Abstract
The incorporation of small-molecule transition state structures into protein design calculations poses special challenges because of the need to represent the added translational, rotational, and conformational freedoms within an already difficult optimization problem. Successful approaches to computational enzyme design have focused on catalytic side-chain contacts to guide placement of small molecules in active sites. We describe a process for modeling small molecules in enzyme design calculations that extends previously described methods, allowing favorable small-molecule positions and conformations to be explored simultaneously with sequence optimization. Because all current computational enzyme design methods rely heavily on sampling of possible active site geometries from discrete conformational states, we tested the effects of discretization parameters on calculation results. Rotational and translational step sizes as well as side-chain library types were varied in a series of computational tests designed to identify native-like binding contacts in three natural systems. We find that conformational parameters, especially the type of rotamer library used, significantly affect the ability of design calculations to recover native binding-site geometries. We describe the construction and use of a crystallographic conformer library and find that it more reliably captures active-site geometries than traditional rotamer libraries in the systems tested.
Keywords: computational protein design, conformer library, enzyme catalysis, rotamer library
As catalysts, enzymes offer advantageous properties, including dramatic rate enhancements, complete control over absolute stereochemistry, and nontoxic biodegradation. Yet a fundamental limiting factor in the use of enzymes for chemical synthesis, bioremediation, therapeutics, and other applications is the availability of enzymes with the required activities, specificities, and tolerances to reaction conditions. It is therefore a major goal of computational protein design to be able to reliably create completely new protein catalysts with specific properties on demand.
A catalyst, by definition, must reduce the energy barrier for formation of the transition state. To design transition-state-stabilizing interactions, computational protein design groups have incorporated transition-state or high-energy intermediate state structures into design calculations. These efforts have yielded experimentally verified new catalytic proteins (1, 2). However, substantial challenges still prevent routine or reliable design of enzymes. One major challenge is finding energy functions fast enough for large calculations but that still provide informative approximations of electrostatic and desolvation effects in the protein environment (3, 4). This paper focuses on another fundamental challenge, the need to represent the large translational, rotational, and conformational freedoms of a small molecule within already astronomically large sequence design calculations.
Here we define protein design as the selection of amino acid sequences, such that the resulting protein occupies a given three-dimensional fold and has desired functional properties. Earlier experiments sought to redesign full-protein sequences or confer increased thermostability (5, 6), but newer work has successfully introduced other properties, including catalytic activity, conformational specificity, ligand affinity, and even novel protein folds (1, 2, 7–9). In these examples, side-chain placement algorithms were used to select from a set of discrete, probable side-chain rotamers by using energy functions tuned to produce thermostable proteins. These calculations represent difficult optimization problems (10), and they can also be large; a sample calculation performed on a typical enzyme active site yields >1065 possible sequence combinations, even when excluding movements of the small molecule.
The computational demands of sequence selection prevent ligand positioning using standard docking procedures, which often approximate or neglect side-chain flexibility (11). Approaches developed specifically for the purpose of enzyme and binding-site design have introduced other schemes to limit the calculation size. Looger et al. (8) used stationary, inflexible ligand poses in a large number of individual protein design calculations and demonstrated experimentally that several of the resulting proteins had high ligand affinity (8). Lilien et al. (12) reported and experimentally validated an ensemble-based method that allows ligand translation and rotation simultaneously with side-chain optimization but permits mutation of only two or three amino acid positions at a time. Chakrabarti et al. (13, 14) described a method for sequence design that neglects conformational and positional ligand flexibility and was not experimentally tested.
To design new enzyme active sites, a ligand placement method must be able to select side chains in many positions and must consider rotational, translational, and conformational freedom of the small molecule. The new catalytic proteins of Bolon and Mayo (1) and Dwyer et al. (2) were designed by treating high-energy-state structures of the reacting molecules as extensions of contacting amino acid side-chain rotamers. In the latter case, a two-step procedure was used, where ligands, anchoring side chains, and other catalytic side chains were placed through a geometric screening procedure, and surrounding side chains were designed in a second step (2, 15). We have developed a process for ligand placement in computational protein design calculations that expands upon previous work, and that allows ligand rotation, translation, and conformational freedom to be explored combinatorially within the sequence design calculation itself. The implementation of ligand-placement procedures within the context of the pairwise-decomposable protein design framework makes it possible to use a single energy function that can be parameterized as needed to reproduce experimental data.
We tested both a simple rotational and translational process for ligand placement as well as the previously used targeted ligand placement approach. A contact-based screening method is described that allows selection of ligand positions and conformations compatible with catalytic contacts. Test calculations in three systems, Escherichia coli chorismate mutase, Saccharomyces cerevisiae triosephosphate isomerase, and Streptomyces avidinii streptavidin, suggest that the success of ligand-placement procedures can be quite sensitive to conformational sampling parameters, including rotational and translational step sizes and the types of rotamer libraries used. We evaluated the efficacy of two standard rotamer libraries and two crystallographic conformer libraries. Traditional rotamers are constructed from canonical χ angles determined by statistical analysis of the Protein Data Bank (16–18), whereas conformers have Cartesian coordinates taken directly from high-resolution structures. Conformer libraries may allow more accurate modeling, because they are not limited to ideal geometries, and their sizes can be tuned more easily and naturally (19, 20). In our tests, a backbone-independent conformer library recovered wild-type-like active-site geometries more successfully than the other libraries, despite smaller size.
Results and Discussion
We have implemented and tested a process for incorporation of small molecules into computational protein design calculations. The procedure is general and may be used to place ground-state ligands or transition-state structures. It is also amenable to multistate design methods that seek to explicitly reflect the energy difference between reactant and transition states or among alternative ligands.
General Calculation Procedure.
Each ligand-placement calculation comprised five steps. In the first step, a large number of discrete variations of ligand coordinates was created. Initial sets of orientations were created by one of two methods, either simple rotation and translation or a targeted placement approach, both of which are discussed in more detail in subsequent sections. In the tests described here, each set of ligand variations contained 106-109 members, reflecting rotational and translational movement as well as internal conformational flexibility.
Next, the large number of substrate orientations was reduced to a manageable number (less than ≈20,000) by using both a simple hard-sphere steric potential to check for backbone clashes and a set of user-defined geometric criteria for side-chain/ligand contacts. In this work, geometric criteria were defined to reflect the distances, angles, and torsions characteristic of important catalytic contacts observed in the crystal structures (Fig. 1and Appendix, which is published as supporting information on the PNAS web site). In designing an enzyme with no naturally existing precedent, ideal contact geometries would be based on chemical intuition and/or quantum mechanical calculations. The geometric criteria were applied as follows. For every ligand variation, each of the geometric criteria was tested for satisfaction by contacts from any possible amino acid side-chain conformation in all designed protein positions. If a ligand variation was not able to make at least one of each type of user-specified contact, that ligand variation was discarded from the set. After geometric and steric pruning, the ligand variations remaining were only those theoretically capable of making each of the user-specified contacts.
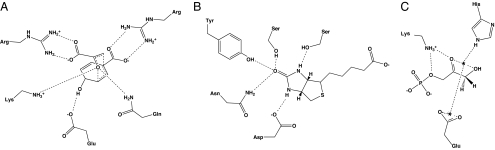
Fig. 1.
Contact geometries specified in small-molecule pruning step. Ranges of distances, angles, and torsions were allowed that included the crystallographic geometries. Exact geometry definitions are included in Appendix. (A) Chorismate mutase. (B) Biotin in streptavidin. (C) Triosephosphate isomerase Michaelis complex, modeled by using an approach similar to that of ref. 2. Asterisks indicate pseudoatoms used in geometry definitions.
In the third step, pairwise energies for all side-chain/side-chain, side-chain/backbone, backbone/ligand, and side-chain/ligand interactions were calculated with the full force field. In our work, this normally includes a scaled van der Waals term (21), hydrogen-bonding and electrostatic terms (22), and a solvation potential (23, 24).
The fourth step is an optional energy biasing that favors side-chain/ligand contacts deemed necessary for catalysis or binding. This energy-biasing step helps to overcome the shortcomings of molecular mechanics energy functions as well as the inherent limitation of treating a multistate design problem, differential stabilization of transition state relative to substrate in protein versus solution, using single-state design algorithms. As methods for modeling electrostatics and solvation and for designing over multiple states improve, the need for this biasing step should be reduced. Previous work used selective application of solvation energy (1) or an additional search algorithm step (8) for the same purpose. We favor the use of adjustable bias energies that can be tailored for specific purposes and investigated as a design variable.
To implement the bias, user-specified energies were added to or subtracted from pairwise side-chain/ligand interaction energies. We use the energy bias under two regimes, one for normal design calculations and another for rapid assessment of catalytic residue arrangements within a protein scaffold. In normal design calculations, a small energy benefit is simply applied to favor specified types of side-chain/ligand contacts. Alternatively, to quickly identify potential catalytic residues, exaggerated energetic benefits and penalties are applied together. A very large energy benefit is given for desired types of pairwise interactions (100 kcal/mol was used in the test cases reported here). An even larger energy penalty (10,000 kcal/mol here) is applied to all other pairwise side-chain/ligand interactions, except when the side chain is alanine or glycine. In other words, the energy penalty forces all designed side chains to alanine or glycine, unless they participate in user-specified catalytic contacts with the ligand. Although this process clearly does not yield physically relevant energetics, it offers a useful tool to investigate the catalytic conformational space within a binding pocket. The tests performed here to study the effect of sampling parameters on calculation results took advantage of this second approach. Calculations performed to demonstrate sequence selection used the normal design approach of applying a simple energy benefit to catalytic contacts.
Finally, in the fifth step, optimal sequences were identified by using the FASTER (25, 26) or HERO (27) search methods. In the test cases described here, the result reported is the lowest-energy sequence with the maximal number of specified contacts.
Rotation/Translation Search.
Simple rotation and translation can be used to fill the active site with an initial set of ligand variations in the first step of the process described. Because discrete steps must be used to rotate and translate the ligand, we evaluated the sensitivity of the calculation results to rotational and translational step sizes. A series of calculations was performed with an alanine-containing active-site background, as discussed in step four above. We first tested different rotational step sizes using the crystallographic translational starting position with three initial random rotations. Backbone-dependent and -independent rotamer and conformer libraries were tested. Each side-chain library was tested with and without inclusion of the specific crystallographic side-chain rotamers from the structure under examination.
As seen in Table 1, the results of these calculations (in terms of both rmsd relative to crystallographic position and number of wild-type contacts) strongly depended on the both the rotational step size and the rotamer library used. In the case of chorismate mutase, only the backbone-independent conformer library was able to find native-like geometry and contacts. Fig. 2shows results from this library with the 5° step size. When the crystallographic rotamers were included in the calculation, however, all four libraries returned native-like results. It should be noted that none of the three test case structures were included in the set of structures used to create the conformer libraries. The backbone-independent conformer library appeared the most consistently successful with the other two test cases as well, although it showed strong dependence on rotational step size in streptavidin.
Table 1.
rmsd and number of wild-type contacts as a function of rotational step size and rotamer library
| Rotamer library* | Rotational step size |
|||||
|---|---|---|---|---|---|---|
| 30° | 20° | 15° | 10° | 5° | ||
| Chorismate | Conformer: bb-ind | — | — | 0.61 ± 0.03 (4.0) | 0.55 ± 0.05 (4.0) | 0.47 ± 0.04 (4.7) |
| mutase | with xtal rotamers | — | — | 0.61 ± 0.03 (4.0) | 0.55 ± 0.05 (4.0) | 0.47 ± 0.04 (4.7) |
| Rotamer: bb-ind | — | — | 3.88 ± 0.37 (0.0) | 2.88 ± 1.44 (0.0) | 3.01 ± 1.61 (0.0) | |
| with xtal rotamers | — | — | 1.57 ± 1.70 (2.7) | 0.51 ± 0.00 (4.0) | 0.52 ± 0.01 (4.0) | |
| Conformer: bb-dep | — | — | 3.66 ± 0.11 (1.0) | 3.59 ± 0.08 (1.0) | 3.60 ± 0.09 (1.0) | |
| with xtal rotamers | — | 1.67 ± 1.78 (3.3) | 1.57 ± 1.83 (3.7) | 0.60 ± 0.08 (4.3) | 0.54 ± 0.06 (5.0) | |
| Rotamer: bb-dep | — | — | — | — | — | |
| with xtal rotamers | — | — | — | 0.49 ± 0.04 (4.3) | 0.52 ± 0.01 (4.0) | |
| Streptavidin-biotin | ||||||
| Conformer: bb-ind | — | — | — | — | 0.27 ± 0.09 (5.0) | |
| with xtal rotamers | — | 0.24 ± 0.09 (5.0) | 0.24 ± 0.07 (5.0) | 0.26 ± 0.06 (5.0) | 0.20 ± 0.13 (5.0) | |
| Rotamer: bb-ind | — | — | 0.77 ± 0.42 (2.3) | 0.60 ± 0.14 (3.0) | 0.60 ± 0.05 (2.7) | |
| with xtal rotamers | 0.37 ± 0.17 (5.0) | 0.24 ± 0.09 (5.0) | 0.24 ± 0.07 (5.0) | 0.26 ± 0.06 (5.0) | 0.30 ± 0.17 (5.0) | |
| Conformer: bb-dep | — | — | — | 0.25 ± 0.12 (5.0) | 0.20 ± 0.07 (5.0) | |
| with xtal rotamers | — | 0.24 ± 0.09 (5.0) | 0.24 ± 0.07 (5.0) | 0.22 ± 0.03 (5.0) | 0.29 ± 0.09 (4.0) | |
| Rotamer: bb-dep | — | — | — | 0.82 ± 0.28 (2.3) | 0.66 ± 0.02 (3.0) | |
| with xtal rotamers | — | 0.24 ± 0.09 (5.0) | 0.24 ± 0.07 (5.0) | 0.26 ± 0.06 (5.0) | 0.16 ± 0.06 (5.0) | |
| Triosephosphate | ||||||
| isomerase | Conformer: bb-ind | — | 1.87 ± 1.07 (0.7) | 3.59 ± 2.28 (1.0) | 0.28 ± 0.07 (3.0) | 0.24 ± 0.05 (3.0) |
| with xtal rotamers | — | 1.31 ± 0.29 (1.0) | 1.95 ± 2.28 (1.3) | 0.27 ± 0.06 (3.0) | 0.15 ± 0.02 (3.0) | |
| Rotamer: bb-ind | 5.09 ± 0.05 (0.3) | 0.60 ± 0.12 (1.7) | 0.55 ± 0.25 (2.3) | 0.34 ± 0.04 (2.3) | 0.25 ± 0.08 (3.0) | |
| with xtal rotamers | 5.06 ± 0.05 (0.3) | 0.60 ± 0.12 (2.0) | 0.37 ± 0.04 (3.0) | 0.25 ± 0.04 (3.0) | 0.15 ± 0.02 (3.0) | |
| Conformer: bb-dep | — | — | — | — | — | |
| with xtal rotamers | — | — | — | — | 0.15 ± 0.02 (3.0) | |
| Rotamer: bb-dep | 3.28 ± 0.73 (1.7) | 0.60 ± 0.12 (1.7) | 0.37 ± 0.05 (2.3) | 0.31 ± 0.04 (2.3) | 0.25 ± 0.08 (3.0) | |
| with xtal rotamers | 3.28 ± 0.73 (2.3) | 0.60 ± 0.12 (2.3) | 0.37 ± 0.05 (3.0) | 0.29 ± 0.03 (3.0) | 0.15 ± 0.02 (3.0) | |
Values are nonhydrogen-atom rmsd in Ångstroms relative to crystallographic ligands or bicyclic ring atom rmsd relative to crystallographic ligand for biotin (i.e., the pentanoic acid moiety was not considered in biotin rmsds). Averages and standard deviations from three random initial positions are reported. Numbers in parentheses are the number of contacts where the amino acid position was the same as in the wild-type structure, averaged over the three trials. Maximum possible number of wild-type contacts: chorismate mutase, five; streptavidin, five; and triosephosphate isomerase, three. Dashes indicate that required contacts were not satisfied in at least one of three trials.
*bb-ind, backbone-independent; bb-dep, backbone-dependent.
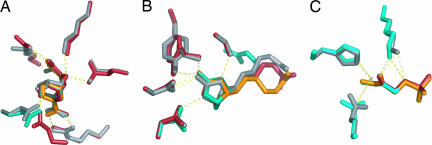
Fig. 2.
Sample results from test calculations presented in Table 1. Crystallographic side chains and ligands are shown in gray. Results from three trials using different initial random rotational positions are shown in red, teal, and orange. In cases where three colors are not visible, the selected rotamers from two or more calculations were identical. Results are shown from calculations with 5° rotation and the backbone-independent conformer library. (A) Chorismate mutase. An alternate backbone position was chosen for a glutamate-hydroxyl contact in one trial (red side chain, lower left). (B) Biotin in streptavidin. Note that the biotin pentanoic acid moiety samples different conformations in the calculation and the surrounding side chains were not designed. (C) Triosephosphate isomerase.
Next, we tested various combinations of rotational and translational step sizes starting from random initial ligand positions and using only the backbone-independent conformer library (Fig. 3and Table 3, which is published as supporting information on the PNAS web site). The crystallographic rotamers from the structures under investigation were not included in these calculations. The results show that, subject to the constraints imposed by the geometries defined in the pruning step and the biasing step, more than one combination of rotational and translational step size is viable for each test case, and the sensitivity of the result to step size varies among the test cases.
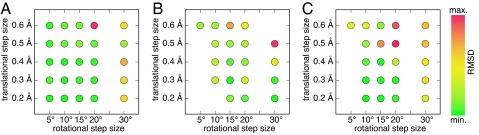
Fig. 3.
Effect of rotational and translational step sizes. Each spot represents the average of three trials with initial random starting positions. Missing points indicate that one or more trials could not identify wild-type-like contacts or else that the calculation was prohibitively large; no calculations were performed with the 25° rotational step size. Colors indicate nonhydrogen atom rmsd, as described in Tables 1–3. (A) Chorismate mutase (minimum, 0.53 Å; maximum, 2.61 Å). (B) Streptavidin–biotin (minimum, 0.57 Å; maximum, 2.05 Å). (C) Triosephosphate isomerase (minimum, 0.44 Å; maximum, 5.64 Å).
The rotation/translation tests were performed by using three initial random starting positions for each system. The starting positions were created by randomly rotating and translating the ligand within a 1-Å3 box around the ligand centroid (or the centroid of the bicyclic ring system in biotin). Using the same atom comparisons as described in Tables 1–3, the nine initial positions had rmsds relative to crystallographic positions of between 2.1 and 4.5 Å, with an average of 3.2 Å. These tests do not provide full unbiased searches of the active sites. Full active-site searches could be conducted by using this method by performing separate calculations for grid points distributed evenly through the active site. Given the time required to perform these smaller calculations (Table 3), searching an entire active site using rotational and translational perturbations would be computationally expensive. For example, examining a 3.6 × 3.6 × 3.6-Å grid using the 10° and 0.3-Å step sizes would require an estimated 324 h on a 16-processor cluster for placement of ligands and catalytic side chains in the chorismate mutase active site. Thus, for initial positioning of a ligand within an active site, rotational and translational placement is inefficient. However, the ability to adjust small-molecule position and conformation simultaneously with side-chain optimization should be extremely valuable for refining an initial position identified from a coarser search method.
Targeted Ligand Placement.
A second approach places the small molecule with reference to a contacting side chain (Fig. 4). In this approach, one or more small molecule variations are placed for every rotamer of the selected contacting side chain in every putative active-site position. This process has the advantage that ligand poses are targeted more efficiently to orientations that are able to make productive side-chain contacts. Previous computational enzyme design work used similar approaches (1, 2). In contrast to previous methods, however, our procedure does not maintain any association between the targeting rotamer and the small molecule; once the set of ligand conformations and orientations is constructed in step one, the ligand variations are all subjected to pruning, pairwise energy calculations, and optimization as independent entities in the calculation. An implication of this procedure is that a ligand may engage in a catalytic contact with a rotamer, amino acid, or protein position that differs from those of the side-chain rotamer that was originally used to place that ligand.
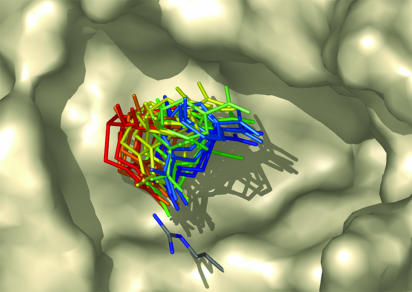
Fig. 4.
Targeted placement procedure. For a given side-chain rotamer, small-molecule ligands are placed such that they are able to meet specified geometric criteria. This is repeated for every possible conformation of the amino acid at every designed position. Shown is a subset of orientations of a chorismate mutase transition-state structure in contact with one conformation of arginine. This figure was created with PyMOL (40).
We tested the effect of four types of side-chain libraries on the ability of a targeted placement process to find wild-type-like ligand positions and contacts. For the three test cases, the following side-chain contacts were used to anchor the ligand: chorismate mutase, C11 carboxylate to arginine; streptavidin, N1 to aspartate; and triosephosphate isomerase, O2 and O3 to histidine. For each contact type, variations were allowed in the geometry of the contact, including the contacting atoms (NH1-NH2 vs. NE-NH1 for arginine) and variations in defined distances, angles, and dihedrals of the contact (see Appendix).
As with the rotational and translational search, success in achieving native active-site conformations depended highly on the side-chain library used (Table 2). Only the backbone-independent conformer library yielded results for all three test cases that were comparable to those with crystallographic rotamers included. Using that library, all three systems returned all wild-type contacts with low ligand rmsd relative to the crystallographic position. As with the rotation/translation search, the chorismate mutase case showed the strongest sensitivity to rotamer library. Inspection of the structures revealed that an arginine side chain (Arg 28) occupies a conformation in the inhibitor-bound crystal structure that was not well approximated in the other rotamer libraries.
Table 2.
Results from targeted placement procedure as a function of rotamer library
| Rotamer library* | log (initial ligand variations) | rmsd, ņ (WT contacts) | Time, hours‡ | ||||
|---|---|---|---|---|---|---|---|
| Chorismate | Conformer: bb-ind | 7.88 | 0.60 (5) | 16 | |||
| mutase | with xtal rotamers | 7.88 | 0.68 (3) | 18 | |||
| Rotamer: bb-ind | 8.18 | 3.61 (0) | 51 | ||||
| with xtal rotamers | 8.18 | 0.66 (4) | 62 | ||||
| Conformer: bb-dep | 7.64 | 3.62 (1) | 8 | ||||
| with xtal rotamers | 7.64 | 0.68 (4) | 9 | ||||
| Rotamer: bb-dep | 7.76 | 2.31 (1) | 14 | ||||
| with xtal rotamers | 7.76 | 0.66 (4) | 16 | ||||
| Streptavidin- | Conformer: bb-ind | 7.07 | 0.64 (5) | 1.4 | |||
| biotin | with xtal rotamers | 7.07 | 0.64 (5) | 1.4 | |||
| Rotamer: bb-ind | 7.20 | 0.54 (4) | 3.5 | ||||
| with xtal rotamers | 7.20 | 0.34 (4) | 3.4 | ||||
| Conformer: bb-dep | 6.35 | 0.37 (5) | 0.2 | ||||
| with xtal rotamers | 6.35 | 0.54 (4) | 0.2 | ||||
| Rotamer: bb-dep | 7.17 | 3.50 (0) | 2.6 | ||||
| with xtal rotamers | 7.17 | 0.19 (5) | 2.8 | ||||
| Triosephosphate | Conformer: bb-ind | 7.31 | 0.49 (3) | 1.3 | |||
| isomerase | with xtal rotamers | 7.31 | 0.49 (3) | 1.3 | |||
| Rotamer: bb-ind | 7.78 | 0.46 (3) | 8.7§ | ||||
| with xtal rotamers | 7.78 | 0.46 (3) | 8.7§ | ||||
| Conformer: bb-dep | 6.82 | 7.51 (0) | 0.3 | ||||
| with xtal rotamers | 6.82 | 0.78 (3) | 0.3 | ||||
| Rotamer: bb-dep | 7.58 | 0.51 (3) | 4.3§ | ||||
| with xtal rotamers | 7.58 | 0.51 (3) | 4.9§ |
*bb-ind, backbone-independent; bb-dep, backbone-dependent.
†rmsds calculated as described in Table 1. Maximum possible number of wild-type contacts: chorismate mutase, five; streptavidin, five; and triosephosphate isomerase, three.
‡Wall clock time; calculations were performed on a 16-processor cluster.
§Calculation was performed as a series of smaller calculations.
The targeted placement approach allowed a thorough and directed search of active-site conformational space, including between 106 and 109 small-molecule orientations and conformations spread throughout the active site. In contrast to the rotation/translation method, a full active-site search took between 1 and 18 hours to complete using the backbone-independent conformer library, and no initial starting position was required. This method offers an efficient first step for defining active-site geometry in a new protein scaffold. One shortcoming is that it may be difficult to sample the many geometrical variations of a flexible hydrogen-bonding interaction. For example, the 972 variations in guanidinocarboxylate contact geometry sampled in the chorismate mutase case are probably adequate to reflect flexibility in this relatively rigid dual hydrogen-bonding interaction (see Appendix). A less-restrained interaction, however, such as a serine hydrogen bonding with a sterically unrestricted ligand carbonyl oxygen, results in a compromise between maintaining a manageable calculation size and modeling contact flexibility. One solution is to use a targeted method to find an initial ligand position within the binding site and then, in a second calculation, optimize both active-site packing and fine rotational and translational placement of the ligand.
Sequence Design.
The computational tests described in the previous sections were designed to evaluate the effects of calculation parameters on recovery of native enzyme geometries, and the design of active-site residues was limited to catalytic side chains. However, the general procedure described here is equally amenable to full active-site design calculations.
In previously published work, 18 active-site residues of E. coli chorismate mutase were redesigned simultaneously with rotational and translational relaxation of the transition-state structure from the starting crystallographic position (28). The six predicted mutations were experimentally investigated, and some were found to confer increased catalytic efficiency (28) or thermostability (J.K.L., J. R. Keeffe, and S.L.M., unpublished results). A detrimental mutation predicted in the study underscored the importance of continued work on energy functions. In the calculation that motivated this experimental work, the initial starting position of the small molecule was taken from the crystal structure, and a limited degree of rotational and translational optimization was used.
We performed a test calculation to demonstrate that small molecules can be placed simultaneously with full active-site side-chain optimization, without reference to any known starting position. In a sample calculation using E. coli chorismate mutase, the targeted placement method was used to identify 107 small-molecule variations. In this example, after the geometric pruning step and elimination of variants with backbone steric clashes, 155 small-molecule variations remained. These variants were evaluated combinatorially with 10 different side-chain identities in 12 active-site positions. Using FASTER for optimization, the calculation took ≈9 h to complete on a 16-processor cluster with ≈70% of the total calculation time consumed in calculating a surface-area-based solvation term.
Conclusions
The described procedures allow the incorporation of small-molecule placement directly into sequence-design calculations. The test calculations performed suggest that the results of computational enzyme design processes can be quite sensitive to calculation parameters, including the rotamer library used and the coarseness of ligand positioning. These results emphasize that the conformational space of a calculation must be explored before meaningful conclusions can be reached about energy functions.
Given that we still have much to learn about the complex relationship between protein structure and catalytic activity (29, 30), luck and choice of system may continue to play a role in the success of de novo computational enzyme design efforts for some time. However, the power of computational enzyme design to stringently evaluate our understanding of the energetics of catalysis should not be overlooked. Experimental feedback gained from both successful and unsuccessful designs will make it possible to critically examine energy functions for modeling active sites. Using quality transition-state structures derived from ab initio calculations and experimental evidence will help computational design experiments to provide more meaningful information about the effectiveness of energy functions. The use of large side-chain structural libraries and fine movements of transition-state structures will help to reduce errors from conformational sampling. Backbone relaxation and multistate design will offer other important tools to improve the value of design calculations. Finally, the construction of gene libraries or large numbers of computationally designed variants has great potential for overcoming the shortcomings of enzyme design models (31), but results from these experiments will be most useful for furthering our understanding of catalysis and design if both active and inactive variants are reported. By critically evaluating current methods for computational enzyme design, we will move closer to a deeper and more practically useful understanding of the sequence determinants of enzyme activity in the future.
Methods
Structures and Charges.
Protein Data Bank files were used without minimization [E. coli chorismate mutase, 1ecm (32); S. avidinii streptavidin, 1mk5 (33); and S. cerevisiae triosephosphate isomerase, 1ney (34)]. Hydrogens were added with Reduce (35).
A library of ligand internal conformations was created for each system as follows.
Chorismate mutase.
An HF/6–31G* ab initio transition-state structure (36) was used with only one variation; the O4 hydroxyl proton was allowed to occupy three positions, 60°, 180°, and −35°, defined by the H-C-O-H dihedral angle. The minima in a torsional profile at the HF/6–31G* level were at ≈180° and −35°, and 60° was included as an option because hydrogen-bonding patterns in chorismate mutases from other species suggested population of that region of torsional space.
Streptavidin.
Four rotatable bonds in biotin were allowed to occupy three positions each (60°, −60°, 180° for sp3-sp3 bonds, and 30°, 90°, 150° for the symmetric carboxylate group). Thirty-four conformations were excluded because of high internal energy calculated by using the van der Waals component of the DREIDING force field (37).
Triosephosphate isomerase.
The Protein Data Bank structure used was the Michaelis complex with the substrate dihydroxyacetone phosphate. In ground-state dihydroxyacetone phosphate, two rotatable bonds (defined by the P-O-C-C and C-C-O-H dihedral angle) were allowed to occupy three positions each (60°, −60°, and 180°). Three conformations were excluded because of high internal DREIDING van der Waals energy.
Ligand atomic charges were obtained by fitting charges to electrostatic potential from HF/6–31G* single-point energy calculations using the transition-state structure (chorismate mutase) or crystallographic ground-state structure (biotin, dihydroxyacetone phosphate). Ab initio calculations and charge determinations were performed by using Spartan (Wavefunction, Irvine, CA) or Jaguar (Schrödinger, San Diego, CA).
Side-Chain Rotamer Libraries.
Standard backbone-dependent and -independent rotamer libraries were used with expansion by 1 SD about χ1 and χ2 (17).
Crystallographic conformer libraries were prepared by using coordinates from 149,813 residue side chains selected from 1,011 unique structures as described in the Appendix and Figs. 5 and 6, which are published as supporting information on the PNAS web site. A clustering algorithm was developed based on ideas described by Shetty et al. (20) and is detailed in Appendix and Figs. 5 and 6. The algorithm allows the construction of both backbone-dependent and -independent libraries to custom sizes by using a granularity factor to define the desired degree of similarity between independent conformers. In this work, granularity factors of 0.3 and 1.0 Å were used for backbone-dependent and -independent rotamer libraries, respectively.
For all calculation types, conformer libraries were smaller than the standard rotamer libraries. As an example, the number of side-chain conformations for the chorismate mutase calculations described in Table 2 were as follows: backbone-independent rotamer, 14,229; backbone-independent conformer, 5,955; backbone-dependent rotamer, 7,945; and backbone-dependent conformer, 5,539.
Calculation Parameters.
All non-Gly and non-Pro residues reasonably within the natural active sites were included in calculations. Residues with any atom within a 5-Å radius from any atom in the crystallographic ligands were included, minus those residues separated from the natural ligand by backbone elements and plus a few adjacent residues not within the 5-Å cutoff. The positions designed were (all in chain A unless otherwise designated): chorismate mutase, 28, 32, 35, 39, 46, 47, 48, 51, 52, 55, 81, 84, 85, 88, 7B, 11B, 14B, and 18B; streptavidin, 23, 24, 25, 27, 43, 45, 46, 47, 49, 50, 79, 86, 88, 90, 92, 108, 110, 112, 128, and 130; and triosephosphate isomerase, 10, 12, 95, 97, 165, 170, 211, and 230.
In ligand-placement test cases, designed residues were restricted to ligand-contacting residues or alanine as follows: Arg, Lys, Gln, Glu, or Ala in chorismate mutase; Ser, Asn, Tyr, Asp, or Ala in streptavidin; and Glu, His, Lys, or Ala in triosephosphate isomerase. Four calculations on triosephosphate isomerase were run as smaller component calculations, as indicated in Table 2, because of prohibitive size as a single calculation.
Energy Functions and Optimization.
Energy functions included scaled van der Waals (21), hydrogen-bonding, and electrostatic terms (22). A surface-area-based solvation potential (23) was used in sequence design calculations but not for ligand placement, where solvation energy would have been heavily outweighed by geometric considerations. Sequences were optimized with respect to the energy function using FASTER (25, 26) or HERO (27). On occasion, a top-ranked sequence contained more than one instance of a given specified geometric contact, because of the energy benefit applied for these contacts. In these cases, Monte Carlo (38, 39) was used to sample around the global minimum energy sequence, and the top-ranked sequence with a single instance of each geometric contact was reported.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, the Ralph M. Parsons Foundation, the Defense Advanced Research Projects Agency, the U.S. Army Research Office Institute for Collaborative Biotechnologies, and an IBM Shared University Research Grant.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bolon DN, Mayo SL. Proc Natl Acad Sci USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwyer MA, Looger LL, Hellinga HW. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 3.Mendes J, Guerois R, Serrano L. Curr Opin Struct Biol. 2002;12:441–446. doi: 10.1016/s0959-440x(02)00345-7. [DOI] [PubMed] [Google Scholar]
- 4.Vizcarra CL, Mayo SL. Curr Opin Chem Biol. 2005;9:622–626. doi: 10.1016/j.cbpa.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Dahiyat BI, Mayo SL. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 6.Malakauskas SM, Mayo SL. Nat Struct Biol. 1998;5:470–475. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 7.Shimaoka M, Shifman JM, Jing H, Takagi J, Mayo SL, Springer TA. Nat Struct Biol. 2000;7:674–678. doi: 10.1038/77978. [DOI] [PubMed] [Google Scholar]
- 8.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 9.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 10.Pierce NA, Winfree E. Prot Eng. 2002;15:779–782. doi: 10.1093/protein/15.10.779. [DOI] [PubMed] [Google Scholar]
- 11.Taylor RD, Jewsbury PJ, Essex JW. J Comput Aided Mol Des. 2002;16:151–166. doi: 10.1023/a:1020155510718. [DOI] [PubMed] [Google Scholar]
- 12.Lilien RH, Stevens BW, Anderson AC, Donald BR. J Comput Biol. 12:740–761. doi: 10.1089/cmb.2005.12.740. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti R, Klibanov AM, Friesner RA. Proc Natl Acad Sci USA. 2005;102:10153–10158. doi: 10.1073/pnas.0504023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakrabarti R, Klibanov AM, Friesner RA. Proc Natl Acad Sci USA. 2005;102:12035–12040. doi: 10.1073/pnas.0505397102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellinga HW, Richards FM. J Mol Biol. 1991;222:763–785. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 16.Ponder JW, Richards FM. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 17.Dunbrack RL, Jr, Cohen FE. Prot Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovell SC, Word JM, Richardson JS, Richardson DC. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 19.Xiang Z, Honig B. J Mol Biol. 2001;311:421–430. doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- 20.Shetty RP, de Bakker PIW, DePristo MA, Blundell TL. Prot Eng. 2003;16:963–969. doi: 10.1093/protein/gzg143. [DOI] [PubMed] [Google Scholar]
- 21.Dahiyat BI, Mayo SL. Proc Natl Acad Sci USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahiyat BI, Gordon DB, Mayo SL. Protein Sci. 1997;6:1333–1337. doi: 10.1002/pro.5560060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Street AG, Mayo SL. Fold Des. 1998;3:253–258. doi: 10.1016/S1359-0278(98)00036-4. [DOI] [PubMed] [Google Scholar]
- 24.Lazaridis T, Karplus M. Prot Struct Funct Genet. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 25.Desmet J, Spriet J, Lasters I. Prot Struct Funct Genet. 2002;48:31–43. doi: 10.1002/prot.10131. [DOI] [PubMed] [Google Scholar]
- 26.Allen BD, Mayo SL. J Comput Chem. 2006;27:1071–1075. doi: 10.1002/jcc.20420. [DOI] [PubMed] [Google Scholar]
- 27.Gordon DB, Hom GK, Mayo SL, Pierce NA. J Comput Chem. 2003;24:232–243. doi: 10.1002/jcc.10121. [DOI] [PubMed] [Google Scholar]
- 28.Lassila JK, Keeffe JR, Oeschlaeger P, Mayo SL. Protein Eng Des Sel. 2005;18:161–163. doi: 10.1093/protein/gzi015. [DOI] [PubMed] [Google Scholar]
- 29.Kraut DA, Carroll KS, Herschlag D. Annu Rev Biochem. 2003;72:517–571. doi: 10.1146/annurev.biochem.72.121801.161617. [DOI] [PubMed] [Google Scholar]
- 30.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 31.Bolon DN, Voigt CA, Mayo SL. Curr Opin Chem Biol. 2002;6:125–129. doi: 10.1016/s1367-5931(02)00303-4. [DOI] [PubMed] [Google Scholar]
- 32.Lee AY, Karplus PA, Ganem B, Clardy J. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 33.Hyre DE, Le Trong I, Merritt EA, Eccleston JF, Green NM, Stenkamp RE, Stayton PS. Protein Sci. 2006;15:459–467. doi: 10.1110/ps.051970306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jogl G, Rozovsky S, McDermott AE, Tong L. Proc Natl Acad Sci USA. 2003;100:50–55. doi: 10.1073/pnas.0233793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Word JM, Lovell SC, Richardson JS, Richardson DC. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 36.Wiest O, Houk KN. J Org Chem. 1994;59:7582–7584. [Google Scholar]
- 37.Mayo SL, Olafson BD, Goddard WA. J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 38.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 39.Voigt CA, Gordon DB, Mayo SL. J Mol Biol. 2000;299:789–803. doi: 10.1006/jmbi.2000.3758. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.