for substrate dissociation from the misfolded ribozyme was eliminated by this substitution (Fig. 12, which is published as supporting information on the PNAS web site), indicating that CYT-19 does not distinguish between the native and misfolded structures but unwinds the P1 duplex less efficiently when it is docked against the ribozyme core. To confirm this conclusion and to explore the degree to which unwinding of P1 is inhibited by docking, we measured unwinding of a P1 duplex containing the oligonucleotide product (CCCUCU), which lacks the A5 tail of the substrate and docks much more strongly (37). This duplex was unwound much less efficiently from the native ribozyme, with a kcat/KM value 900-fold lower than from the misfolded ribozyme (Fig. 4B). This difference is the same within error as the equilibrium constant for docking of the product under these conditions, suggesting that CYT-19 possesses little or no unwinding activity for the docked P1 duplex and unwinds P1 only when it transiently undocks.
Specificity Determinants of the Unwinding Reaction.
We next investigated whether the efficient P1 duplex unwinding is specific to the Tetrahymena ribozyme and CYT-19 or a more general feature of the action of DExD/H-box proteins. We first tested whether the entire ribozyme is necessary or whether smaller extensions would also stimulate unwinding of P1 (Fig. 13, which is published as supporting information on the PNAS web site). Thus, we bound the substrate to an RNA oligonucleotide that includes only the internal guide sequence of the Tetrahymena ribozyme and the adjacent P2 helix (see Fig. 3A). Strikingly, CYT-19 unwound this P1 duplex 10-fold more efficiently than the minimal duplex. RNase T1 footprinting confirmed that the P2 base pairs are formed (Fig. 14, which is published as supporting information on the PNAS web site), suggesting that the increased efficiency arises from binding to the double-stranded P2 helix by the second RNA binding site on CYT-19. This binding is apparently not specific for the P2 sequence, because attachment of the unrelated P9.2 helix gave a significant enhancement. Attachment of 20-nt single-stranded extensions A20 or A4U16 gave slightly less efficient unwinding than attachment of P2. This result provides strong evidence against an alternative model in which CYT-19 first unwinds the P2 helix and then translocates into P1 to continue unwinding, because this model would predict that single-stranded extensions would give much larger enhancements of activity than double-stranded extensions (14). To provide further evidence against this model, we measured P1 unwinding from a construct that retains the 5′ sequence of P2 but has the 3′ sequence scrambled, eliminating the P2 base pairs. This construct did not give a significant enhancement of P1 unwinding relative to the double-stranded P2 extension, providing further evidence against obligatory unwinding of P2 and subsequent translocation. This conclusion is further supported by independent experiments demonstrating that DEAD-box proteins can unwind short duplexes without translocating (E. Jankowsky, personal communication). On the other hand, all of these single-stranded extensions gave significant enhancements relative to the P1 duplex alone, most simply suggesting that the second site on CYT-19 can bind to either single-stranded or double-stranded RNA. All of these model duplexes were unwound less efficiently than P1 in the context of the entire ribozyme, raising the possibility that the interaction between the ribozyme and the second site of CYT-19 extends beyond the structure immediately adjacent to P1.
We next investigated whether the efficient unwinding of the P1 duplex was specific to the Tetrahymena ribozyme or whether CYT-19 possesses similar activity with an RNA on which it functions in vivo. We designed and constructed a ribozyme from the minimal active form of the Neurospora mtLSU intron (38) by eliminating the 5′ and 3′ exons (39–41) (Fig. 15A, which is published as supporting information on the PNAS web site). CYT-19 unwound the P1 duplex from this ribozyme with the same efficiency as from the Tetrahymena ribozyme (kcat/KM = 1.5 × 107 M−1 min−1) (Fig. 15B), suggesting that the activity is a general property of CYT-19. Interestingly, the efficiency was unaffected by addition of the CYT-18 protein (Fig. 15B), which binds this intron RNA tightly and is required for efficient splicing (42–44), as well as for efficient substrate cleavage by this ribozyme version (H.B. and R.R., unpublished results). The lack of an effect of bound CYT-18 on P1 duplex unwinding by CYT-19 suggests that the RNA provides the primary recognition elements for CYT-19 action.
Finally, we tested whether a DExD/H-box protein that does not function with group I introns, but does interact functionally with multiple RNAs, is also enhanced for unwinding activity by the attachment of RNA structure. The Saccharomyces cerevisiae DED1 protein is involved in translation initiation of a range of mRNAs (45, 46), suggesting that it may function as a general RNA chaperone. In vitro, it has robust duplex unwinding activity (15) and can displace a protein from single-stranded RNA (20). DED1 also accelerated unwinding of the P1 duplex, and its efficiency was enhanced by attachment of the Tetrahymena ribozyme to approximately the same extent as that of CYT-19 (Fig. 16, which is published as supporting information on the PNAS web site). This result suggests that, like CYT-19, DED1 possesses an additional RNA binding site that can localize its action to structured RNAs.
Discussion
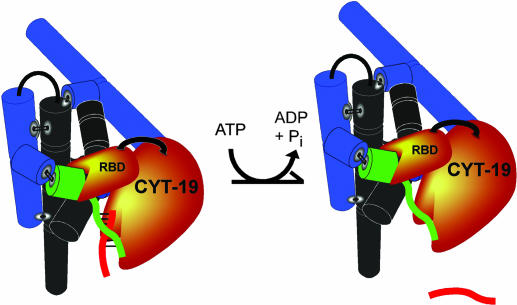
It is increasingly clear that many structured RNAs require DExD/H-box proteins to facilitate folding transitions. The CYT-19 protein was shown previously to accelerate splicing of several group I introns in vivo and in vitro (21) and to be able to facilitate folding of group II introns as well (23, 24). Here we used CYT-19 and a long-lived misfolded species of the Tetrahymena ribozyme to explore further how DExD/H-box proteins mediate RNA refolding reactions and how they are targeted to their physiological substrates. As described below and shown in Fig. 5, our results suggest a model for the general chaperone activity of CYT-19, features of which are likely to be general for the action of DExD/H-box proteins.
Fig. 5.
Model for general chaperone activity. CYT-19 binds to structured RNAs, including the Tetrahymena ribozyme (shown as black and blue cylinders representing the core and peripheral helices, respectively), via a binding site (labeled RBD) that is distinct from the one responsible for RNA unwinding activity. Binding of this site to the structured RNA brings the active site into proximity of the structured RNA, allowing it to unwind loosely associated structural elements like the P1 duplex (green and red strands). In general, unwinding of these duplexes gives the strands an opportunity to refold, but for P1, unwinding leads to dissociation of the noncovalently bound substrate (red). As described in Discussion, the additional RNA binding site on CYT-19 is likely to reside in a separate domain from the helicase motor domain as shown, but it is also possible that the site is formed by a loop or surface within the motor domain, or by the unwinding active site of a second CYT-19 monomer if CYT-19 functions as a multimer. The site apparently binds double-stranded RNA as shown (the P2 element is shown in green), and, because the efficiency of unwinding is increased by the presence of the rest of the ribozyme (see Results), there may be additional contacts.
Chaperone Action of CYT-19.
As a first step for its chaperone action, CYT-19 must bind to the structured RNA. We suggest that this initial binding occurs through an RNA binding domain (RBD) or surface that is distinct from the active site used for unwinding because attachment of the ribozyme, or even a simple duplex, to the isolated P1 duplex increases the efficiency of its unwinding by CYT-19. Presumably this initial binding functions to tether the unwinding active site in proximity to the RNA. Because the interaction appears to be at least as strong to double-stranded RNA, or even highly structured RNA, as single-stranded RNA, it is apparently distinct from the single-strand loading activity that is characteristic of conventional DNA and RNA helicases (16, 47).
Next, while remaining bound through this RBD to its initial attachment point on the RNA, CYT-19 can rearrange the RNA by unwinding nearby double-stranded regions. The unwinding activity is apparently much more efficient for duplex regions that are loosely associated with the RNA structure, because tertiary docking of the P1 duplex essentially abolishes unwinding (Fig. 4B). Presumably docking of P1 protects it from CYT-19 action by making it inaccessible to the unwinding active site. Binding by this active site may require diffusive entry of the duplex, because formation of tertiary contacts by P1 would, by definition, severely restrict motion relative to the RNA body. Although the undocked P1 duplex may form transient nonnative contacts with other regions of the ribozyme (36, 48), the substrate binds with the affinity of a simple duplex (35, 36), suggesting that the contacts are not stable and are not likely to severely restrict motion of the undocked P1 duplex.
The P1 base pairs are native base pairs, and thus unwinding P1 is not expected to be a productive reaction for CYT-19. However, we suggest that CYT-19 is likely to employ a similar mechanism in refolding RNAs to their native states, except that it may disrupt long-range tertiary contacts instead of, or in addition to, local secondary structure. The CYT-19-mediated refolding reaction of the misfolded Tetrahymena ribozyme is much less efficient than P1 duplex unwinding, perhaps because the structure that must be disrupted is packed tightly against the misfolded core or because more extensive unfolding is required. The action of CYT-19 in folding of its natural group I RNA substrates may also include displacement of the CYT-18 protein, which binds tightly to the RNAs and could increase the lifetimes of misfolded species (21, 49). Although previous results suggested, as one of several possibilities, that CYT-18 could provide a critical targeting function for CYT-19 (21), our results suggest that target recognition is mediated instead by RNA structure, as also suggested by recent findings that CYT-19 can chaperone folding of group I and group II introns in yeast in the absence of CYT-18 (23, 24).
Although our data do not establish what region of CYT-19 harbors the RNA binding site that enhances duplex unwinding (RBD in Fig. 5), a likely candidate is the C-terminal region, which includes ≈175 aa past the conserved motifs of the helicase motor domain. Such activity would be analogous to that of the accessory C-terminal domain of the bacterial protein DbpA and its ortholog YxiN (6–9), which recognizes a specific site within the 23S rRNA. Although the C-terminal region of CYT-19 does not include sequences that are readily identified as RNA binding motifs, it is highly basic, containing arginine-rich sequences reminiscent of those found in other RBDs (50). These features are shared by sequences at both the N and C termini of DED1, which displays a similar enhancement of unwinding activity upon attachment of the group I RNA structure (see Fig. 16).
The nonspecific targeting of CYT-19 to structured RNA and unwinding of a loosely associated duplex provide a physical model that may underlie prior observations that CYT-19 assists folding of a diverse set of RNAs in vitro and in vivo (21, 23, 24). On the other hand, this model begs the question of why CYT-19 was identified genetically as a factor required by group I introns (51). First, it is quite possible that CYT-19 functions in folding of additional RNAs. It was noted in the early work that cyt-19 mutants are also defective in 5′ end processing of certain pre-mRNAs (21, 51), suggesting additional functions for CYT-19. It is also possible that some functions were not detected because a second mitochondrial DExD/H-box protein may have overlapping activities. Finally, localization of CYT-19 to the mitochondria probably restricts the number of RNAs that it encounters and therefore limits its natural range of substrates below its in vitro capabilities. The Neurospora genome encodes ≈25 DEAD box proteins, including a putative ortholog of DED1, raising the possibility that other proteins function analogously as general RNA chaperones in other subcellular compartments.
Implications: A Model for General RNA Chaperone Activity.
The diversity of structured RNAs and their propensity to misfold suggest that there are many misfolded RNA species that require assistance from RNA chaperones in vivo (2, 52). Indeed, there may be a general necessity for chaperones during folding of RNAs, conceptually analogous to the general role of chaperones during the folding of proteins (reviewed in ref. 53). Some misfolded RNA species may be sufficiently difficult to resolve, or sufficiently important to resolve quickly, that a DExD/H-box protein is dedicated to that task alone (6–9). However, it would presumably not be feasible to dedicate a unique chaperone to each structured RNA, much less to each misfolded conformation of each RNA. Even some DExD/H-box proteins that were initially identified with a unique substrate have been found subsequently to interact functionally with additional RNAs (5, 54–56), and there is almost certainly a need for general chaperones that can facilitate folding of multiple RNAs, as demonstrated for CYT-19 and its yeast ortholog MSS116 (21, 23, 24).
But how do these chaperones interact with misfolded RNAs, and how do they resolve them? Two features of CYT-19 action observed here are likely to underlie its general chaperone activity, and other DExD/H-box proteins may share these properties. First, recognition of RNA structure, with limited specificity beyond the presence of secondary or tertiary structure, would be expected to allow an RNA chaperone protein to interact with multiple RNAs. A second challenge for a general chaperone is to determine which parts of the many RNA species it binds are misfolded and therefore need to be disrupted. We suggest that the strong preference of CYT-19 for unwinding double-stranded regions that are not stably associated with the core RNA structure is likely to be critical because nonnative structure is likely, in general, to be less able than native structure to form stable tertiary contacts with other regions of structured RNAs. Thus, this preference is expected to bias a chaperone protein toward disrupting nonnative structure, allowing it an opportunity to refold. The central features of this mode of action may also be applicable to DExD/H-box proteins that function on specific complexes, except that these proteins possess more specific RNA or protein binding domains and are therefore localized to a particular misfolded structure or assembly intermediate. The experimental system of the group I ribozyme and the model duplexes derived from it should be very useful for probing further which DExD/H-box proteins have properties similar to those of CYT-19 and thus may be well suited to function as general RNA chaperones.
Materials and Methods
Materials.
Ribozymes (L-21/ScaI Tetrahymena and Neurospora LSU) were prepared by in vitro transcription (39) and purified by using RNeasy columns (Qiagen, Valencia, CA) (57). RNA oligonucleotides (Dharmacon Research, Lafayette, CO) were 5′ end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase and purified by nondenaturing PAGE (58). CYT-19 was expressed and purified as described (21) with minor modifications (see Purification of CYT-19 in Supporting Text).
Activity Assay to Monitor CYT-19-Mediated Ribozyme Refolding.
Unless otherwise indicated, reactions were at 25°C with 50 mM Na-Mops (pH 7.0), 5 mM Mg2+, 50 mM KCl, 2 mM ATP-Mg2+, and 5% glycerol. Misfolded ribozyme (≈90%) was generated by incubation with 10 mM Mg2+ (10 min, 25°C). The Mg2+ concentration was then decreased, as desired, and CYT-19 was added to initiate refolding. Aliquots were quenched for refolding by adding MgCl2 to 50 mM, which dramatically increases the lifetime of the misfolded conformation (31). A series of control experiments demonstrated that CYT-19 is also inactivated under these conditions (Fig. 7), such that no significant refolding occurs after the addition of 50 mM MgCl2. The fraction of native ribozyme at each time was determined by adding trace 32P-labeled substrate (S*) and 500 μM G for 1 min. This time is sufficient for S* to bind to the native and misfolded ribozyme (kon = 108 M−1 min−1, ribozyme concentration = 30 nM) and for the native ribozyme to cleave S*, but not for S* to dissociate from the misfolded ribozyme (koff = 0.02 min−1). Thus, the fraction of S* cleaved reflects the fraction of native ribozyme (31) (Fig. 7D). S* was separated from its shorter cleavage product by 20% denaturing PAGE and quantitated by using a PhosphorImager (GE Healthcare, Fairfield, CT), and the time course was fit by a single exponential equation (Kaleidagraph; Synergy Software, Reading, PA).
Native Gel Mobility Shift Assay to Monitor P1 Duplex Unwinding.
Unless otherwise indicated, reactions were at 25°C with 50 mM Na-Mops (pH 7.0), 10 mM Mg2+, 50 mM KCl, 2 mM ATP-Mg2+, and 5% glycerol. The ribozyme was prefolded to the native or misfolded states by incubating with 10 mM Mg2+ at 50°C for 30 min (28) or at 25°C for 5 min (30, 59), respectively. Trace S* was added and allowed 5 min to bind. CYT-19 was then added with excess unlabeled S to initiate rapid and irreversible dissociation of S* (10 nM ribozyme, 2.5 μM unlabeled S, and ≥10 nM CYT-19), which was monitored by 20% native PAGE (30 mM Tris, 60 mM Mes, 0.1 mM EDTA, 10 mM MgCl2, and 300 mM KCl) at 5°C (see Fig. 2A) and quantitated as above. Unwinding of model P1 duplexes was followed identically except that higher concentrations and longer times were used to form the duplex (600 nM unlabeled ribozyme strand, 100 nM S, trace S*, ≥60-min incubation at 5°C). The duplex was diluted upon addition of excess S and CYT-19 so that its concentration was the same as that in experiments measuring S dissociation from the ribozyme (10 nM). All other solution conditions were identical.
Supplementary Material
Acknowledgments
We thank Sabine Mohr, Georg Mohr, Roland Saldanha, and Alan Lambowitz (University of Texas) for plasmids encoding CYT-19 and the Neurospora mtLSU intron and for advice on purification of CYT-19; Jacob Grohman for purification of CYT-19; Robert Coon for purification of CYT-18; Eckhard Jankowsky (Case Western Reserve University, Cleveland, OH) for the gift of the DED1 protein; and Sabine Mohr, Alan Lambowitz, and Eckhard Jankowsky for helpful comments on the manuscript. This work was funded by Welch Foundation Grant F-1563 (to R.R.) and National Institutes of Health Grant R01-GM070456 (to R.R.).
Abbreviations
- S
the oligonucleotide substrate CCCUCUA5
- S*
(5′-32P)-labeled S
- RBD
RNA binding domain
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
‡An alternative to weak binding would be slow and rate-limiting binding (kon = 9 × 104 M−1 min−1), which could produce the observed concentration dependence even if binding were tight. However, CYT-19 unwinds the P1 duplex of the ribozyme >100-fold faster than it refolds the ribozyme (see Fig. 2). This result sets a lower limit on the rate constant for CYT-19 binding the ribozyme of ≈107 M−1 min−1, and thus weak binding is indicated by the concentration dependence in the refolding reaction.
§The P1 duplex contains only 6 bp, so the ability of CYT-19 to unwind it does not indicate that CYT-19 is a processive helicase. Furthermore, the unwinding activity is decreased significantly upon lengthening the helix to 11 bp (R.R., unpublished results), suggesting a lack of processivity by CYT-19. Thus, the term “unwinding activity” is used here, rather than helicase activity, to avoid suggesting mechanistic or functional analogies with DNA or RNA helicases (60). It should be noted that higher processivity would probably not be useful for RNA chaperones in general, because RNAs of typical length (500 nt) are unlikely even to have potential to form any fortuitous, intramolecular Watson–Crick duplexes longer than 7 or 8 bp (see Fortuitous Helix Probabilities in Supporting Text).
¶The refolding assays of Fig. 1 were performed in the presence of 5 mM Mg2+, whereas the P1 unwinding experiments used 10 mM Mg2+. At 10 mM Mg2+, the kcat/KM value for ribozyme refolding was ≤2 × 103 M−1 min−1, 5,000-fold smaller than the kcat/KM value for P1 helix unwinding from the ribozyme (see Fig. 6).
References
- 1.Tanner NK, Linder P. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Herschlag D. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 3.Tinoco I, Jr, Bustamante C. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Treiber DK, Williamson JR. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 5.Silverman E, Edwalds-Gilbert G, Lin RJ. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 6.Nicol SM, Fuller-Pace FV. Proc Natl Acad Sci USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsu CA, Kossen K, Uhlenbeck OC. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kossen K, Karginov FV, Uhlenbeck OC. J Mol Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 9.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. J Biol Chem. 2005;280:35499–505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 10.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuman S. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DW, Gwack Y, Han JH, Choe J. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 13.Schwer B, Gross CH. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers GW, Jr, Richter NJ, Merrick WC. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 15.Iost I, Dreyfus M, Linder P. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 16.Jankowsky E, Gross CH, Shuman S, Pyle AM. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 17.Henn A, Medalia O, Shi SP, Steinberg M, Franceschi F, Sagi I. Proc Natl Acad Sci USA. 2001;98:5007–5012. doi: 10.1073/pnas.071372498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorsch JR, Herschlag D. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 19.Jankowsky E, Gross CH, Shuman S, Pyle AM. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 20.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 21.Mohr S, Stryker JM, Lambowitz AM. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 22.Lorsch JR. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- 23.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarrinkar PP, Williamson JR. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 26.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 27.Walstrum SA, Uhlenbeck OC. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 28.Herschlag D, Cech TR. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 29.Pan J, Woodson SA. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 30.Russell R, Herschlag D. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 31.Russell R, Herschlag D. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 32.Treiber DK, Williamson JR. J Mol Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999. [Google Scholar]
- 34.Herschlag D. Biochemistry. 1992;31:1386–1399. doi: 10.1021/bi00120a015. [DOI] [PubMed] [Google Scholar]
- 35.Bevilacqua PC, Kierzek R, Johnson KA, Turner DH. Science. 1992;258:1355–1358. doi: 10.1126/science.1455230. [DOI] [PubMed] [Google Scholar]
- 36.Narlikar GJ, Herschlag D. Nat Struct Biol. 1996;3:701–710. doi: 10.1038/nsb0896-701. [DOI] [PubMed] [Google Scholar]
- 37.Narlikar GJ, Gopalakrishnan V, McConnell TS, Usman N, Herschlag D. Proc Natl Acad Sci USA. 1995;92:3668–3672. doi: 10.1073/pnas.92.9.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo QB, Akins RA, Garriga G, Lambowitz AM. J Biol Chem. 1991;266:1809–1819. [PubMed] [Google Scholar]
- 39.Zaug AJ, Grosshans CA, Cech TR. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 40.Testa SM, Haidaris CG, Gigliotti F, Turner DH. Biochemistry. 1997;36:15303–15314. doi: 10.1021/bi9713097. [DOI] [PubMed] [Google Scholar]
- 41.Kuo LY, Davidson LA, Pico S. Biochim Biophys Acta. 1999;1489:281–292. doi: 10.1016/s0167-4781(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 42.Collins RA, Lambowitz AM. J Mol Biol. 1985;184:413–428. doi: 10.1016/0022-2836(85)90291-8. [DOI] [PubMed] [Google Scholar]
- 43.Majumder AL, Akins RA, Wilkinson JG, Kelley RL, Snook AJ, Lambowitz AM. Mol Cell Biol. 1989;9:2089–2104. doi: 10.1128/mcb.9.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Q, Lambowitz AM. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 45.de la Cruz J, Iost I, Kressler D, Linder P. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang RY, Weaver PL, Liu Z, Chang TH. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 47.Lohman TM, Bjornson KP. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 48.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 49.Webb AE, Rose MA, Westhof E, Weeks KM. J Mol Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]
- 50.Burd CG, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 51.Bertrand H, Bridge P, Collins RA, Garriga G, Lambowitz AM. Cell. 1982;29:517–526. doi: 10.1016/0092-8674(82)90168-4. [DOI] [PubMed] [Google Scholar]
- 52.Karpel RL, Miller NS, Fresco JR. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 53.Young JC, Agashe VR, Siegers K, Hartl FU. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 54.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Combs DJ, Nagel RJ, Ares M, Jr, Stevens SW. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russell R, Herschlag D. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaug AJ, Been MD, Cech TR. Nature. 1986;324:429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- 59.Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Proc Natl Acad Sci USA. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staley JP, Guthrie C. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 61.Herschlag D, Eckstein F, Cech TR. Biochemistry. 1993;32:8312–8321. doi: 10.1021/bi00083a035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.