Abstract
Estrogen receptor (ER)-mediated gene expression plays an essential role in mammary gland morphogenesis, function, and carcinogenesis. The repressor of ER activity (REA) is an ER-interactive protein that counterbalances estrogen-induced ER transcriptional activity. Our previous study showed that genetic deletion of both REA alleles resulted in embryonic lethality. This study demonstrates that REA and ERα are coexpressed in mammary epithelial cells. REA heterozygous (REA+/−) mutant mice exhibit faster mammary ductal elongation in virgin animals, increased lobuloalveolar development during pregnancy, and delayed mammary gland involution after weaning. These morphological phenotypes of REA+/− mice are associated with significantly increased cell proliferation and ER transcriptional activities, as indicated by the estrogen response element (ERE)-luciferase reporter in the WT/ERE-Luc and REA+/−/ERE-Luc bigenic mice and by the higher expression levels of estrogen-responsive genes such as progesterone receptor and cyclin D1 in the mammary gland. Our analysis also revealed that REA is an important repressor of ER transcriptional activity in the mammary gland under natural, as well as ovariectomized and estrogen-replaced, hormonal conditions. Our results indicate that REA is a physiological modulator of ER function in the mammary gland and that its correct gene dosage is required for maintenance of normal ER activity and normal mammary gland development. Consequently, a reduction or loss of REA function may cause overactivation of ER and increase breast cancer risk in humans.
Keywords: prohibitin 2, nuclear receptor, coregulator, breast, gene expression
The mammary gland is a dynamic tissue in which morphogenesis, epithelial differentiation, and physiological function are tightly regulated by estrogen and progesterone in accordance with pubertal development and reproductive cycles (1). Estrogen regulates mammary gland development and function through binding to the estrogen receptor α (ERα) and inducing the expression of ERα target genes such as progesterone receptor (PR) and cyclin D1. The phenotype of ER knockout mice has clearly shown that ERα plays a mandatory role in promoting mammary epithelial proliferation and mammary ductal growth after birth (2). Recent studies have demonstrated that the transcriptional activity of nuclear receptors, including ERα and PR, is determined not only by hormone binding but also by relative activities of nuclear receptor-associated coactivators and corepressors (3–5). Repressor of ER activity (REA) is a recently identified corepressor that interacts with select nuclear receptors such as ER (6, 7) and the orphan nuclear receptors chicken ovalbumin upstream promoter transcription factors I and II (COUP-TFI and II) (8). Unlike most other known corepressors that are recruited to nuclear receptors mainly in the absence of ligand or in the presence of antagonists, REA belongs to a subclass of corepressors that interacts dynamically with both agonist- and antagonist-occupied ER (6). REA and other ER corepressors such as ligand-dependent nuclear receptor corepressor (LCoR) (9) and receptor interacting protein-140 (RIP140) (10) attenuate ER activity by (i) recruiting class I and II histone deacetylases to modify chromatin into a transcriptionally silent status (8) and (ii) competing with coactivators such as SRC-1 for binding to ER in the presence of estrogens (11).
To date, only a few studies have explored the expression pattern of REA in tissues and the role of REA in ER function. REA is expressed in normal estrogen target tissues such as uterus and breast (12, 13). Interestingly, REA expression levels in breast cancer are positively correlated with ER, but inversely correlated with tumor grade, suggesting a potential role of REA in repression of breast carcinogenesis (14). In cultured cells, the reduction of REA levels increases ER transcriptional activity (7). In mice, inactivation of both REA alleles (REA−/−) results in embryonic lethality, making it impossible to investigate the function of REA in adult animals (15). Interestingly, although REA+/− mice exhibit normal development, their uteri show enhanced growth and hyperproliferation in response to estrogen compared with WT mice; cell proliferation is higher and the expression of estrogen-responsive genes is up-regulated in the REA+/− uterus. Genes that normally are down-regulated by estrogen in WT mice are no longer efficiently down-regulated in REA+/− uteri (15). Taken together, these studies suggest that REA is a significant modulator of estrogen action in normal tissues and breast cancer cells.
To date, haploinsufficiency phenomena have only been observed in heterozygous mutant mice of essential coregulators that are required for functions of all transcription factors, such as p300, Brg1, and Trap220 (16–18). This striking feature of REA haploinsufficiency highlights the importance of its gene dosage in the regulation of important physiological functions. In this study, we investigated REA function in normal mammary gland development and in the regulation of ER activation. We demonstrate that the reduced REA level in REA+/− mice promotes faster growth of the mammary ductal tree during puberty and an increase in ductal side-branches and alveoli during pregnancy and lactation. Furthermore, we show that these mammary phenotypes are attributable to an enhanced ER function in the mammary glands of REA+/− mice under conditions of either normal or depleted estrogen levels.
Results
REA Is Coexpressed with ERα in Mammary Epithelial Cells.
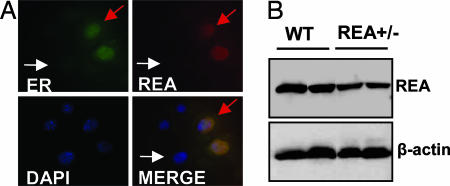
Although REA expression was detected in the breast (12–14), the spatial distribution relationship between REA and ERα in mammary compartments is unclear. To assess the cell type-specific expression pattern of REA, we first performed immunohistochemistry (IHC) on sections of mammary tissue and found that the available REA antibodies could not reliably recognize the REA protein because of either epitope masking or low levels of REA. Therefore, we performed double immunocytofluorescence (ICF) staining of REA and ERα in cultured PN2 cells, which have been developed from the ERα-positive mouse mammary epithelial population (19). Our analysis revealed that PN2 cells express different levels of ERα and that REA is expressed primarily in mammary epithelial cells with higher ERα concentrations (Fig. 1A). Both REA and ERα proteins were localized mainly in the nucleus, which is consistent with previous findings (8). These results indicate that REA and ERα are coexpressed in mammary luminal epithelial cells, providing a spatial basis for REA to serve as ERα corepressor in the mammary gland.
Fig. 1.
REA colocalization with ER and REA levels in REA+/− mammary gland. (A) Double ICF staining for ERα (green) and REA (red) in PN2 mouse mammary epithelial cells. Red arrows indicate a cell expressing high levels of both ERα and REA proteins; white arrows indicate a cell that expresses undetectable levels of REA and ERα. DAPI stains for DNA. Lower Right shows the superimposition of the other three panels. (B) REA protein levels in the mammary glands of pregnant WT and REA+/− mice (five animals per group) were analyzed by Western blotting. β-actin was analyzed as the loading control.
Next, we measured REA protein levels in the mammary glands of pregnant WT and REA+/− mice by immunoblotting. As shown in Fig. 1B, the REA levels were significantly reduced in REA+/− mammary glands compared with WT glands. Our results validate that inactivation of one of the two REA alleles indeed reduces REA protein levels to ≈50% in the mammary gland and that the REA+/− mouse line is a useful model for studying the effect of REA gene dosage on estrogen-regulated mammary gland development and function.
REA Haploinsufficiency Leads to an Accelerated Development of Mammary Ducts.
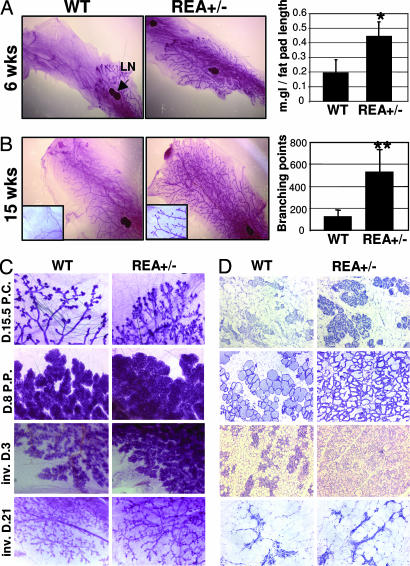
Rapid mammary gland ductal elongation and branching occur at puberty because of the pubertal estrogen surge. To assess the effect of REA haploinsufficiency on mammary gland development, we compared mammary gland morphogenesis at different developmental stages in virgin WT and REA+/− mice by whole-mount staining. At the age of 4 weeks, mammary ducts showed only slight growth and the morphology of WT and REA+/− mammary glands appeared similar (data not shown). However, at 6 weeks, the mammary ducts were greatly elongated in REA+/− mice compared with WT mice, as measured from the center of the lymph node to the front edge of the terminal end buds (Fig. 2A). At 15 weeks, the mammary ducts of both WT and REA+/− mice reached the dorsal end of the fat pads (Fig. 2B Left). However, the number of mammary ductal branches in REA+/− mice was ≈4-fold higher than in WT mice (Fig. 2B). These results demonstrate that REA haploinsufficiency accelerates mammary ductal elongation during puberty and promotes mammary ductal branching.
Fig. 2.
REA haploinsufficiency accelerates mammary gland morphogenesis. (A) Whole-mount staining of mammary glands from 6-week-old WT and REA+/− virgin mice. The length of the mammary duct tree was measured from the lymph node to the upper boundary of the ductal tree and normalized to the length of the fat pad. The relative mammary gland length was determined from five mice for each genotype and is presented as mean ± SD. ∗, P < 0.05. (B) Whole-mount staining of mammary glands from 15-week-old WT and REA+/− virgin mice. Secondary and tertiary branching points of the inguinal glands from five mice were counted. ∗∗, P < 0.01. (C and D) Whole-mount analysis (C) or H&E-stained tissue sections (D) of mammary glands in WT and REA+/− mice at the midpregnant (D15.5 p.c.), lactating (D.8 p.p.), and involuting (inv. D.3 and inv.D.21) stages.
REA Haploinsufficiency Enhances Pregnancy-Associated Mammary Ductal and Alveolar Morphogenesis and Delays Mammary Gland Involution.
To evaluate the effect of REA haploinsufficiency on mammary gland morphogenesis during pregnancy, we compared the mammary gland morphologies of pregnant WT and REA+/− mice at day 15.5 postcoitum (p.c.). Staining of both whole-mount (Fig. 2C) and histological sections (Fig. 2D) showed that REA+/− mammary glands possess significantly higher numbers of ductal branches and lobuloalveoli compared with WT mammary glands. At day 8 postpartum (p.p.), the number and density of alveoli in the lactation mammary glands of REA+/− mice also were increased compared with WT mice (Fig. 2 C and D). To determine whether the enhanced abnormal morphogenesis of the REA+/− mammary glands was due to an intrinsic mammary gland problem or to anomalous hormonal status, age-matched WT and REA+/− mice were ovariectomized and treated with estradiol and progesterone for 25 days to create equal ovarian hormone conditions similar to pregnant stages in both WT and REA+/− mice, as described in ref. 20. After this treatment procedure, more extensive mammary ductal branching and a greater number of mammary alveoli were observed in REA+/− mice compared with WT mice (data not shown). These results demonstrate that REA haploinsufficiency promotes pregnant and lactation hormone-stimulated mammary gland morphogenesis by increasing the responsiveness of mammary gland tissue to estrogen and progesterone.
Considering the increased ductal and alveolar morphogenesis in the REA+/− mammary gland, we wished to know whether, during the involution stage, the gland could correctly revert to its normal structure. At involution day 3 (inv. D.3), WT mammary glands began to regress and their alveoli started to collapse. In contrast, REA+/− mammary glands still showed dense alveoli, although with a different morphology than lactational glands (Fig. 2 C and D). However, at inv. D.21, both WT and REA+/− mammary glands completed the remodeling process and their tissue structures appeared indistinguishable from each other (Fig. 2 C and D). Taken together, these observations demonstrate that REA haploinsufficiency results in a delay in mammary gland involution, probably due to the greater degree of growth during pregnant and lactation stages.
REA Haploinsufficiency Increases Mammary Epithelial Proliferation.
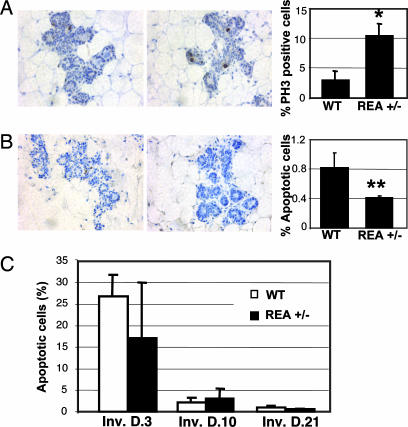
To understand the cellular mechanisms responsible for the enhanced mammary morphogenesis in REA+/− mice, cell proliferation and apoptosis rates at day 15.5 p.c. were measured by detection of PH3- and TUNEL-positive mammary epithelial cells, respectively (Fig. 3A and B). The ratio of mitotic to total mammary epithelial cells in REA+/− mice was 10%, which was more than three times higher than the 3% observed in WT mammary glands (Fig. 3A). On the other hand, the apoptosis rate of epithelial cells in REA+/− mammary glands decreased to 0.4%, which was approximately half of the 0.8% observed in WT mammary glands (Fig. 3B). Thus, the mammary morphogenesis enhanced by the REA haploinsufficiency results from a combined increase in epithelial proliferation and decrease in epithelial apoptosis.
Fig. 3.
Cell proliferation and apoptosis in WT and REA+/− mammary epithelium. (A) IHC for PH3 in WT and REA+/− mammary tissue sections. The average percentage of PH3-positive epithelial cells to total epithelial cells is represented in the bar graph. ∗, P < 0.05. (B) Detection of apoptotic cells in the pregnant mammary glands of WT and REA+/− mice by TUNEL assay. TUNEL-positive epithelial cells and total examined epithelial cells were counted. The percentage of apoptotic cells is presented in the bar graph. ∗∗, P < 0.01. (C) Apoptosis rate analyzed in WT (open bars) and REA+/− (filled bars) mammary epithelium at the indicated involution stages. For each mouse (four or five animals per group), 2,500 cells were counted over two independent mammary gland sections.
To determine whether the delayed mammary gland involution in REA+/− mice was also a result of reduced apoptosis, we measured the apoptosis rates at inv. D.3, D.10, and D.21. As expected, in WT mammary glands the cell apoptosis rates were as high as 26% on inv. D.3 and quickly reduced to as low as 1% on inv. D.21. Interestingly, the apoptosis rates in REA+/− mammary glands were similar to those observed in WT mammary glands throughout all of the evolution stages examined (Fig. 3C). Our results suggest that REA haploinsufficiency does not affect cell apoptosis during mammary gland involution. Delayed involution of the REA+/− mammary glands appears to be a consequence of increased alveolar development during pregnancy and lactation, wherein the excess number of alveolar epithelial cells requires a longer time to revert to prepregnancy status.
REA Haploinsufficiency Leads to ER Overactivation in the Mammary Gland.
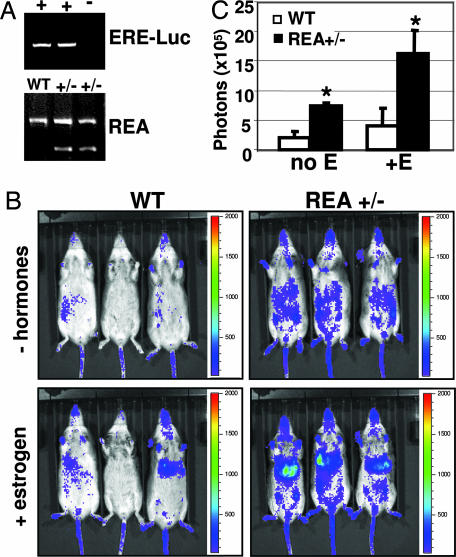
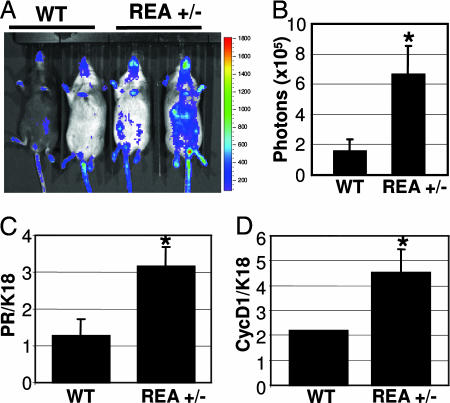
Because the estrogen signaling pathway is essential for mammary gland development and REA is characterized as an ER corepressor in biochemical experiments and in the uterus, we performed a series of experiments to assess the effects of REA haploinsufficiency on the estrogen signaling process in the mammary gland. We measured the 17β-estradiol concentrations in serum of 8-week-old mice in the proestrous phase of the estrous cycle and the ERα protein levels in mammary gland tissue extracts and found that they were similar in WT and REA+/− mice (see Fig. 6, which is published as supporting information on the PNAS web site). These results indicate that REA haploinsufficiency does not affect the circulating estrogen concentration in the blood or ERα expression in the mammary gland. We next quantitatively measured the ER transcriptional activities by in vivo imaging of the luciferase activity from the ER-responsive reporter ERE-Luc in WT/ERE-Luc and REA+/−/ERE-Luc mice. The mice were generated by cross-breeding REA+/− mice (15) with ERE-Luc mice (21) (Fig. 4A). After endogenous ovarian hormones were depleted by ovariectomy, luciferase-emitted photons were detectable, but their intensities were very low in WT/ERE-Luc mice. As expected, the light generated by luciferase was enhanced in WT/ERE-Luc mice after ER stimulation with estrogen for 4 h (Fig. 4B Left). Surprisingly, the photon signals were quite high in ovariectomized REA+/− mice even before estrogen treatment and became more intense after estrogen treatment (Fig. 4B Right). The imaged luciferase activity in REA+/− mice was particularly strong in the area corresponding to inguinal mammary glands and liver, compared with WT mice. Previous studies also detected strong ER activity in the liver by in vivo luciferase imaging (21, 22). Quantitative analysis showed that photons emitted from the entire abdominal area of REA+/− mice were 3.5-fold higher before estrogen treatment and 4-fold higher after estrogen treatment, compared with photons from the same area of WT mice. The differences are statistically significant (Fig. 4C). These results suggest that REA haploinsufficiency significantly enhances ER transcriptional activity in ovariectomized mice either with or without estrogen treatment.
Fig. 4.
ER activity in ovariectomized WT/ERE-Luc and REA+/−/ERE-Luc mice. (A) Identification of WT/ERE-Luc and REA+/−/ERE-Luc mice by PCR analysis. (B) Imaging of luciferase activity in WT/ERE-Luc and REA+/−/ERE-Luc mice. Ovariectomized mice were anesthetized, injected with luciferase substrate solution, and imaged either without hormonal treatment or after estrogen treatment for 4 h. The color scale represents the intensity of light coming from tissues, expressed as photons flux. (C) Luciferase counts captured from the abdomen of five WT/ERE-Luc mice (open bars) and five REA+/−/ERE-Luc mice (filled bars) before (no E) and after (+E) estrogen treatment. Data are presented as average counts ± SD. ∗, P < 0.05.
We examined ER activities in the mammary glands of WT/ERE-Luc and REA+/−/ERE-Luc mice under physiological hormone conditions. Because estrogen regulates ER activity and estrogen levels fluctuate during the estrous cycle (22), we monitored the estrous cycle and performed luciferase imaging in the diestrous phase. The luciferase activity in REA+/− mice was also much stronger than in WT mice (Fig. 5A). The average of photons emitted from the abdominal area of REA+/− mice was approximately three times higher than that coming from the same area of WT mice (Fig. 5B). Again, these results demonstrate that REA haploinsufficiency indeed promotes increased ER activity under physiological levels of estrogen.
Fig. 5.
ER activity in virgin female mice with physiological hormone conditions. (A) Luciferase activity in adult WT/ERE-Luc and REA+/−/ERE-Luc mice in diestrous phase was monitored by in vivo imaging. The color scale indicates the intensity (photons flux) of the light captured from the abdominal area of the indicated mice. Note that the color of the animals' fur, is either black or brown because of the genetic background. (B) Luciferase activity was quantified, and the average luciferase counts (mean ± SD) obtained from five WT/ERE-Luc and five REA+/−/ERE-Luc mice are shown. (C and D) Relative expression levels of PR (C) and cyclin D1 (D) mRNAs in WT and REA+/− mammary glands. The relative mRNA concentrations of PR, cyclin D1, and K18 in total RNA prepared from the mammary glands of six animals for genotype were measured by qPCR. The PR and cyclin D1 expression levels were normalized to the K18 levels in the same RNA sample. Data are presented as mean ± SD. ∗, P < 0.05.
Finally, we validated our findings from the ERE-Luc reporter mice by analyzing the expression levels of endogenous estrogen-responsive genes (PR and cyclin D1) in the mammary gland (23–25). PR and cyclin D1 mRNA concentrations in total mammary gland RNAs prepared from pregnant mice were measured by quantitative real-time PCR (qPCR) and normalized to the mRNA concentration of cytokeratin 18, a widely used epithelial cell marker. Our assay revealed that both PR mRNA and cyclin D1 mRNA were significantly up-regulated in REA+/− mammary glands compared with WT mammary glands (Fig. 5 C and D). Together, our findings indicate that REA haploinsufficiency significantly increases ER activation in mammary glands and substantiate that REA is indeed a physiological corepressor of ER function in the mammary gland.
Discussion
This study demonstrates that REA is a gene dosage-dependent physiological corepressor of ER function during mammary gland development. In our study, we investigated REA localization in mammary epithelial cells and REA corepressor function during estrogen-regulated mammary gland development. We found that REA protein is coexpressed with ERα in mammary epithelial cells, suggesting that REA and ERα potentially can interact with each other to regulate ERα-dependent gene expression and mammary gland development. This notion is clearly validated by multiple lines of evidence. Disruption of one of the two REA alleles resulted in an abnormal phenotype during mammary gland development and altered ERα-regulated transcription. REA haploinsufficiency led to an ER overactivation in the mammary gland, as measured by the ERE-Luc reporter and by the expression levels of the ER target genes PR and cyclin D1. Because estrogen signaling is known to stimulate mammary epithelial proliferation through ER-mediated up-regulation of cyclin D1 (24), the greater ER activity resulting from REA haploinsufficiency is likely responsible for the enhanced mammary epithelial proliferation, the accelerated ductal elongation and branching in virgin mice, and the increased lobuloalveolar development that we observed in REA+/− pregnant and lactating mice.
The correlation of REA expression levels with ERα levels in the ERα-positive mammary epithelial cells raises the possibility that REA might be an estrogen-responsive gene. If this is the case, the estrogen-induced REA expression would form a negative feedback regulatory mechanism to control ER-dependent gene expression in normal cells. In REA+/− cells, reduction of REA may weaken this negative feedback inhibition and result in a constant increase in ER activity.
In the mammary gland, PR is a known target gene of ERα (23, 26). Not surprisingly, our data showed that REA haploinsufficiency enhances PR expression. Because PR is an essential regulator of mammary gland side-branching and lobuloalveolar development (27), it is likely that the increased number of mammary ductal branches and alveoli in REA+/− mice are a consequence of the elevated PR expression induced by the REA-dependent overactivation of ERα. Similarly, the elevation of cyclin D1 expression also may be the combined result of both ERα overactivation and PR overproduction, inasmuch as both receptors have been shown to up-regulate cyclin D1 expression (24).
In the mammary glands of pregnant REA+/− mice, the reduced cell apoptosis observed may partially contribute to the enhanced mammary morphogenesis caused by REA haploinsufficiency. Because estrogen signaling is required for mammary epithelial survival, the extremely low rate of cell apoptosis in REA+/− mammary glands at this stage may be a result of increased epithelial cell survival resulting from ER overactivity, rather than defective apoptotic pathways. After weaning, the survival signaling pathways are reduced and most of the mammary alveolar epithelial cells are programmed for death (28). This notion supports the fact that although we observed a morphological delay during the involution of REA+/− mammary glands, the apoptotic rate was similar between WT and REA+/− mammary glands. Thus, the morphological delay in involution should be a simple consequence of the higher alveoli number present in REA+/− mammary glands at the lactation stage. This is also supported by the fact that WT and REA+/− mammary glands are morphologically indistinguishable after an extended involutional period. Results suggest that REA either does not play a role in alveolar epithelial apoptosis during involution or that haploid reduction of REA is insufficient to affect this involution process.
Our data demonstrate that REA haploinsufficiency significantly enhances ER activity in the mammary glands of REA+/− mice with physiological estrogen levels and with or without estrogen treatment after ovariectomy. This suggests that, in the mammary gland, REA corepressor function for ER action is independent of estrogen levels. The results showing more active ER function in intact REA+/− mice with normal estrogen levels, or in ovariectomized REA+/− mice after estrogen treatment, are consistent with the model proposed from previous in vitro experiments. In fact, in the presence of estrogen, REA has an increased affinity for ER and, on equilibrium, association of REA with estrogen-activated ER may competitively counterbalance ER transcriptional activity that is enhanced by estrogen-induced recruitment of coactivators to ER (6). Recently, a similar result was found in the uteri of REA+/− mice treated with estrogen (15). However, the overactivity of ER in the mammary glands of ovariectomized REA+/− mice without estrogen treatment is a surprising finding because the estrogen level is extremely low after ovariectomy. Several explanations are possible. First, when REA is reduced by half in the mammary glands of ovariectomized mice, ER function may become much more sensitive in response to the very low levels of estrogen. Second, in addition to estrogen, ER can be ligand independently activated by growth factors and certain other pathways (22, 29, 30). It is possible that REA interacts with ER and represses activation resulting from such estrogen-independent pathways in the mammary glands of ovariectomized mice. Finally, recent studies have revealed that ER is present in the nucleus and binds to DNA even in the absence of ligands (31, 32). REA might be associated with DNA-bound ER and repress its basal activity. When the repression is relieved by REA reduction or inactivation, ER basal activity may increase and drive target gene expression.
ER-dependent gene expression is essential for the development of several tissues and for the normal function of many physiological processes. Abnormal ER function has been associated with the onset of numerous pathological conditions, such as breast and endometrial cancers, osteoporosis, atherosclerosis, and Alzheimer's disease (33). Because REA is a crucial physiological modulator of ER activity, alterations in REA levels or in its repressor function could be involved in the initiation and progression of a variety of these pathological processes. Furthermore, the increase in ER activity observed with REA haploinsufficiency under conditions of extremely low estrogen concentrations, as suggested previously in our studies in REA heterozygous mouse embryo fibroblast cells (15), may have a particularly important implication for breast cancer resistance to drug therapy or chemotherapy. Aromatase inhibitors have been widely used in breast cancer treatment to block estrogen synthesis (34). REA expression levels have been found to be reduced in certain human breast tumors (13, 14). Because reduction of REA sensitizes ER to activation by low estrogen concentrations, low REA levels may make breast cancers more resistant to aromatase inhibitor therapy. In this scenario, REA may be useful as a potential prognostic indicator for response to aromatase inhibitors.
Materials and Methods
Animals.
All animals were maintained in accordance with the National Institutes of Health directives, and all experimental procedures were approved by the Baylor College of Medicine Animal Care and Use Committee. The REA heterozygous and the ERE-Luc transgenic mouse lines were described previously (15, 21). The REA+/−/ERE-Luc bigenic mice were produced by crossing REA+/− mice with ERE-Luc mice in the C57BL/6J and 129 mixed background. Mouse genotypes were analyzed by PCR, using genomic DNA and specific primers as described in refs. 15 and 21.
Cell Culture and ICF.
For ICF, PN2 cells were cultured on coverslips in DMEM/F12 medium with 2% FCS, 50 μg/ml gentamicin, 50 μg/ml insulin, 50 μg/ml epidermal growth factor and 1 mg/ml BSA. Cells were fixed with 4% paraformaldehyde for 30 min on ice, washed three times with ice-cold PBS and permeabilized with 0.5% Triton X-100 in PBS for 30 min at room temperature. Next, cells were washed and incubated in Tris-buffered 5% powdered milk for 1 h and then with primary antibodies against ER (clone H226; Lab Vision, Fremont, CA) or REA (Upstate, Lake Placid, NY) overnight at 4°C. Cells were washed with blocking solution and then incubated with fluorescent secondary antibodies against rat or rabbit IgG for 30 min at room temperature. After washing, stained cells on the coverslips were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) containing DAPI.
Western Blotting.
Breast tissue from pregnant animals was pulverized with liquid nitrogen, resuspended in a lysis buffer with 50 mM Tris·HCl (pH 7.4), 0.2% SDS, and proteinase inhibitors and sonicated. Supernatant with 30 μg of protein was separated in a 7.5% acrylamide gel for Western blotting. The blots were reacted with antibodies specific to REA (7), ERα (MC-20; Santa Cruz Biotechnology, Santa Cruz, CA), β-actin (AC-74; Sigma, St. Louis, MO), and cytokeratin endo A (35) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). The experiment was independently repeated twice using a total of five animals, and the images of a representative experiment are shown in Figs. 1B and 6B.
Mammary Gland Whole Mount, Histology, and IHC.
Whole-mount staining of inguinal mammary glands was performed as described previously (20, 36). For histological analysis, mammary tissue was fixed in 4% paraformaldehyde at 4°C overnight, embedded in paraffin, cut at 5-μm thickness, and stained with H&E. To detect proliferating cells, IHC was performed on deparaffinized sections using the antibody against Ser10-phosphorylated histone 3 (PH3), as described previously (37). Sections were counterstained with hematoxylin. Apoptosis was assessed by using the TdT-FragEL DNA fragmentation detection kit (Calbiochem, Darmstadt, Germany), in accordance with the manufacturer's instructions. IMAGE TOOL software (University of Texas Health Science Center, San Antonio, TX) was used to count mammary gland branching points, PH3-positive cells, and apoptotic cells. Five mice were analyzed for each genotype group. For each mouse, the secondary and tertiary ductal branches of the inguinal mammary glands were counted. To calculate the proliferative and apoptotic rate, 2,500 cells per gland were counted over two independent sections and four or five mice were used per group. Results are expressed as the ratio of proliferative or apoptotic cell number to total cell number. Statistical analysis was performed using the unpaired Student's t test; P < 0.05 was considered significant.
In Vivo Imaging of Luciferase Activity.
Adult WT/ERE-Luc and REA+/−/ERE-Luc female mice were ovariectomized. On day 15 after ovariectomy, mice were anesthetized with an oxygen/isoflurane mixture using an inhalation anesthesia system (VetEquip, Pleasanton, CA) and injected with the d-luciferin solution (1 mg per mouse, i.p.). Ten minutes later, the luciferase activity in each live mouse was imaged using the IVIS imaging system (Xenogen, Alameda, CA). After imaging, mice were treated with 17β-estradiol (50 μg/kg body weight, s.c.) in corn oil for 4 h and imaged again after a second injection of the d-luciferin solution. For intact mice, the phase of the estrous cycle was monitored by examination of vaginal smears, and the luciferase activity was imaged at the diestrous phase. Luciferase activity was quantified using Living Image software (Xenogen), and the average luciferase counts (photons per second) of five mice per group were represented as mean ± SD in bar graphs. Statistical analysis was performed with the unpaired Student's t test; P < 0.05 was considered significant.
qPCR.
For each genotype group, mammary glands were isolated from six pregnant mice and immediately frozen on dry ice. Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA). RNA was treated with DNase I and reverse-transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen). qPCR was performed using 20 ng of cDNA and the TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA). For PR mRNA/cDNA measurement, the forward primer 5′-GGGAGCTGCAAGGTCTTCTTT, the reverse primer 5′-CGAATTTTATCAACAATGCAGTCAT, and the fluorescence-labeled probe 5′-AAATAGTTATGCTGCCCTTCCATTGCCCT were used. For cyclin D1 mRNA/cDNA analysis, the forward primer 5′-CGGCCCGAGGAGCTG, the reverse primer 5′-GGCCAGGTTCCACTTGAGC, and the probe 5′-TGTTCACCAGAAGCAGTTCCATTTGCA were used. The standard curve method was used for data analysis. Data are presented as mean ± SD. Statistical difference was calculated by the unpaired Student's t test; P < 0.05 was considered significant.
Estradiol Measurement.
Blood was collected from 8-week-old female mice at the proestrous phase of the estrous cycle (12 mice for each genotype), and serum was prepared from the blood clot. The 17β-estradiol concentration in the serum was measured by using an RIA kit (DSL4400; Diagnostic Systems Laboratories, Webster, TX), as described in ref. 38. Results are expressed as mean ± SD. Data were statistically analyzed by the unpaired Student's t test.
Supplementary Material
Acknowledgments
We thank Dr. D. Medina (Baylor College of Medicine) for providing the PN2 cells; the Developmental Studies Hybridoma Bank for the monoclonal cytokeratin endo A antibody; Ms. S. Zhou for mouse genotyping; and Drs. B. Mulac-Jericevic, O. M. Conneely, T. Thompson, and R. Lanz for technical assistance and helpful discussions. This work was partially supported by National Institutes of Health (NIH) Grants HD07857 and DK59820; by a Welch Foundation grant (to B.W.O.); by NIH Grants CA119689, CA112403, and DK59820-CoreA (to J.X.); and by NIH Grant CA18119 (to B.S.K.).
Abbreviations
- ER
estrogen receptor
- ERE
estrogen response element
- ICF
immunocy-tofluorescence
- IHC
immunohistochemistry
- inv.
involution
- p.c.
postcoitum
- p.p.
postpartum
- PR
progesterone receptor
- REA
repressor of estrogen receptor activity.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hennighausen L, Robinson GW. Dev Cell. 2001;1:467–475. doi: 10.1016/s1534-5807(01)00064-8. [DOI] [PubMed] [Google Scholar]
- 2.Couse JF, Korach KS. Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- 3.McKenna NJ, O'Malley BW. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 4.Glass CK, Rosenfeld MG. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 5.Xu J, Li Q. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 6.Montano MM, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen BS. Proc Natl Acad Sci USA. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS. J Biol Chem. 2000;275:35848–35856. doi: 10.1074/jbc.M001327200. [DOI] [PubMed] [Google Scholar]
- 8.Kurtev V, Margueron R, Kroboth K, Ogris E, Cavailles V, Seiser C. J Biol Chem. 2004;279:24834–24843. doi: 10.1074/jbc.M312300200. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, et al. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 10.Miyata KS, McCaw SE, Meertens LM, Patel HV, Rachubinski RA, Capone JP. Mol Cell Endocrinol. 1998;146:69–76. doi: 10.1016/s0303-7207(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 11.Martini PG, Delage-Mourroux R, Kraichely DM, Katzenellenbogen BS. Mol Cell Biol. 2000;20:6224–6232. doi: 10.1128/mcb.20.17.6224-6232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dotzlaw H, Leygue E, Watson PH, Murphy LC. Cancer Res. 1999;59:529–532. [PubMed] [Google Scholar]
- 13.Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH. Cancer Res. 2000;60:6266–6271. [PubMed] [Google Scholar]
- 14.Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH, Murphy LC. Cancer Res. 2000;60:2796–2799. [PubMed] [Google Scholar]
- 15.Park SE, Xu J, Frolova A, Liao L, O'Malley BW, Katzenellenbogen BS. Mol Cell Biol. 2005;25:1989–1999. doi: 10.1128/MCB.25.5.1989-1999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 17.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 18.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 19.Medina D, Kittrell FS, Shepard A, Stephens LC, Jiang C, Lu J, Allred DC, McCarthy M, Ullrich RL. FASEB J. 2002;16:881–883. doi: 10.1096/fj.01-0885fje. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 21.Ciana P, Di Luccio G, Belcredito S, Pollio G, Vegeto E, Tatangelo L, Tiveron C, Maggi A. Mol Endocrinol. 2001;15:1104–1113. doi: 10.1210/mend.15.7.0658. [DOI] [PubMed] [Google Scholar]
- 22.Ciana P, Raviscioni M, Mussi P, Vegeto E, Que I, Parker MG, Lowik C, Maggi A. Nat Med. 2003;9:82–86. doi: 10.1038/nm809. [DOI] [PubMed] [Google Scholar]
- 23.Nardulli AM, Greene GL, O'Malley BW, Katzenellenbogen BS. Endocrinology. 1988;122:935–944. doi: 10.1210/endo-122-3-935. [DOI] [PubMed] [Google Scholar]
- 24.Said TK, Conneely OM, Medina D, O'Malley BW, Lydon JP. Endocrinology. 1997;138:3933–3939. doi: 10.1210/endo.138.9.5436. [DOI] [PubMed] [Google Scholar]
- 25.Altucci L, Addeo R, Cicatiello L, Dauvois S, Parker MG, Truss M, Beato M, Sica V, Bresciani F, Weisz A. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- 26.Katzenellenbogen BS, Nardulli AM, Read LD. Prog Clin Biol Res. 1990;322:201–211. [PubMed] [Google Scholar]
- 27.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, Shyamala G, Conneely OM, O'Malley BW. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 28.Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, Hennighausen L, Furth PA. Proc Natl Acad Sci USA. 1997;94:3425–3430. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klotz DM, Hewitt SC, Ciana P, Raviscioni M, Lindzey JK, Foley J, Maggi A, DiAugustine RP, Korach KS. J Biol Chem. 2002;277:8531–8537. doi: 10.1074/jbc.M109592200. [DOI] [PubMed] [Google Scholar]
- 30.Power RF, Mani SK, Codina J, Conneely OM, O'Malley BW. Science. 1991;254:1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- 31.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 32.Reese JC, Katzenellenbogen BS. Mol Cell Biol. 1992;12:4531–4538. doi: 10.1128/mcb.12.10.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deroo BJ, Korach KS. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell A. Trends Endocrinol Metab. 2005;16:420–428. doi: 10.1016/j.tem.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Brulet P, Babinet C, Kemler R, Jacob F. Proc Natl Acad Sci USA. 1980;77:4113–4117. doi: 10.1073/pnas.77.7.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuang SQ, Liao L, Wang S, Medina D, O'Malley BW, Xu J. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Liao L, Kuang SQ, Xu J. Endocrinology. 2003;144:1435–1443. doi: 10.1210/en.2002-0018. [DOI] [PubMed] [Google Scholar]
- 38.Yuan Y, Liao L, Tulis DA, Xu J. Circulation. 2002;105:2653–2659. doi: 10.1161/01.cir.0000018947.95555.65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.