Abstract
Depletion of intracellular calcium (Ca2+) stores induces store-operated Ca2+ (SOC) entry across the plasma membrane (PM). STIM1, a putative Ca2+ sensor in the endoplasmic reticulum (ER), has been recently shown to be necessary for SOC channel activation. Here we show that STIM1 dynamically moves in tubulovesicular shape on the ER and its subcompartment in resting living cells, whereas, upon Ca2+ store depletion, it is rapidly redistributed into discrete puncta that are located underneath, but not inserted into the PM. Normal constitutive movement of STIM1 is mediated through the coiled-coil and Ser/Thr-rich C-terminal domains in the cytoplasmic region of STIM1, whereas subsequent inducible puncta formation further requires the sterile α motif domain protruding into the ER lumen. Each of these three domains (coiled-coil, Ser/Thr-rich, and sterile α motif) was essential for activating SOC channels. Hence, our findings based on structure–function experiments suggest that constitutive dynamic movement of STIM1 in the ER and its subcompartment is obligatory for subsequent depletion-dependent redistribution of STIM1 into puncta underneath the PM and activation of SOC channels.
Keywords: B cell receptor, calcium signaling, DT40, store-operated calcium
Cytosolic Ca2+ signals are a key to the regulation of various physiological events (1, 2). Two stages of calcium mobilization have been distinguished in lymphocytes and other nonexcitable cells (3–5). The first stage involves activation of phospholipase C by trimeric G protein- or tyrosine kinase-coupled receptors. This enzyme hydrolyzes phosphatidylinositol bisphosphate to release the second messenger inositol-1,4,5-trisphosphate, which binds to its receptor in the endoplasmic reticulum (ER) membrane, thereby causing rapid but transient release of Ca2+ from ER stores. The second stage involves a sustained influx of extracellular Ca2+ across the plasma membrane (PM) in a process termed store-operated Ca2+ (SOC) entry. In this process, depletion of Ca2+ within the ER lumen serves as the primary trigger to open SOC channels residing in the PM.
STIM1 has recently emerged to play a critical role in coupling the first and second stages of calcium mobilization (6, 7). The STIM1 protein is thought to function primarily as a sensor of Ca2+ within the ER stores, because a single N-terminal EF-hand Ca2+ binding motif is located within the ER lumen (7, 8). The activation mechanism of STIM1, however, has remained elusive. For instance, Zhang and colleagues (9, 10) proposed that insertion of STIM1 from the ER to the PM, presumably through vesicular transport, would be a prerequisite for subsequent SOC channel activation. Furthermore, STIM1 in the PM has been reported to play a role for SOC activation (11). But others have shown that STIM1 redistributed into puncta near the PM without inserting into the PM and proposed that this aggregated STIM1 might activate SOC channels (7, 12).
To elucidate the mechanisms by which STIM1 activates SOC channels, we have constructed STIM1 mutants and assessed their dynamic translocation and functional consequences. Here we report that STIM1 undergoes dynamic steady-state movement in the ER and its subcompartment, and we suggest that this process is obligatory for subsequent depletion-mediated redistribution of STIM1 into puncta juxtaposed to the PM, leading to SOC activation.
Results
Impaired SOC Influx and Ca2+ Oscillation in the Absence of STIM1.
To examine the action mechanisms of STIM1, we first established STIM1-deficient DT40 B lymphocytes by a gene-targeting method. Lack of STIM1 was verified by Southern blotting, RT-PCR (data not shown), and immunoblotting analyses (Fig. 6A, which is published as supporting information on the PNAS web site). Expression of B cell receptor (BCR) in various mutant DT40 B cell lines in this study is presented in Fig. 6B. To determine the effect of STIM1 on SOC entry, we performed BCR or thapsigargin (TG) stimulation in the presence of 0.5 mM EGTA to deplete Ca2+ stores, and then 2 mM Ca2+ was added back to monitor Ca2+ influx. In STIM1-deficient DT40 B cells, a significant suppression of Ca2+ influx after BCR or TG stimulation was observed, whereas the Ca2+ influx was enhanced, more than WT DT40 cells, by an introduction of Flag-tagged WT STIM1 into STIM1-deficient cells (Fig. 1A). This exaggerated Ca2+ influx is presumably due to overexpression of Flag-STIM1. Because internal Ca2+ elevation was comparable between WT and STIM1-deficient DT40 B cells, STIM1 has no effect on Ca2+ pool size in the ER and the release of Ca2+ from stores after stimulation (Fig. 1A).
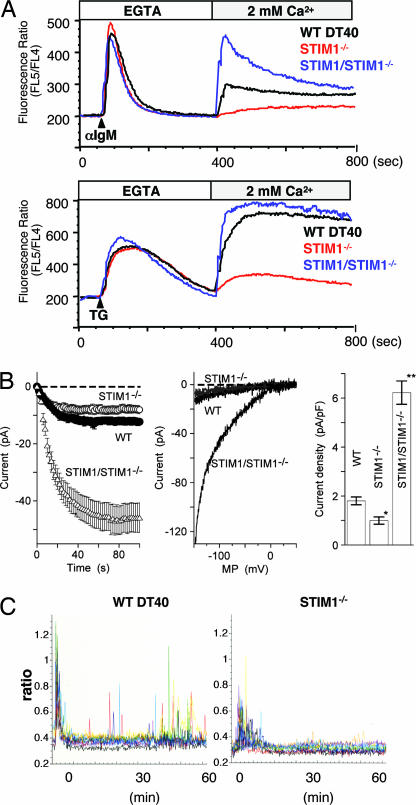
Fig. 1.
Suppression of SOC influx, Icrac, and Ca2+ oscillation in STIM1-deficient DT40 B cells. (A) The Ca2+ mobilization profiles were monitored by Ca2+ add-back using Indo-1 imaging. Ca2+ release was first evoked by the stimulation of BCR [anti-IgM mAb (M4)] (Upper) or TG (Lower) in Ca2+-free conditions (0.5 mM EGTA), and Ca2+ influx was induced by restoring the extracellular Ca2+ concentration to 2 mM in WT (black lines), STIM1-deficient (STIM1−/−; red lines), or Flag-tagged STIM1 transduced STIM1-deficient (STIM1/STIM1−/−; blue lines) DT40 B cells. FL4, 500–520 nm; FL5, 400–420 nm. (B) Average time courses of ionic currents evoked by 10 μM IP3 at −130 mV in WT DT40 cells (•; n = 10), STIM1−/− cells (○; n = 24), and STIM1/STIM1−/− cells (△; n = 14). Cytosolic calcium was clamped to near zero with 10 mM EGTA. Data are means ± SE. The whole-cell configuration of the patch-clamp recording was established at time 0 (Left). Representative leak-subtracted current–voltage relationships of WT DT40 (black), STIM1−/− (gray), and STIM1/STIM1−/− (black) are shown (Center). A comparison of current density was also done at −130 mV. Columns are the means ± SE (Right). (C) Ca2+ oscillation in STIM1-deficient DT40 B cells after BCR stimulation. Ca2+ responses in single cells upon BCR stimulation with anti-IgM mAb (M4) (1 μg/ml) were monitored in Fura-2 loaded WT DT40 (Left) and STIM1−/− (Right) cells.
The importance of STIM1 in SOC influx was further investigated by electrophysiological analyses. The best-studied SOC channels in terms of electrophysiological properties are Ca2+-release-activated Ca2+ channels, and they are clearly operational in hematopoietic cells (13). As shown in Fig. 1B, the patch-clamp assay revealed a reduction of Icrac (Ca2+-release-activated Ca2+ current) in STIM1-deficient DT40 cells in comparison with WT DT40 cells. Furthermore, introduction of WT STIM1 into STIM1-deficient DT40 B cells resulted in a considerable increase in Ca2+-release-activated Ca2+ channel activity.
In the presence of 2 mM Ca2+, WT DT40 B cells exhibited BCR-mediated Ca2+ oscillation, which decayed in a few minutes and resumed thereafter, lasting at least 1 h, whereas this Ca2+ oscillation in STIM1-deficient cells was almost completely inhibited (Fig. 1C). Based on these results, we conclude that STIM1 plays an essential role for Ca2+ influx through Ca2+-release-activated Ca2+ channels, thereby contributing to generation of long-lasting Ca2+ oscillation in B cells.
STIM1 Utilizes Multiple Functional Domains for Ca2+ Influx.
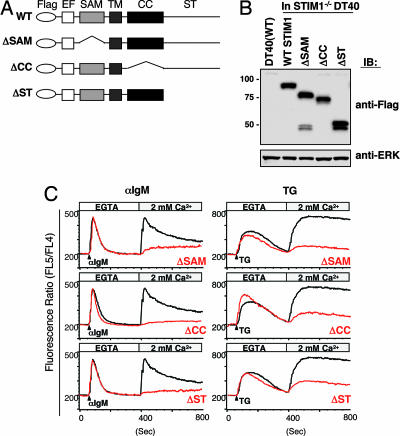
The above data validated the DT40 B cell system for elucidating mechanisms by which STIM1 activates SOC channels. STIM1 has several functional domains, depicted in Fig. 2A. To clarify which domains are required for SOC influx, we established STIM1-deficient DT40 B cells stably expressing Flag-tagged STIM1 lacking the sterile α motif [SAM (ΔSAM)], coiled-coil (ΔCC), or Ser/Thr-rich C-terminal (ΔST) domains (Fig. 2B) and examined whether these mutants have an ability to restore Ca2+ influx. The intensive restoration of BCR- or TG-evoked Ca2+ influx was observed in STIM1-deficient DT40 B cells expressing WT Flag-STIM1, but not Flag-STIM1 lacking the coiled-coil or Ser/Thr-rich domains (Fig. 2C). Introduction of Flag-STIM1ΔSAM slightly restored Ca2+ influx particularly in the BCR-evoked situation. These results indicate that STIM1 utilizes the SAM, coiled-coil, and Ser/Thr-rich domains for activating SOC channels.
Fig. 2.
STIM1 utilizes its multiple functional domains for Ca2+ influx. (A) Schematic representation of STIM1 mutants and the functional domains, including Flag-tagged, SAM, coiled-coil (CC), and Ser/Thr-rich C-terminal (ST) domains. Flag-tagged STIM1 cDNAs encoding deletion mutations were transfected into STIM1-deficient DT40 B cells. EF, EF-hand motif; TM, a single transmembrane. (B) Expression of Flag-STIM1 in various mutant DT40 cells. Whole-cell lysates were fractionated by SDS/PAGE and immunoblotted with anti-Flag mAb (Upper) or anti-ERK Ab (Lower). (C) STIM1 utilizes the multiple functional domains for Ca2+ influx. Intracellular Ca2+ release and influx in response to BCR with anti-IgM mAb (Left) and TG stimulation (Right) in STIM1-deficient DT40 cells expressing WT Flag-STIM1 (black lines) or mutants (red lines) were measured by Ca2+ add-back methods.
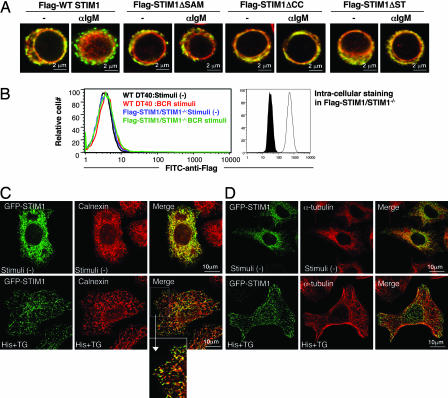
To determine why these mutants cannot activate SOC channels, the distribution of these mutants was investigated by immunofluorescence staining before and after BCR stimulation. At steady state, a large portion of the WT Flag-STIM1 colocalized with calnexin, an ER marker, and BCR stimulation induced its puncta formation near or within the PM (Fig. 3A). Similar results were obtained in the case of TG stimulation (data not shown). In significant contrast, Flag-STIM1 mutants did not accumulate into puncta after BCR (Fig. 3A) or TG treatment (data not shown). These results strongly suggest that redistribution of STIM1 into puncta near or within the PM is critical for activating SOC channels.
Fig. 3.
Requirement of multiple functional domains for redistribution of STIM1 to punctate subcellular compartments. (A) Flag-STIM1 (green) and calnexin (red) immunofluorescence staining of DT40 mutants before and after BCR stimulation (10 min) in the absence of extracellular Ca2+. All images were taken with confocal microscopy. (B) Extracellular and intracellular staining of Flag-STIM1. Flow cytometry using FITC-anti-Flag mAb was conducted in nonpermeabilized WT DT40 B cells before (black line) and after (red line) BCR stimulation or STIM1-deficient DT40 B cells expressing Flag-STIM1 before (blue line) and after (green line) BCR stimulation (Left). Permeabilized cells were stained with FITC-anti-Flag mAb. Filled and open histograms represent WT DT40 B cells and STIM1-deficient DT40 B cells expressing Flag-STIM1, respectively (Right). (C and D) Subcellular distribution of STIM1 in HeLa cells. HeLa cells expressing GFP-STIM1 were stimulated with 100 μM histamine (His) and 2 μM TG for 10 min and then fixed and processed for immunofluorescence with anti-calnexin Ab (C) or anti-α-tubulin mAb (D). An enlargement of the boxed region in the merge images from C is also shown. All images were taken with confocal microscopy.
Puncta Formation Occurs in the ER or Its Subcompartment near the PM.
It remains controversial whether STIM1 puncta, induced by store depletion, is located beneath the PM (7, 12) or is inserted into it (9, 10). To address this question, flow cytometry was conducted with or without permeabilization of the PM by using anti-Flag mAb that recognizes the extracellular region of Flag-STIM1 (Fig. 2A). In nonpermeabilized B cells, the fluorescence intensity of WT Flag-STIM1 was background level before stimulation and was not affected by BCR stimulation (Fig. 3B Left). In contrast, once permeabilized by saponin, substantial staining of Flag-STIM1 was observed (Fig. 3B Right). This observation was further substantiated by confocal microscope analyses; puncta formation was observed only in permeabilized cells (Fig. 3A), but not in nonpermeabilized cells (data not shown). In further support of the noninsertion model, we found that brefeldin A, which inhibits constitutive exocytosis through suppression of vesicular transport from the transitional ER (14), had no effect on either puncta formation or Ca2+ influx triggered by store depletion in DT40 B cells (data not shown). Based on these results, we conclude that the BCR-induced puncta formation of STIM1 occurs inside B cells close to the PM, but not within the PM.
B lymphocytes have limitations for defining the precise localization of STIM1 due to their tiny cytosols. Therefore, this point was further investigated with HeLa cells transiently transfected with WT GFP-STIM1. Although most GFP-STIM1 colocalized with ER markers, calnexin (Fig. 3C) or PDI (data not shown), a small fraction of GFP-STIM1 did not, suggesting that STIM1 distributes in a specialized subcompartment, in addition to the bulk ER, in resting cells. Store depletion induced by histamine and TG resulted in redistribution of GFP-STIM1 into puncta that concentrated inside the cells in the immediate vicinity of the ER near the PM (Fig. 3C). These spots, however, appear not to correspond to Golgi, vesicular–tubular transport carrier, or mitochondrial compartments, because GFP-STIM1 did not colocalize with such markers (anti-GM130 mAb for Golgi, anti-KDEL receptor Ab for Golgi and vesicular–tubular transport carrier, or MitoFluor Red for mitochondria) (data not shown). Hence, these spots most likely correspond to specialized subcompartments of the ER that are separated from the bulk ER.
Distinct Requirement for STIM1 Functional Domains in Its Constitutive Movement and Depletion-Mediated Puncta Formation.
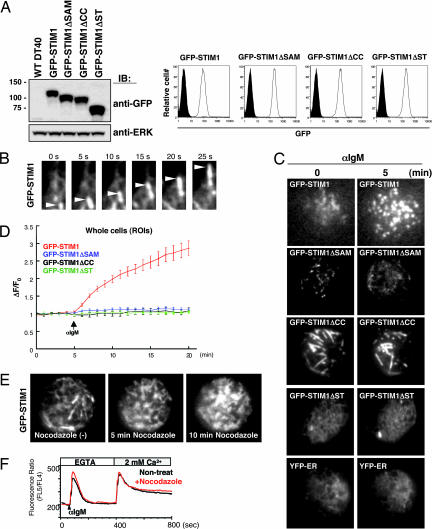
To further investigate the dynamic processes of STIM1 translocation near the PM, we used total internal reflection fluorescence microscopy, which selectively excites fluorescence within 100 nm of the PM (15). To visualize the dynamics of STIM1 in living cells, we established STIM1-deficient DT40 B cells expressing GFP-tagged WT STIM1 and their mutants. The expression of transfected proteins was confirmed by immunoblotting and flow cytometry (Fig. 4A). WT GFP-STIM1, but not its mutants, has the same ability to activate SOC influx as Flag-STIM1, judged by “Ca2+ add-back” experiments (Fig. 7, which is published as supporting information on the PNAS web site). Intriguingly, at steady state, WT GFP-STIM1 moved in tubulovesicular shape (Fig. 4B and Movie 1, which is published as supporting information on the PNAS web site). After BCR stimulation, the constitutive movement was rapidly decreased and much GFP-STIM1 drastically accumulated as puncta near the PM (Fig. 4C and Movie 2, which is published as supporting information on the PNAS web site). As shown in Fig. 4D, the kinetics of GFP-STIM1 accumulation near the PM is consistent with the previously shown time required for SOC activation (4). Very similar results were obtained with TG stimulation (data not shown). As was the case with WT GFP-STIM1, GFP-STIM1 lacking the SAM domain moved in a tubulovesicular shape (Movie 3, which is published as supporting information on the PNAS web site), but BCR stimulation resulted in no increase of accumulation near the PM (Fig. 4C and Movie 4, which is published as supporting information on the PNAS web site). GFP-STIM1 lacking the coiled-coil domain formed large tubular structures (Fig. 4C and Movie 5, which is published as supporting information on the PNAS web site); however, unlike WT GFP-STIM1, the coiled-coil domain mutation significantly decreased the velocity of the movement in unstimulated cells and BCR-induced puncta accumulation was impaired (Fig. 4C and Movie 6, which is published as supporting information on the PNAS web site). Furthermore, we found that GFP-STIM1 lacking the Ser/Thr-rich domain exhibited no movement in tubulovesicular shape in unstimulated cells (Movie 7, which is published as supporting information on the PNAS web site), and no puncta were formed in BCR-stimulated cells (Fig. 4C and Movie 8, which is published as supporting information on the PNAS web site). The movement pattern of STIM1ΔST was very similar to the distribution of YFP tagged with KDEL, an ER retention signal (YFP-ER) (Fig. 4C and Movie 9, which is published as supporting information on the PNAS web site), suggesting that the Ser/Thr-rich C-terminal domain is needed for transport from bulk ER to tubular granules. Hence, we conclude that STIM1 constitutively moves in a tubulovesicular shape through its coiled-coil and Ser/Thr-rich domains and that stimulation-mediated puncta accumulation further requires the SAM domain.
Fig. 4.
Microtubule-dependent dynamic behavior of GFP-STIM1 in living B cells. (A) cDNAs of GFP-tagged STIM1, STIM1 lacking SAM, CC, or ST domains were transduced in STIM1-deficient DT40 B cells, and these proteins were detected by immunoblotting with anti-GFP Ab (Left Upper). ERK protein was detected as a loading control (Left Lower). GFP expression in various DT40 mutants was detected by flow cytometry (Right). Filled and open histograms represent WT DT40 B cells and GFP-STIM1-transfected cells, respectively. (B) GFP signals of WT STIM1 near the PM were monitored by total internal reflection fluorescence microscopy at steady state. The movement of GFP-STIM1 was detected as tubular granules (arrowheads). Stills from the associated time-lapse Movies 1 and 2, taken from 410 s, are shown. (C) The movement of WT GFP-STIM1 and mutants were monitored before and after BCR stimulation in the absence of extracellular Ca2+. Stills from the associated time-lapse total internal reflection fluorescence movies are shown (Movies 1 and 2 for WT GFP-STIM1, Movies 3 and 4 for GFP-STIM1ΔSAM, Movies 5 and 6 for GFP-STIM1ΔCC, and Movies 7 and 8 for GFP-STIM1ΔST). The dynamics of YFP-ER are also shown (Movie 9). (D) Kinetic analysis of the average region of interest intensities of GFP-WT STIM1 (red line; n = 14), GFP-STIM1ΔSAM (blue line; n = 12), GFP-STIM1ΔCC (black line; n = 6), and GFP-STIM1ΔST (green line; n = 8). Cells were stimulated with anti-IgM mAb after 5 min in the absence of extracellular Ca2+. ΔF/Fo, the ratios of average intensities of regions of interest at indicated times after stimulation (F) and at steady state (Fo). Regions of interest are for whole cells. Error bars represent standard deviations of mean. (E) Dynamic behavior of GFP-STIM1 in DT40 cells treated with nocodazole (10 μM). Stills from the associated time-lapse Movie 10 are shown. (F) After 1 h of incubation of nocodazole, intracellular Ca2+ release and influx in response to BCR stimulation in DT40 B cells expressing Flag-STIM1 were monitored by Ca2+ add-back methods.
The immunofluorescence staining in HeLa cells (Fig. 3C) and movement of GFP-STIM1 in DT40 cells (Movies 1 and 2) suggested that tubulovesicular appearance of STIM1 aligned along linear structures, reminiscent of microtubule arrays. Indeed, as shown in Fig. 3D, WT GFP-STIM1 colocalized with α-tubulin in HeLa cells. Moreover, when DT40 cells were treated with nocodazole, which depolymerizes microtubules (data not shown), the tubular behavior of GFP-STIM1 disappeared dramatically and polarized granular structures of GFP-STIM1 were gradually observed near the PM (Fig. 4E and Movie 10, which is published as supporting information on the PNAS web site). Subsequent BCR stimulation of nocodazole-pretreated B cells resulted in a little increase of puncta formation under this condition (data not shown). Consistent with previous reports using basophilic cell lines (16) or lung cancer cells (17), BCR-mediated Ca2+ influx was, however, indistinguishable between nocodazol-treated and nontreated DT40 B cells (Fig. 4F). Assuming that both anterograde (from the bulk ER to the juxtamembrane ER regions) and retrograde (back to the bulk ER) movements of STIM1 are blocked by nocodazole, one of the explanations for these results is that nocodazole treatment may place STIM1 into the juxtamembrane regions by changing the balance between anterograde and retrograde movements in favor of the former. This mislocalization in the juxtamembrane regions may allow STIM1 to still activate SOC channels.
Cell Death Was Induced by Expression of the EF-Hand Mutant of STIM1 but Rescued by Introducing Additional STIM1 Mutations.
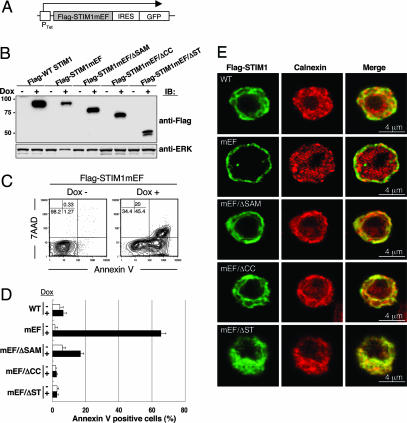
The data presented above suggested that STIM1, after sensing Ca2+ depletion by the EF hand, redistributes near the PM through its other functional domains, which in turn activates SOC channels. This hypothesis predicts that constitutive Ca2+ influx induced by mutation of the EF hand, mimicking Ca2+ depletion, would be suppressed by deletion of the coiled-coil, Ser/Thr-rich, or SAM domains. Because Flag-tagged STIM1 EF-hand mutant (STIM1mEF) might cause cell death, because of elevated Ca2+ influx before stimulation (9), we used a doxycycline-inducible expression system, with GFP linked via an internal ribosome entry site sequence to monitor protein expression by flow cytometry in DT40 B cells (Fig. 5A). The induced proteins were detected by immunoblotting (Fig. 5B). Induction of Flag-STIM1mEF protein led to significant detection of annexin V-positive (Fig. 5C), an early apoptosis marker, in a large population of GFP-expressing cells, whereas doxycycline-induced WT Flag-STIM1 had no effect on cell death (Fig. 5D). Because we observed extremely high Ca2+ concentrations in resting DT40 cells expressing Flag-STIM1mEF (data not shown), the cell death is most likely due to abnormally high Ca2+ concentrations through constitutive activation of SOC channels. Strikingly, introduction of deletions of the coiled-coil or Ser/Thr-rich domains into the Flag-STIM1mEF mutant resulted in no apoptosis (Fig. 5D). The Flag-mEF/ΔSAM mutant exhibited a slight induction of cell death; this leaky effect might correlate with a small restoration of Ca2+ influx induced by the STIM1ΔSAM mutation as shown in Fig. 2C. The Flag-STIM1mEF mutant was distributed near the PM, whereas double mutants had no effect on such constitutive localization near the PM (Fig. 5E).
Fig. 5.
Expression of STIM1 EF-hand mutants, but not double mutants of EF hand and its functional domains, induces cell death. (A) Schematic representation of the doxycycline-inducible construct of STIM1 EF-hand mutants. cDNA of Flag-tagged STIM1 EF-hand mutants (mEF; D76A/D78A/N80A/E87A) and double mutation of mEF in addition with ΔSAM (mEF/ΔSAM), ΔCC (mEF/ΔCC), and ΔST (mEF/ΔST) linked by an internal ribosome entry site (IRES) to a cDNA encoding EGFP were transfected into STIM1-deficient DT40 B cells. Ptet, doxycycline responsible promoter. (B) Expression of STIM1 mEF mutants in DT40 cells after doxycycline induction for 24 h was detected by immunoblotting with anti-Flag mAb (Upper). ERK protein was detected as a loading control (Lower). (C) The expression of STIM1 EF-hand mutants induced cell death. STIM1-deficient DT40 cells expressing Flag-STIM1mEF were stained with phycoerythrin-annexin V and 7-amino-actinomycin D before and after 24 h doxycycline (Dox) induction and were analyzed by flow cytometry. The induction of STIM1 mutants was monitored by GFP (data not shown), and doxycycline-induced cell death was detected in the GFP-positive gate. (D) Cells were stained with phycoerythrin-annexin V before and after doxycycline induction for 24 h and were analyzed by flow cytometry. Bar graphs show the percentage of annexin V-positive cells among total (Dox−) and GFP+ (Dox+) cells. These results are representative of three independent experiments (means ± SE). (E) Localization of various STIM1 EF-hand mutants. Various Flag-STIM1 (green) and calnexin (red) immunofluorescence staining was shown after 24 h of doxycycline induction.
Discussion
We now have shown that STIM1 multiple domains contribute to the dynamic movement in the ER and its subcompartment and to subsequent SOC entry. In contrast to the current belief that, in cells with replete Ca2+ stores, STIM1 is distributed uniformly throughout the bulk ER membrane, our results demonstrate that at least a small fraction of STIM1 distributes in the ER subcompartment under steady-state conditions. The existence of such vesicle-like specialized subcompartments of the ER (18) that move along microtubules (19–22) and have an ability to take and release Ca2+ (22) was also reported. Thus, our data further support the contention that the ER is a highly heterogenous compartment (23) and that this heterogeneity is at least partly established by nonuniform distribution of ER Ca2+-handling proteins (24, 25).
The importance of the cytoplasmic domains of STIM1 in both constitutive movement and depletion-dependent redistribution of STIM1 suggests a close relationship between these two processes. Thus, normal constitutive movement is likely to be obligatory for subsequent depletion-mediated redistribution of STIM1. Moreover, both processes might use similar or overlapping mechanisms. In the case of PM proteins, constitutive cycling requires a balance between constitutive exocytosis and endocytosis (26). Also, a stimulant is thought to change this balance, thereby modulating the expression of protein at the cell surface. Similarly, if the juxtamembrane ER region for activating SOC channels preexists, constitutive anterograde (from the bulk ER to this specified juxtamembrane ER region) and retrograde (back to the bulk ER) movements of STIM1 might take place. According to this model, in resting cells, the amount of STIM1 on the juxtamembrane ER region might not be sufficient to activate SOC channels. Upon Ca2+ store depletion, STIM1 could accumulate in the juxtamembrane ER regions by decreasing retrograde and/or increasing anterograde movement, or by generating a retention signal for STIM1 in this specialized ER region. Another possibility is that the specified juxtamembrane ER regions might be newly formed by store depletion, thereby allowing STIM1 to be redistributed to this region.
The issue of whether STIM1 translocates to the PM and functions on cell surface remains controversial. For example, Zhang et al. (9) showed that store depletion resulted in insertion of STIM1 within the PM. Overexpressed or native STIM1 was detected on the cell surface (11, 27, 28). In contrast, others demonstrated that STIM1 was not expressed on the cell surface, even if after store depletion (7, 12). The discrepancy of these observations might be due to the cell types used for experiments. Here in DT40 B cells we could not detect cell surface expression of WT Flag-STIM1 at steady state and after store depletion using flow cytometry and immunofluorescence methods. Thus, our results with STIM1-null background strongly argue that STIM1 in the PM is not essential for activating SOC influx. However, it should be mentioned that STIM1 in the PM in other cells such as HEK293 might play a modulatory role in activating SOC channels. Because we used Flag-tagged STIM1, it is still possible that exogenous small epitope-tag addition in our system may affect the distribution of STIM1. Even if so, the Flag-STIM1 protein is functional and sufficient for activating SOC channels (Fig. 1).
In line with a previous report (29), we found that STIM1 formed a homo-complex through the coiled-coil domain, but neither SAM nor Ser/Thr-rich domains, and that this complex was independent of store depletion in reconstituted 293T cells and DT40 B cells (Fig. 8, which is published as supporting information on the PNAS web site). Therefore, it is possible that the oligomeric structure is required for its rapid constitutive movement along microtubules. Because the coiled-coil domain in STIM1 contains the FERM (four-point one, ezrin, radixin, moesin) domain known as a binding site with several proteins and lipids in the PM (30, 31), it is also possible that this domain might have a role not only for the homo-complex formation but also for binding to the lipids or the cytoplasmic region of membrane proteins including SOC channels.
The Ser/Thr-rich domain contains many potential phosphorylation sites, the status of which might affect constitutive STIM1 movement. Indeed, STIM1 was initially purified as a phosphoprotein (27). Because most of the Ser/Thr-rich domain is missing in Drosophila STIM (9), it has been thought that this domain might not be of prime importance. However, considering that conserved Ser/Thr residues still exist in both Drosophila and mice, these residues might play a crucial role. Alternatively, the machinery needed to promote constitutive movement of STIM might differ between species.
In contrast to the coiled-coil and Ser/Thr domains, the SAM domain in the luminal side of STIM1 is required only for depletion-mediated puncta formation near the PM. From structural insights, the SAM domain in STIM1 is thought to function in heteromolecular interactions with other SAM domains or non-SAM domain-containing proteins (32). Hence, the unknown binding partner of the STIM1 SAM domain would be a key molecule for puncta formation as well as redistribution of STIM1 into the juxtamembrane ER regions.
One of the mechanisms for ensuring spatiotemporally restricted Ca2+ entry through SOC channels, most likely Orai families (12, 33–35), in the PM is that Ca2+ sensors in the ER are sequestered into the contact region between the ER and PM when and where they are needed. Our findings demonstrate the importance of STIM1 domains for its retention in this contact region and subsequent SOC influx. Further work should be aimed at defining the mechanism of how STIM1 moves in and out this specialized region by these domains; this information could be important as attempts are made to exploit the signaling pathway to activate SOC channels.
Materials and Methods
Detailed descriptions of all materials and methods are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Expression Constructs.
Flag- or GFP-tagged mouse WT STIM1 and its mutant cDNAs were generated by PCR. Each cDNA was cloned into the pApuro expression vector or the doxycycline-inducible vector pcDNA4/TO (Invitrogen). Flag or GFP was inserted immediately downstream of the predicted signal sequence of the STIM1 gene (36). The four point mutations in EF hand (D76A/D78A/N80A/E87A) were made by using QuikChange (Stratagene) according to manufacturer protocols. The deletion mutant of the SAM domain (ΔSAM: amino acids 122–199), the coiled-coil domain (ΔCC: 249–390), or the Ser/Thr-rich C-terminal domain (ΔST: 391 to stop codon) was generated by PCR. Nucleotide sequences of these constructs were verified by sequencing. These constructs were transfected into STIM1-deficient DT40 B cells by electroporation as described (37). Transient transfection using HeLa or 293T cells was performed by FuGENE 6 reagent (Roche Applied Science) according to manufacturer protocols.
Calcium Measurements.
For Ca2+ add-back method, 1 × 107 DT40 cells were loaded with Indo-1-AM and 0.015% Pluronic F127 (Invitrogen) in RPMI medium 1640 containing 1% FCS at 37°C for 45 min. Subsequently, cells were washed three times with Hanks' balanced salt solutions (Invitrogen) composed of 10 mM Hepes (pH 7.0) and 1% FCS. Before measurements, cells were resuspended in the above solution containing 0.5 mM EGTA. Cells were stimulated with 5 μg/ml anti-chicken IgM mAb (clone M4) or 2 μM TG at 60 s. The extracellular Ca2+ level was restored to 2 mM after 400 s. Changes in fluorescence intensity were monitored on an LSR (Becton Dickinson). Ca2+ oscillations were measured as described (38). In some experiments, cells were pretreated with nocodazole (10 μM) or brefeldin A (5 μg/ml) for 1 h at 37°C.
Supplementary Material
Acknowledgments
We thank our colleagues M. Kurosaki and T. Murakami for expert technical assistance and H. Ohno and P. W. Kincade for valuable discussions and a critical reading of the manuscript. This work was supported by a grant to T.K. from the Ministry of Education, Culture, Sports, Science, and Technology in Japan.
Abbreviations
- BCR
B cell receptor
- PM
plasma membrane
- TG
thapsigargin
- ER
endoplasmic reticulum
- SAM
sterile α motif
- SOC
store-operated Ca2+.
Footnotes
Conflict of interest statement: A.W. is on the advisory board of the RIKEN Institute and encouraged T.K. to pursue this work.
References
- 1.Berridge MJ, Lipp P, Bootman MD. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Lewis RS. Annu Rev Immunol. 2001;19:497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Gallo EM, Cante-Barrett K, Crabtree GR. Nat Immunol. 2006;7:25–32. doi: 10.1038/ni1295. [DOI] [PubMed] [Google Scholar]
- 4.Parekh AB, Putney JW., Jr Physiol Rev. 2005;85:757–810. doi: 10.1152/physrev.00057.2003. [DOI] [PubMed] [Google Scholar]
- 5.Venkatachalam K, van Rossum DB, Patterson RL, Ma HT, Gill DL. Nat Cell Biol. 2002;4:E263–E272. doi: 10.1038/ncb1102-e263. [DOI] [PubMed] [Google Scholar]
- 6.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putney JW., Jr J Cell Biol. 2005;169:381–382. doi: 10.1083/jcb.200503161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spassova MA, Soboloff J, He LP, Xu W, Dziadek MA, Gill DL. Proc Natl Acad Sci USA. 2006;103:4040–4045. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW., Jr J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parekh AB, Penner R. Physiol Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- 14.Klausner RD, Donaldson JG, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steyer JA, Almers W. Nat Rev Mol Cell Biol. 2001;2:268–275. doi: 10.1038/35067069. [DOI] [PubMed] [Google Scholar]
- 16.Bakowski D, Glitsch MD, Parekh AB. J Physiol. 2001;532:55–71. doi: 10.1111/j.1469-7793.2001.0055g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padar S, Bose DD, Livesey JC, Thomas DW. Biochem Pharmacol. 2005;69:1177–1186. doi: 10.1016/j.bcp.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Tsukita S, Ishikawa H. J Cell Biol. 1980;84:513–530. doi: 10.1083/jcb.84.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C, Chen LB. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 20.Dailey ME, Bridgman PC. J Neurosci. 1989;9:1897–1909. doi: 10.1523/JNEUROSCI.09-06-01897.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterman-Storer CM, Salmon ED. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- 22.Bannai H, Inoue T, Nakayama T, Hattori M, Mikoshiba K. J Cell Sci. 2004;117:163–175. doi: 10.1242/jcs.00854. [DOI] [PubMed] [Google Scholar]
- 23.Levine T, Rabouille C. Curr Opin Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Papp S, Dziak E, Michalak M, Opas M. J Cell Biol. 2003;160:475–479. doi: 10.1083/jcb.200207136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatti G, Trifari S, Mesaeli N, Parker JM, Michalak M, Meldolesi J. J Cell Biol. 2001;154:525–534. doi: 10.1083/jcb.200103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royle SJ, Murrell-Lagnado RD. BioEssays. 2003;25:39–46. doi: 10.1002/bies.10200. [DOI] [PubMed] [Google Scholar]
- 27.Manji SS, Parker NJ, Williams RT, van Stekelenburg L, Pearson RB, Dziadek M, Smith PJ. Biochim Biophys Acta. 2000;1481:147–155. doi: 10.1016/s0167-4838(00)00105-9. [DOI] [PubMed] [Google Scholar]
- 28.Soboloff J, Spassova MA, Hewavitharana T, He LP, Xu W, Johnstone LS, Dziadek MA, Gill DL. Curr Biol. 2006;16:1465–1470. doi: 10.1016/j.cub.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 29.Williams RT, Senior PV, Van Stekelenburg L, Layton JE, Smith PJ, Dziadek MA. Biochim Biophys Acta. 2002;1596:131–137. doi: 10.1016/s0167-4838(02)00211-x. [DOI] [PubMed] [Google Scholar]
- 30.Bretscher A, Edwards K, Fehon RG. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 31.Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, et al. Trends Biochem Sci. 1998;23:281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 32.Kim CA, Bowie JU. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 34.Peinelt C, Vig M, Koomoa DL, Beck A, Nadler MJ, Koblan-Huberson M, Lis A, Fleig A, Penner R, Kinet JP. Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soboloff J, Spassova MA, Tang XD, Hewavitharana T, Xu W, Gill DL. J Biol Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 36.Oritani K, Kincade PW. J Cell Biol. 1996;134:771–782. doi: 10.1083/jcb.134.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba Y, Hashimoto S, Matsushita M, Watanabe D, Kishimoto T, Kurosaki T, Tsukada S. Proc Natl Acad Sci USA. 2001;98:2582–2586. doi: 10.1073/pnas.051626198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyakawa T, Maeda A, Yamazawa T, Hirose K, Kurosaki T, Iino M. EMBO J. 1999;18:1303–1308. doi: 10.1093/emboj/18.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.