Abstract
The dissociable σ subunit of bacterial RNA polymerase is required for the promoter-specific initiation of transcription. When bound to RNA polymerase, σ makes sequence-specific promoter contacts and plays a crucial role in DNA melting. In isolation, however, σ lacks significant promoter binding activity. In the crystal structure of the flagellar σ factor, σ28, bound to the anti-σ factor, FlgM, σ28 adopts a compact conformation in which the promoter binding surfaces are occluded by interdomain contacts. To test whether σ28 adopts this conformation in the absence of FlgM, we engineered a set of double cysteine mutants predicted to form interdomain disulfides in the conformation observed in the FlgM complex. We show that these disulfides form in both the presence and absence of FlgM. For two of the mutants, quantitative measurements of disulfide formation under equilibrium conditions suggest that the major solution conformation favors disulfide formation. The results indicate that the compact conformation of σ28 observed in the σ28/FlgM structure is similar to the predominant conformation of free σ28 in solution. This finding suggests that autoinhibition of DNA binding in free σ28 is accomplished by steric occlusion of the promoter binding surfaces by interdomain interactions within the σ factor as well as by a suboptimal distance between the promoter −10 and −35 element binding determinants in σ2 and σ4, respectively.
Keywords: protein conformation, sigma factor, transcription
In bacteria, the 450-kDa RNA polymerase (RNAP) holoenzyme, comprising the evolutionarily conserved catalytic core (subunit composition α2ββ′ω) combined with the initiation-specific σ subunit, directs transcription initiation (1). The σ subunit recruits RNAP to promoters through sequence-specific binding of the promoter elements and mediates formation of the transcription competent open complex. The principal control point of gene expression in bacteria is transcription initiation, and a major mechanism by which bacteria regulate transcription initiation is through regulation of σ activity.
Almost all σ's comprise a homologous family (the σ70 family) with defined regions of conserved amino acid sequence (2, 3). Structural analysis reveals that Group I (primary) σ's comprise four flexibly linked domains, σ1.1, σ2, σ3, and σ4, containing conserved regions 1.1, 1.2–2.4, 3.0–3.1, and 4.1–4.2, respectively (4). Binding to core RNAP induces large movements of the σ domains with respect to each other (5, 6), converting σ into an active conformation in which the DNA-binding determinants in σ2 and σ4 are exposed and appropriately spaced to recognize the −10 and −35 promoter elements, respectively (7–11). The σ's only perform their initiation activities in the RNAP holoenzyme. The structural basis for the lack of promoter binding and melting activity in free σ is not understood. To date, the structure of an intact σ in the absence of a binding partner has not been determined.
X-ray crystal structures of the flagellar σ, σ28, from the motile hyperthermophile Aquifex aeolicus (Aae) in complex with its anti-σ, FlgM, explain the inhibition mechanism of FlgM (6). In the σ28/FlgM complex, a previously unrecognized conformation of the σ is observed in which the structural domains, σ282, σ283, and σ284, pack together in a compact unit. The extended FlgM wraps around the outside and occludes the core RNAP binding determinants on σ282 and σ284. Formation of the holoenzyme requires a helix-coil transition in the σ3-σ4 linker as well as large rearrangements of the σ domains with respect to each other, exposing and properly spacing the promoter binding determinants.
The extensive interdomain interactions within σ28 in the σ28/FlgM structure suggest that this conformation may be stable in solution even in the absence of FlgM. In this inactive conformation of the σ (Fig. 1 A and B), the promoter binding determinants are buried in interdomain interfaces, preventing DNA binding, whereas the core binding surfaces, in the absence of FlgM, are exposed, allowing σ to readily interact with core RNAP, consistent with the behavior of free σ.
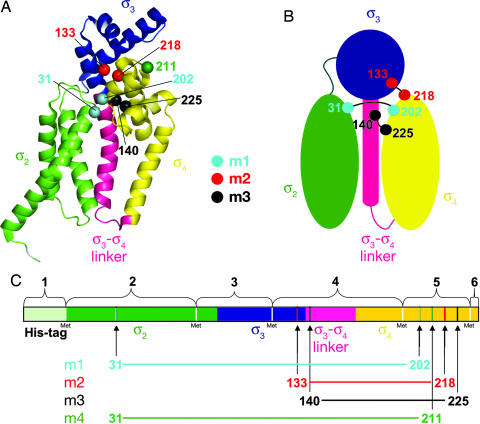
Fig. 1.
Location of introduced cysteines in σ28. (A) Aae σ28 from the σ28/FlgM cocrystal structure [1RP3 (6)] is shown as a ribbon diagram. FlgM has been omitted. Structural elements are color-coded as follows: σ2, green; σ3, blue; σ3-σ4 linker, pink; and σ4, yellow. The Cα atoms of sites mutated to cysteine are shown as spheres and color-coded as follows: m1, C31 and C202, cyan; m2, C133 and C218, red; m3, C140 and C225, black; C211 (control), green. (B) Schematic diagram showing the interdomain connections made by the disulfides. The color coding is the same as in A. (C) Linear representation of σ28, color-coded as in A. The CNBr cleavage fragments are denoted by numbered brackets. Cysteine sites are indicated by vertical lines and numbered arrows (color-coded as in A).
Here we use a disulfide cross-linking approach to investigate the solution conformation of σ28. The results indicate that the compact conformation of σ28 observed in the σ28/FlgM structure is similar to the predominant conformation of free σ28 in solution. This finding suggests that autoinhibition of DNA binding in free σ28 is accomplished by steric occlusion of the promoter binding surfaces by interdomain interactions within the σ factor as well as by a suboptimal distance between the promoter −10 and −35 element binding determinants in σ2 and σ4, respectively.
Results
Disulfide Mutant Design and Preparation.
To investigate the conformation of free Aae σ28, we sought to crystallize the protein in the absence of FlgM. The σ28 was prepared from a pET-based expression vector. Expression levels were high, and the protein was well behaved and purified to homogeneity. Nevertheless, we have been unable to obtain crystals even after extensive crystallization trials. This result did not surprise us because a free, intact σ factor has never been crystallized, despite much effort. We presume that the structural architecture of the protein, with three or four structural domains linked by flexible linkers (4, 6, 8, 9, 11–14), makes crystallization difficult. Although σ in solution likely adopts a preferred conformation (in terms of the disposition of the individual domains with respect to each other), this architecture may lead to conformational heterogeneity that is recalcitrant to crystallization.
We then considered using NMR spectroscopy to characterize the σ28 conformation in solution. At 27.4 kDa, σ28 is the smallest σ factor [excluding Group IV σ's, which lack σ3 (14)]. Despite new technology facilitating the analysis of larger proteins (15), this target is still a challenging NMR problem. In addition, gel filtration, dynamic light scattering, and glutaraldehyde cross-linking all indicated that free σ28 is predominantly a dimer at high concentration, with an effective mass of ≈55 kDa, making it a very challenging target for NMR (data not shown). For this reason, we chose to pursue biochemical probes of the σ28 solution conformation.
Disulfide cross-linking has been used successfully as a probe of protein solution conformation and interactions (16, 17). This approach is particularly convenient in the case of Aae σ28, because the native sequence lacks Cys residues. We analyzed the σ28 structure [all four copies in the asymmetric unit of the 2.3-Å-resolution structure, Protein Data Bank ID code 1RP3 (6)] with the program MODIP (18), which assesses all possible residue pairs for proximity and geometry consistent with disulfide formation (assuming the residues were mutated to Cys). Five potential interdomain disulfide pairs were identified. Two pairs were not considered further because they involved mutating an absolutely conserved residue among σ28 homologs (corresponding to Aae σ28 P34). This left us with three pairs of residues that, if mutated to Cys, were predicted to form interdomain disulfide bonds under appropriate conditions, provided the conformation of σ28 in solution (in the absence of FlgM) samples the conformation observed in the σ28/FlgM complex (Fig. 1). The three pairs are as follows.
Mutant 1 (m1): K31/P202 (cyan in Fig. 1). This pair would form a disulfide connecting σ282 with σ284. Both positions are poorly conserved (in fact, Aae σ28 is the only σ28 with a Pro at this position).
Mutant 2 (m2): L133/S218 (red in Fig. 1). This pair would form a disulfide connecting σ283 with σ284. L133 is highly conserved. S218 is highly conserved, but mutation to a Cys would be a conservative substitution. Moreover, Cys occurs at this position in some σ28 homologs.
Mutant 3 (m3): Y140/L225 (black in Fig. 1). This pair would form a disulfide connecting the σ283-σ284 linker with σ284. Both positions are poorly conserved.
Four double-Cys mutants of Aae σ28 were constructed beginning with a triple-Met mutant [I77M/L122M/L191M, originally constructed for phasing of the crystal structure and shown to have the same structure as the wild-type σ28 (6)] as the parent strain (Fig. 1C), to facilitate detection of disulfide bonds by CNBr cleavage (specific to peptide bonds C-terminal of Met residues). Comparison of the wild-type and σ28-3M crystal structures revealed that the structure is unperturbed by the Met substitutions (6). The three mutant pairs described above, predicted to form interdomain disulfides, as well as a negative control, σ2-31/σ4-211 [mutant 4 (m4), green in Fig. 1], not predicted to form a disulfide in the FlgM-bound conformation, were constructed. The parent protein σ28-3M and the four double-Cys mutants were each purified both in the absence and the presence of FlgM. The purifications were performed without reducing agent to allow the formation of disulfide bonds. The FlgM binding of all of the mutants was unperturbed. The only mutant that differed noticeably in its behavior from σ28-3M was the negative control, σ2-31/σ4-211, which was toxic to the expression host and had a tendency to form intermolecular disulfides leading to aggregation during purification.
Qualitative Identification of Disulfide Bonded Fragments by CNBr Cleavage and MALDI-TOF MS.
Disulfides were detected qualitatively in m1, m2, and m3 by using CNBr cleavage and MALDI-TOF MS. The σ28-3M contains five Mets, whereas FlgM lacks any internal Mets, therefore complete CNBr cleavage is predicted to yield six σ28-peptide fragments (Fig. 1C). Because the Cys residues of each mutant pair are separated by at least one Met residue, disulfides can be detected after cleavage by MALDI-TOF MS as covalently connected CNBr fragments which disappear upon reduction.
The CNBr cleavage pattern of σ28-3M was recorded as a reference, and the expected peaks were observed in the MALDI-TOF spectrum (Fig. 2A; For all of the MALDI-TOF experiments, the measured and expected masses for relevant fragments are listed in Table 1). In addition, a peak corresponding to CNBr cleavage products 4–5 together was observed, indicating partial cleavage at Met-191 separating CNBr fragments 4 and 5 (Fig. 2A). SDS/PAGE analysis of the cleavage peptides confirmed incomplete CNBr cleavage (data not shown). Modified cleavage protocols were attempted (including the addition of denaturants and addition of CNBr to saturation), but complete cleavage could not be achieved, which may be due to the presence of oxidized Mets, which are not recognized by CNBr.
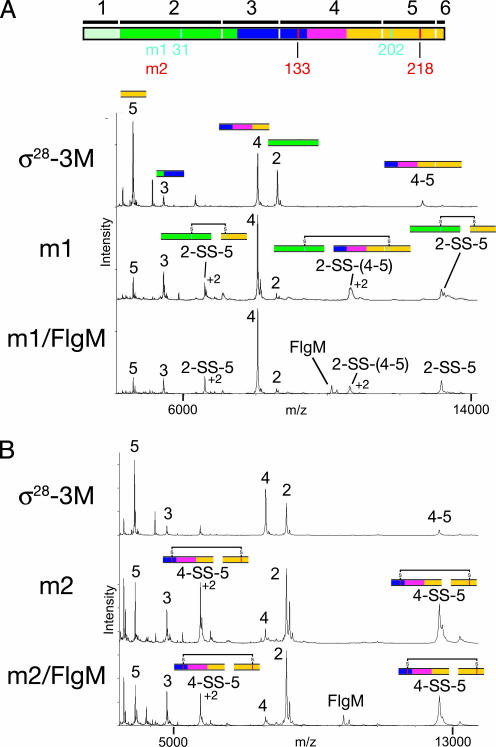
Fig. 2.
MS of CNBr-digested disulfide mutants. A linear diagram of Aae σ28, color-coded as in Fig. 1, is shown at the top of the figure. Methionines (CNBr cleavage sites) are denoted by vertical white lines, with the expected fragments from complete CNBr cleavage denoted by black bars and numbered on top. The positions of the cysteine mutants for m1 (cyan) and m2 (red) are denoted underneath the diagram. (A) Mass spectra of CNBr digests of σ28-3M (top) and mutant m1 without (middle) and with (bottom) FlgM. Peaks are labeled with the fragment number and with colored segments corresponding to the linear diagram at the top of the figure. Peaks with charge state greater than +1 are also labeled with the charge state. (B) Mass spectra of CNBr digests of σ28-3M (top) and mutant m2 without (middle) and with (bottom) FlgM. Peaks are labeled with the fragment number and with colored segments corresponding to the linear diagram at the top of the figure. Peaks with charge state greater than +1 are also labeled with the charge state.
Table 1.
Observed (MALDI-TOF) and theoretical masses of CNBr fragments
| Mutant | CNBr fragment no.* (residues) | Theoretical mass, Da | Observed mass, Da | |Δmass|,† Da |
|---|---|---|---|---|
| m1 (σ2-31/σ4-202) | 2 (2–77) | 8,574.1 | 8,573.3 | 0.8 |
| 5 (192–231) | 4,588.5 | 4,588.3 | 0.2 | |
| 2-SS-5 | 13,160.6 | 13,162.0 | 1.4 | |
| m2 (σ3-133/σ4-218) | 4 (123–191) | 8,041.1 | 8,040.3 | 0.8 |
| 5 (192–231) | 4,598.5 | 4,598.1 | 0.4 | |
| 4-SS-5 | 12,637.6 | 12,637.8 | 0.2 | |
| m3 (σlinker-140/σ4-225) | 4 (123–191) | 7,991.1 | 7,990.5 | 0.6 |
| 5 (192–231) | 4,572.4 | 4,572.3 | 0.1 | |
| 4-SS-5 | 12,561.5 | 12,562.0 | 0.5 |
*See Fig. 1C.
†|Δmass| = |(theoretical mass) − (observed mass)|.
The MALDI-TOF spectra of the CNBr cleavage peptides for mutant m1 in the absence (middle) and presence (bottom) of FlgM are shown in Fig. 2A. The peaks for the individual peptides 2 and 5 had significantly weakened signals compared with the spectrum of σ28-3M (top), whereas a previously unobserved species corresponding to disulfide-linked peptides 2 and 5 (2-SS-5) appeared, confirming the formation of a C31-C202 disulfide. In addition, a disulfide bonded species containing 2 and the partially cleaved 4–5 fragment was detected [2-SS-(4–5)]. Upon reduction, the disulfide-linked peaks disappeared (data not shown), and the intensity of the individual fragments 2 and 5 increased.
The MALDI-TOF spectra of the CNBr peptides from mutant m2 without (middle) and with (bottom) FlgM are shown in Fig. 2B. The σ28-3M CNBr spectrum is also shown as a reference (shown above A). A C133-C218 disulfide connects CNBr fragments 4 and 5 (4-SS-5). The mass of fragments 4 and 5 connected by a disulfide (4-SS-5) differs from that of the partially cleaved fragment (4–5) by only 16 Da, and these species are not completely resolved in the MALDI-TOF MS spectrum. The increase in the peak height for this species in the m2 spectrum compared with σ28-3M is consistent with the appearance of the disulfide-linked species. Moreover, as this peak, height increases significantly, and the peak heights for the individual fragments 4 and 5 are reduced.
The expected disulfide for mutant m3 (linking C140-C255) also connects fragments 4 and 5, and the MALDI-TOF spectra for this mutant are very similar to those of m2 (data not shown). Because of the presence of intermolecular disulfides in the control mutant, m4, MALDI-TOF analysis was not expected to give an informative result.
Quantitative Disulfide Analysis by SDS/PAGE.
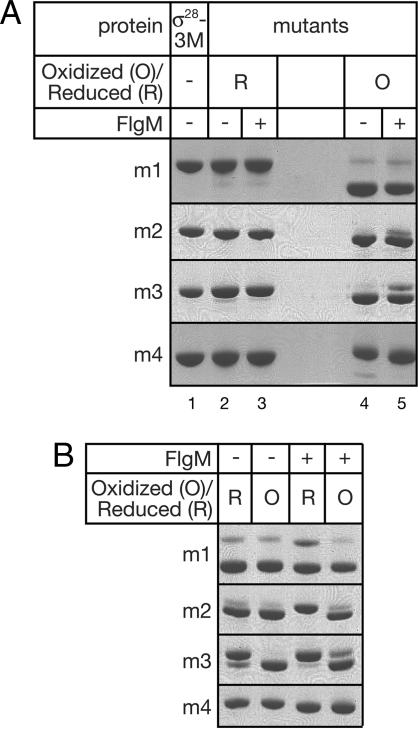
The mobility of each disulfide mutant on a denaturing, nonreducing gel increased significantly upon disulfide formation, allowing resolution and quantitative analysis of the oxidized versus the reduced species. Each of the disulfide mutants migrated at the same rate as σ28-3M when reduced (Fig. 3A, lanes 1–3) but showed increased mobility when oxidized (Fig. 3A, lanes 4 and 5), with the longest range mutant (m1) giving rise to the largest shift. For disulfide mutants m1, m2, and m3, disulfide formation under oxidizing conditions was essentially complete in the absence of FlgM (Fig. 3A, lane 4). The mutant m1 forms a disulfide link equally efficiently in the presence of FlgM (Fig. 3A, lane 5). The disulfide-bonded species predominate for both m2 and m3 in the presence of FlgM, but significant amounts of reduced protein are detectable. The negative control mutant, m4, does not form significant amounts of intramolecular disulfide under any condition, although a very faint band corresponding to this species is visible in the absence of FlgM.
Fig. 3.
SDS/PAGE analysis of disulfide mutants. Samples (oxidized or reduced as indicated) were separated on a 10% NuPAGE Novex Bistris denaturing gel. (A) Mobility shift of disulfide-bonded mutants (lanes 4–5) compared with the parent protein (σ28-3M, which contains no cysteines, lane 1) or reduced mutants (lanes 2–3). (B) Analysis showing proportion of disulfide-bonded species after incubation under equilibrium conditions (see Materials and Methods). “R” indicates incubations that started with reduced samples, and “O” indicates incubations that started with oxidized samples.
Quantitative Analysis of Equilibrium Populations.
Equilibrium populations of disulfide-bonded versus reduced species can be estimated by incubating the sample in buffer containing a 1:1 mixture of reduced and oxidized glutathione (19). Under these conditions, an equilibrium distribution of disulfide-bonded versus reduced species will be maintained, provided the reactive Cys residues (or S-S bond) are accessible to the glutathione in solution on the time scale of the experiment. The mutants were analyzed in this way, using both reduced and oxidized samples as the starting material (Fig. 3B). Because the same equilibrium populations should be obtained regardless of the oxidation state of the starting material, this procedure served as a control for buried sites. The mutant m1 was mostly oxidized both in the presence and absence of FlgM, and this result did not depend on the oxidation state of the starting material. Similarly, the disulfide-bonded species predominated for m2 in the absence of FlgM, both when starting with reduced protein and when starting with oxidized protein. In the presence of FlgM however, the amount of oxidation of this mutant was similar to that of the starting material, suggesting that the cysteines were inaccessible to the glutathione catalyst. The same was true for the m3 mutant with or without FlgM, and consequently this mutant was not amenable to this type of analysis. No disulfide formation was detected upon glutathione incubation in the control mutant m4.
Discussion
We have demonstrated that engineered, long-range, interdomain disulfide bridges predicted to form in σ28 bound to FlgM also form in free σ28, indicating that the solution conformation of σ28 is similar to that observed in the crystal structure of the σ28/FlgM complex. This conformation differs significantly from the σA holoenzyme structures, in which σA adopts an elongated conformation in which the domains are spread across one face of the enzyme, with each domain engaged with the core RNAP and with no interdomain contacts (8, 11). In the holoenzyme, the σ3-σ4 linker, which mostly lacks secondary structure, passes from σ3 through the interior of the enzyme near the active site and threads out through the RNA exit channel to finally connect with σ4. It is reasonable to assume that σ28 adopts a similar holoenzyme conformation, because the structures of the individual domains of Aae σ28 are very similar to the individual domains of Taq σA and because both recognize −10 and −35 promoter elements, thus requiring the same interdomain spacing. The σ3-σ4 linker of σA, however, bears no structural resemblance to the α-helical linker conformation in the σ28/FlgM complex. Because detectable sequence conservation is also absent, and because a similar function has not been established for the σ3-σ4 linker of σ28 (i.e., it is not known whether the σ3-σ4 linker of σ28 passes through the RNAP active site channel like the linker of σA), it is not possible to predict whether the structural differences are strictly context dependent (i.e., due to binding to RNAP). However, it is reasonable to assume that σ3 and σ4 bind to the holoenzyme in the same manner in both σ factors. The relative positions of σ3 and σ4 in the holoenzyme constrain the structure of the σ3-σ4 linker, and modeling indicates that the helical conformation found in the σ28/FlgM complex is incompatible with holoenzyme formation.
We have directly shown that σ2 and σ4 can be disulfide-linked in free σ28, and equilibrium measurements suggest that this represents a favored conformation. The maximum distance between the Cα atoms of two cysteines to form a disulfide is 7 Å (20), and in the σ28/FlgM complex, the Cα atoms of R31 and P202 are 5.36 Å apart. In the Taq holoenzyme (8), the corresponding residues of σA, G219 and T397, are 63.26 Å apart, appropriately spaced for −10 and −35 recognition (Table 2). The close proximity of these domains in free σ28 is thus incompatible with simultaneous recognition of these promoter elements, providing one explanation for autoinhibition and suggesting that these residues should move apart by ≈58 Å upon binding to core RNAP. A study using luminescence resonance energy transfer between introduced probes has shown that the distance between σ2 and σ4 increases by 15 Å in Escherichia coli σ70 upon holoenzyme formation (5). Although qualitatively this is consistent with our results, our data suggest a much greater magnitude. Alternatively, the average distance between these domains in free σ may be greater than that trapped by the disulfide, giving rise to a smaller net movement upon binding to core, but the equilibrium measurements argue against this. Another possibility is that the conformation of Escherichia coli σ70 is altogether different from that of σ28. Comparison of the Cα–Cα distances between the Taq σA residues corresponding to the other two disulfide pairs (m2 and m3) with those in the σ28/FlgM complex also reveals large differences in the two conformations (Table 2).
Table 2.
Distances (Cα–Cα) between σ28 residue pairs in the FlgM- and holoenzyme-bound conformations
We have also shown that σ3 and σ4 can be disulfide-linked in the m2 mutant and that this disulfide is favored at equilibrium in free σ28. The larger proportion of disulfide bonded species in free σ28 (compared with the FlgM complex) indicates that disulfide formation in this mutant is more rapid in the absence of FlgM (Fig. 3). This observation suggests that the proximity of the domains is similar but that rigidity imposed by FlgM decreases the rate of disulfide formation. Under equilibrium conditions, mostly oxidized protein was obtained for free σ28, regardless of whether the protein was reduced or oxidized at the beginning of the experiment (Fig. 3B). For the FlgM-bound species, in contrast, the population after incubation under equilibrium conditions resembled the starting material, suggesting that the site was inaccessible to the catalyst. FlgM does not directly occlude this site in the structure, but it may constrain interdomain flexibility which otherwise would allow access of the catalyst. S218 of Aae σ28 corresponds to R413 of Taq σA, which has been observed in a cocrystal structure to interact with the −35 element (4). Although the −35 binding surface of σ28 has not been established, it is reasonable to infer that σ28 binds to the −35 element using the same surface of σ4. Interaction with L133 of σ3 thus buries the −35 binding surface, directly occluding it, and providing a role for σ3 in autoinhibition of promoter binding. L133 is located on an acidic face of σ3, and the acidity of this surface is conserved in Group I σ factors, as noted in ref. 6. Thus, this mode of autoinhibition may be more general.
In Group I σ's, the N-terminal region 1.1 (σ1.1) contributes to the autoinhibition mechanism, because σ's with N-terminal truncations of σ1.1 show detectable, promoter-specific binding (21, 22). The autoinhibition was proposed to occur through a direct interaction between σ1.1 and conserved region 4.2 that would occlude the −35 element recognition determinants in region 4.2 (21). However, NMR studies (22) ruled out a direct σ1.1/σ4.2 interaction, indicating that σ1.1 acts indirectly to inhibit promoter binding. Our finding reported here lead us to suggest that in Group I σ's, σ1.1 may act indirectly to inhibit promoter binding by stabilizing a compact conformation of σ that is incompatible with promoter recognition.
Finally, we have demonstrated by using mutant m3 that a cysteine introduced into the σ3-σ4 linker cross-links to a cysteine in the C-terminal helix of σ4, both in the FlgM complex and in free σ28. In the σA-holoenzyme, the corresponding part of the linker is unfolded. Because the cross-link requires this residue to be on the appropriate side of the linker helix, its formation supports the idea that the σ3-σ4 linker is α-helical even in free σ28. The disulfide formation was more rapid in free σ28 than in the FlgM/σ28 complex for mutant m3 as well, again suggesting greater flexibility but a similar conformation of free σ28.
Together these data suggest that the conformation of free σ28 resembles that observed in the FlgM complex but that it is less rigid. This conformation provides an explanation for the autoinhibition of DNA binding and provides insight into the conformational change that accompanies binding to core RNAP. Autoinhibition is accomplished both by suboptimal interdomain distances and by steric occlusion of both promoter element binding surfaces. We suggest that these principles of σ factor regulation may apply to Group I σ factors as well.
Materials and Methods
Mutagenesis.
The program MODIP (18) provided a list of pairs of residues which, if mutated to cysteine, are predicted to form disulfides, based on the crystal structure of Aae σ28/FlgM (Protein Data Bank ID code 1RP3). Four double cysteine mutants of Aae σ28 were constructed in a triple methionine mutant I77M/L122M/L191M (σ28-3M) background. The cysteine mutations were introduced by site-directed mutagenesis, giving the quintuple mutants: m1, σ28-3M/R31C/P202C (σ2-31/σ4-202); m2, σ28-3M/R31C/E211C (σ2-31/σ4-211); m3, σ28-3/L133C/S218C (σ3-133/σ4-218); and the negative control m4, σ28-3M/Y140C/L225C (σlinker-140/σ4-225). The correct DNA sequence was confirmed for each construct.
Expression and Purification.
σ28-3M and each of the four disulfide mutants were expressed in BL21-CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA) except for m4, which was expressed in 4L Rosetta (DE3)pLysS (Novagen, San Diego, CA). The cells were grown in LB with shaking at 37°C. At OD600 of 0.4–0.6 expression was induced by adding 1 mM isopropyl β-d-thiogalactoside, and the temperature was lowered to 19°C. The cells were harvested by centrifugation after 14–16 h. Aae FlgM in pET21a (without any tag) was expressed separately in the same way. The pellets were resuspended in 20 mM Tris·HCl (pH 8.0), 0.5 M NaCl, and a protease inhibitor mixture (final concentration of protease inhibitors of 174 μg/ml PMSF, 312 μg/ml benzamidine, 5 μg/ml chymostatin, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 10 μg/ml aprotinin). The cells were lysed with a French press, and the soluble fraction was removed by centrifugation. One-half of the soluble lysate of each mutant was mixed with soluble FlgM lysate (corresponding to 2L culture), and the other half was kept separate. The lysates were then incubated at 65°C for 45 min to precipitate heat-sensitive Escherichia coli proteins. Precipitated contaminants were removed by centrifugation, and the soluble protein was purified by using Ni2+-chelating chromatography. For the FlgM complexes, this step removes excess FlgM, yielding a 1:1 σ28/FlgM complex. The Ni2+ eluates were further purified on a Superdex 75 (GE Healthcare, Piscataway, NJ) gel filtration column in 20 mM Tris·HCl (pH 8.0) and 0.5 M NaCl. The pure protein was concentrated to between 8 and 70 mg/ml in the same buffer. No reducing agent was added at any time during the purification, and disulfides formed during purification without the aid of a catalyst.
CNBr Cleavage and MALDI-TOF MS.
CNBr crystals were dissolved in 20 μl of 1 mg/ml protein in 70% TFA and then incubated for 19 h in the dark at room temperature. The samples were subsequently dried in a SpeedVac (Savant, Irvine, CA) and redissolved in 20 μl of water. For MALDI-TOF MS, the samples were first diluted to 1 mg/ml in 67% acetonitrile and 0.1% TFA and then diluted to 0.2 mg/ml in the same solvent saturated with α-cyano-4-hydroxycinnamic acid and immediately spotted on a MALDI sample plate as described (23).
SDS/PAGE Analysis.
Concentrated protein stock was diluted in 20 mM Tris·HCl (pH 8.0) and 0.5 M NaCl to 0.25–0.5 mg/ml. The protein was then mixed 4:1 with 5× SDS sample buffer (with or without β-mercaptoethanol, as noted), then heated to ≈100°C for 1 min before loading. NuPAGE 10% Bistris precast gels (Invitrogen, Carlsbad, CA) were used with Mops running buffer (50 mM Mops, 50 mM Tris base, 0.1% SDS, 1 mM EDTA, pH 7.7). The gels were run at 200 V for 1–2 h and stained with Coomassie brilliant blue.
Glutathione Catalyzed Disulfide Equilibrium Measurements.
The procedure was essentially as described in ref. 19. Concentrated protein was diluted to 1 mg/ml in 20 mM Tris (pH 8), 0.5 M NaCl, and 100 mM DTT and incubated at 37°C for 90 min. Fifty microliters of reduced protein was then dialyzed against >1 L of 10 mM Tris (pH 7.5), 0.5 M NaCl, and 0.1 mM EDTA for 4 h with a buffer change after 2 h, at 4°C. Ten microliters of dialyzed protein was removed and alkylated by adding 2.5 μl of 50 mM iodoacetamide and incubated for 1 h in the dark at room temperature. Twenty microliters of the remaining protein was mixed with 5 μl of 250 mM Tris·HCl (pH 9.0), 0.5 M NaCl, and 2.5 mM reduced glutathione and 2.5 mM oxidized glutathione and incubated for 16 h at room temperature, and then alkylated with iodoacetamide as described above.
Acknowledgments
We thank B. T. Chait (Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, The Rockefeller University) for access to MS facilities and Soumya S. Ray for helpful discussions. This work was supported by a National Science Foundation Graduate Research Fellowship (to M.K.S.) and National Institutes of Health Grant GM53759 (to S.A.D.).
Abbreviations
- RNAP
RNA polymerase
- Aae
Aquifex aeolicus
- m1, -2, -3, and -4
mutants 1, 2, 3, and 4.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 2.Gruber TM, Bryant DA. J Bacteriol. 1997;179:1734–1747. doi: 10.1128/jb.179.5.1734-1747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lonetto M, Gribskov M, Gross CA. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Mol Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 5.Callaci S, Heyduk E, Heyduk T. Mol Cell. 1999;3:229–238. doi: 10.1016/s1097-2765(00)80313-5. [DOI] [PubMed] [Google Scholar]
- 6.Sorenson MK, Ray SS, Darst SA. Mol Cell. 2004;14:127–138. doi: 10.1016/s1097-2765(04)00150-9. [DOI] [PubMed] [Google Scholar]
- 7.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 8.Murakami K, Masuda S, Darst SA. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 9.Murakami K, Masuda S, Campbell EA, Muzzin O, Darst SA. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 10.Murakami K, Darst SA. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 11.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 12.Malhotra A, Severinova E, Darst SA. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EA, Masuda S, Sun JL, Muzzin O, Olson CA, Wang S, Darst SA. Cell. 2002;108:795–807. doi: 10.1016/s0092-8674(02)00662-1. [DOI] [PubMed] [Google Scholar]
- 14.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Mol Cell. 2003;11:1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 15.Pervushin K, Riek R, Wider G, Wuthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony LC, Burgess RR. J Biol Chem. 2002;277:46433–46441. doi: 10.1074/jbc.M208205200. [DOI] [PubMed] [Google Scholar]
- 17.Anthony LC, Dombkowski AA, Burgess RR. J Bacteriol. 2002;184:2634–2641. doi: 10.1128/JB.184.10.2634-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sowdhamini R, Srinivasan N, Shoichet B, Santi DV, Ramakrishnan C, Balaram P. Protein Eng. 1989;3:95–103. doi: 10.1093/protein/3.2.95. [DOI] [PubMed] [Google Scholar]
- 19.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 20.Dani VS, Ramakrishnan C, Varadarajan R. Protein Eng. 2003;16:187–193. doi: 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- 21.Dombroski AJ, Walter WA, Record MT, Siegele DA, Gross CA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 22.Camarero JA, Shekhtman A, Campbell EA, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Proc Natl Acad Sci USA. 2001;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadene M, Chait BT. Anal Chem; 2000. pp. 5655–5658. [DOI] [PubMed] [Google Scholar]