Abstract
The formation of the dorsoventral (DV) boundary is central to establishing the body plan in embryonic development. Although there is some information about how limbs are positioned along the DV axis and how DV skin color pattern is determined, the way in which mammary glands are positioned is unknown. Here we focus on Bmp4 and Tbx3, a gene associated with ulnar-mammary syndrome, and compare their expression along the DV axis in relation to mammary gland initiation in mouse embryos. Tbx3 is expressed in the mammary gland-forming region with Tbx15, a gene involved in a DV coat color being expressed more dorsally and Bmp4 being expressed more ventrally. When Tbx3 was overexpressed, formation of mammary gland epithelium was extended along the DV axis. In contrast, overexpression of Bmp4 inhibited both Tbx3 and Tbx15 expression. In addition, when BMP signaling was inhibited by NOGGIN, Lef1 expression was lost. Thus, we propose that mutual interactions between Bmp4 and Tbx3 determine the presumptive DV boundary and formation of mammary glands in early mouse embryogenesis. 1,19-Dioctadecyl-3,3,39,39-tetramethyl indocarbocyanine perchloride labeling experiments showed that cells associated with mammary glands originate more dorsally and then move ventrally. This finding, together with previous findings, suggests that the same DV boundary may not only position limbs and determine coat color but also position mammary glands. Furthermore, Bmp signaling appears to be a fundamental feature of DV patterning.
Keywords: dorsoventral patterning, ulnar-mammary syndrome
A key event in vertebrate embryogenesis is establishment of the main body axes, anteroposterior (head to tail) and dorsoventral (DV; back to front), and specifying cell position along them to give the body plan. One mechanism for specifying cell position is through the response to gradients of various extracellular signaling molecules (1). Positional information is then encoded by expression of transcription factors that control subsequent development of that region of the embryo, and this ensures that organs are initiated in the correct locations. Striking examples of organs that develop at a particular DV level are the mammary glands (2). These arise along mammary lines that form at the boundary between anterior and lateral cutaneous nerve branches (3, 4) and run in an anteroposterior direction between forelimb and hindlimb (5, 6). These mammary lines are morphologically evident in the flank (interlimb region) of rabbit embryos and are marked by expression of several different genes, including Lef1 and Wnt10b, in mouse embryos (7). Here we examine mammary gland initiation and positioning with respect to DV body patterning in mouse embryos and examine the roles of Bmp signaling and genes that encode Tbx transcription factors.
Several aspects of DV body patterning have already been well documented, and some of the key molecules have been identified. DV patterning of the mesoderm in early embryos leads to tissue-specific differentiation. For example, dorsal explants from early frog embryos differentiate into muscle, and ventral explants form blood (8). In the embryo, mesoderm becomes regionalized to give somites, intermediate mesoderm, and lateral plate mesoderm, going from dorsal to ventral (9). DV body patterning is also crucial for positioning the limbs at the sides of the body. This positioning is accomplished by formation of the apical ectodermal ridge, the thickened epithelium required for limb bud outgrowth, at a DV compartment boundary in the body ectoderm (10). Yet another striking outcome of DV patterning is the difference between back and belly skin or coat color (11). Bmp signaling has been implicated in several of these examples. Thus, graded Bmp signaling specifies mesoderm pattern in early Xenopus embryos with high levels specifying ventral mesoderm, which differentiates into blood (12). Mesodermal regionalization in chicken embryos is also controlled by Bmp signaling with high levels of Bmp4 signaling specifying ventral lateral plate mesoderm (9). Finally, in ventral limb ectoderm, Bmp signaling acts upstream of the gene encoding the transcription factor Engrailed, which is required for proper DV patterning of the limb (13).
Several members of the T-box transcription factor family have been implicated in encoding position in embryos, and Tbx15 has been shown to play a role in DV specification of skin or coat color. In the absence of Tbx15 there is dorsal displacement of yellow belly hair in agouti black and tan mice (11). Interestingly, another Tbx gene, Tbx3, is associated with mammary gland development. Haploinsufficiency of Tbx3 has been associated with ulnar-mammary syndrome (UMS) in human patients (14). UMS is an inherited disorder characterized by deficiencies in the ulnar ray in the upper limb and hypoplasia of the mammary glands. In Tbx3−/− mouse embryos there is almost complete failure of initiation of mammary gland development (15).
One attractive possibility is that molecular mechanisms similar to those involved in other aspects of DV patterning are used in formation of the mammary glands. Therefore, we first compared expression patterns of Bmp4 and Tbx3 with respect to the DV boundary of the body where mammary gland development is initiated and then tested their involvement in mammary gland positioning. The results have implications for understanding how UMS arises during embryonic development and for the hypothesis that Bmp signaling and Tbx gene transcription factors are fundamental to DV patterning of the body.
Results
Gene Expression in Relation to the DV Position of Mammary Gland Development.
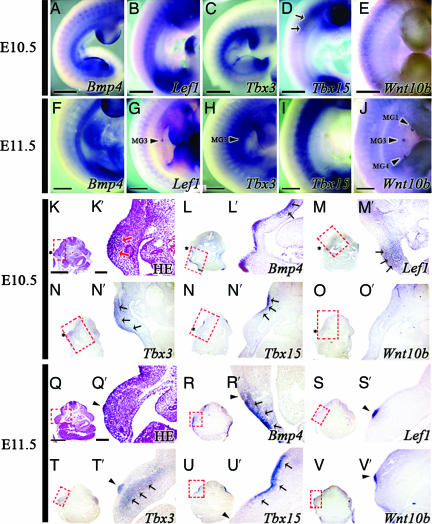
In mice, five individual mammary glands (numbered 1–5 from anterior to posterior) form along two mammary lines running down each side of the ventral region of the body. In embryonic day 10.5 (E10.5) embryos, an epithelial thickening forms in the flank between the forelimb bud and the posterior hindlimb bud (Fig. 1 K and K′). At E11.5 the third mammary placode is first detected as epithelial thickening (Fig. 1 Q and Q′), and over the next day the remaining four mammary placodes form. These epithelial thickenings then grow down into the underlying mesenchyme to form bud-like structures (6, 16).
Fig. 1.
Gene expression patterns of Lef1, Bmp4, Tbx3, Tbx15, and Wnt10b in the flank around the time of mammary gland initiation at E10.5 and E11.5. (A–J) Whole-mount in situ hybridization. (K–V and K′–V′) Section in situ hybridization showing the expression of each gene in serial sections of the same embryo. Histological sections of the mammary gland-forming area at E10.5 (K and K′) and E11.5 (Q and Q′) mouse embryos are shown. (A–E, K–P, and K′–P′) E10.5. (A) Bmp4 expressed in the ventral flank. (L and L′) Bmp4 expression detected in the flank mesenchyme. (B) Broad Lef1 expression seen in the flank. (M and M′) Lef1 was also expressed in the ventral and dorsal regions of the flank. (C) Tbx3 was strongly expressed in the anterodorsal region and was expressed in a broad band in the flank. (N and N′) A Tbx3 section showing expression in both the epithelium and mesenchyme. (D) Tbx15 expressed in the anterodorsal part of the body. (O and O′) Tbx15 expressed in the mesenchyme. (E, P, and P′) Wnt10b expression was not detected in the flank. (F–J, Q–V, and Q′–V′) E11.5. (F) Bmp4 was strongly expressed in the ventral region. (R and R′) Bmp4 was strongly expressed in both the epithelium and underlying mesenchyme of the ventral flank. (G, S, and S′) Restricted Lef1 expression was observed in the epithelium of the third mammary bud (arrowheads) at E11.5. (H, T, and T′) Tbx3 was expressed in the epithelium and mesenchyme underlying the third mammary bud (arrows). (I) Tbx15 expressed in the dorsal band. (V and V′) Tbx15 expressed in the dorsal flank beneath the mesenchyme at E11.5. (J, V, and V′) Wnt10b was expressed in the epithelium of the mammary buds. Red dotted boxes indicate areas of higher magnification. Black arrowheads indicate mammary glands (MG1, MG3, and MG4). Asterisks indicate epithelial thickening in a region where the third mammary gland (MG3) will form. Red arrows indicate mesenchymal condensation. Black arrows indicate strong mRNA expressions. (Scale bars: 125 μm.)
Bmp4, Lef1, Tbx3, Tbx15, and Wnt10b gene expression patterns were documented with respect to mammary gland development and the DV boundary in E10.5 and E11.5 mouse embryos using whole-mount in situ hybridization (Fig. 1 A–J). To determine the precise boundaries of expression of individual genes and to compare precisely expression patterns of different genes, section in situ hybridization methods were used and adjacent serial sections were examined (Fig. 1 L–P, L′–V′, R–V, and R′–V′). At E10.5 Lef1 was detected in both ventral and dorsal regions of the flank (Fig. 1 B, M, and M′) whereas Bmp4 expression in the ectoderm and underlying mesenchyme was observed ventrally (Fig. 1 A, L, and L′). Tbx3 was expressed in a broad band all along the anteroposterior axis of the flank between forelimb and hindlimb (Fig. 1C), with strong expression in the thickened epithelium that forms the mammary line and underlying mesenchyme (Fig. 1 N and N′). Tbx15 was expressed in the dorsal region of the flank between forelimb and hindlimb in epithelium and underlying mesenchyme (Fig. 1 D, O, and O′). Wnt10b was not detected in the flank region of the E10.5 embryos (Fig. 1 E, P, and P′). At E11.5 Lef1 was expressed in the discrete epithelial thickening, which is the earliest sign of the third mammary bud (Fig. 1 G, S, and S′) whereas Bmp4 was still expressed in both ventral epithelium and underlying mesenchyme (Fig. 1F). Compared with E10.5, Bmp4 expression was much stronger in the epithelium and weaker in the mesenchyme (Fig. 1 R and R′). At E11.5 the intensity of Tbx3 expression was much higher than E10.5 (Fig. 1H). Section in situ hybridization showed that Tbx3 is expressed in both epithelium and mesenchyme in the area in which the third mammary bud is forming (Fig. 1 T and T′) whereas Tbx15 expression was dorsal and restricted to mesenchyme just beneath epithelium (Fig. 1 I, U, and U′). At E11.5 Wnt10b expression was observed not only in the third mammary gland (MG3) but also in the first (MG1) and fourth (MG4) mammary glands (Fig. 1 J, V, and V′).
Bmp4 and Tbx3 Play Key Roles in DV Patterning of Mammary Glands.
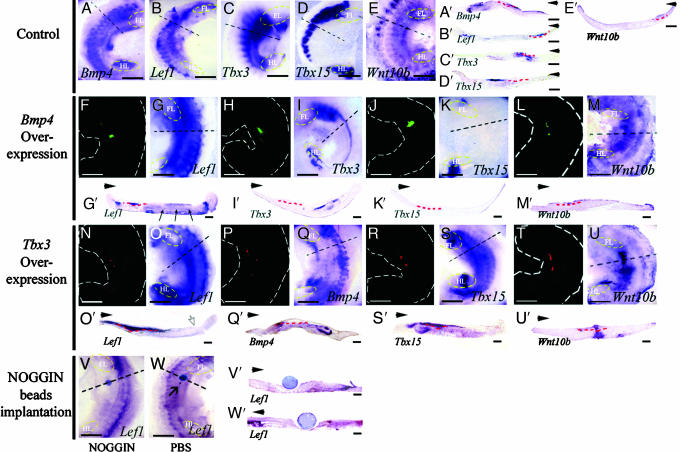
To test the interactions between Bmp4 and Tbx3 we electroporated expression constructs containing either Bmp4 or Tbx3 together with a fluorescent reporter protein into the mouse flank posterior to the forelimb at E10.0, and the electroporated mouse flank was then placed in in vitro organ culture for 48 h. Expression patterns of Lef1, Bmp4, Tbx3, Tbx15, and Wnt10b were then examined to elucidate the signaling network that specifies the formation of mammary glands. Fig. 2 A–E and A′–E′ shows expression of these genes in controls in which only the vector expressing fluorescent protein was electroporated.
Fig. 2.
Mammary gland initiation after manipulation of Bmp4 signaling and Tbx3. (A–E) Whole mount in situ hybridization after electroporation of E10.0 mouse embryo with a vector containing only fluorescent protein into the right flank. (F–U) Overexpression of Bmp4–pEGFP-N1 and Tbx3–pIRES-DsRed in the ventral flank posterior to the regions of the forelimb bud. The left flank of each E10.0 embryo was used as the experimental side, and the right flank was used as the control. (A–U) In vitro organ culture for 48 h and whole-mount in situ hybridization after electroporation. (A′–U′) Transverse sections after in situ hybridization of in vitro organ cultured tissue. (F–U) Dark-field views of ectopic GFP expression (F–M) and of DsRed (N–U). (F–M and F′–M′) Overexpression of Bmp4. (G and G′) Lef1 expression in the dorsal mesenchyme induced by Bmp4 overexpression in the flank. (I, I′, K, and K′) Expression of Tbx3 and Tbx15 was reduced by Bmp4 overexpression. (M and M′) Wnt10b expression; no change after Bmp4 overexpression. (N–U and N′–U′) Tbx3 overexpression. (O and O′) Lef1 expression was increased and was more widely expressed, and the epithelium was thickened. (Q and Q′) Bmp4 expression was reduced by Tbx3 overexpression. (S and S′) Tbx15 expression extended more ventrally in the flank when Tbx3 was overexpressed. (M, M′, U, and U′) Wnt10b was expressed throughout the whole depth of the mesenchyme in the mammary gland-forming area after Tbx3 overexpression. (V, V′, W, and W′) Effect of NOGGIN on gene expression in the developing flank at E10.0. (V and W) In vitro organ cultures 48 h after implanting NOGGIN (V) and PBS-soaked beads (W) to the flank posterior to the forelimb in E10.0 embryos after whole-mount in situ hybridization for Lef1. (V′ and W′) Section through beads. (V and V′) Lef1 was inhibited in the region around the NOGGIN bead and in the third mammary bud. (W and W′) No changes in the Lef1 expression pattern in the flank or third mammary bud were observed (arrow). The yellow dotted line indicates the limb. FL, forelimb; HL, hindlimb. The red dotted line indicates the basement membrane of epithelial thickening. The black dotted line indicates the section level. The white dotted line indicates the outlining of embryo. The point of each arrowhead indicates dorsal direction. The open arrows indicate the ventral margin of the somite region. The filled arrows in G′ indicate the mesenchymal Lef1 ectopic expression after Bmp4 overexpression. (Scale bars: 150 μm.)
After overexpression of both Bmp4 (Fig. 2 F, H, J, and L) and Tbx3 (Fig. 2 N, P, R, and T), Lef1 expression increased in the flank (Fig. 2 G and O). Transverse sections showed that, after Tbx3 overexpression in the flank, the thickened epithelium expressing Lef1 was dorsally extended toward the somite-forming region and dorsal midline (n = 26/30; 86.7%) (Fig. 2O′) rather than being a discrete epithelial thickening expressing Lef1 as in the control third mammary bud. In four cases the thickened epithelium appeared to be extended bidirectionally. The sections also confirmed that the intensity of Lef1 expression was more pronounced than in controls (compare Fig. 2 B′, G′, and O′). In contrast, Bmp4 overexpression did not alter the morphogenesis of the mammary gland-forming epithelium, and only the discrete thickening associated with the third mammary bud was present (n = 30/30; 100%) (Fig. 2G′).
To examine further the regulation of Lef1 by Bmp4, beads soaked in the Bmp antagonist NOGGIN were implanted posterior to the forelimb in E10.0 mouse embryo flank and then cultured for 48 h. This application of NOGGIN resulted in local inhibition of Lef1 expression (n = 30; frequency of inhibition 70%) (Fig. 2 V and V′). As controls, beads soaked in PBS were implanted (n = 30), and in these cultures Lef1 expression was unaffected (Fig. 2 W and W′). These data confirm that Bmps regulate Lef1 expression. Thus both Tbx3 and Bmp4 appear to play key roles in mammary gland initiation by regulating Lef1 expression.
Lef1 and Wnt10b expression patterns were similar in early mammary gland development, with the genes being expressed in a raised streak of lateral body wall epithelium at E11.5 (Fig. 1 G, J, S, S′, V, and V′). However, after Tbx3 and Bmp4 overexpression, expression of Wnt10b did not respond in the same way as expression of Lef1. Thus, Wnt10b expression was not changed by Bmp4 overexpression (n = 26/30; 86.7%) (Fig. 2 M and M′) whereas, in contrast, after Tbx3 overexpression Wnt10b was expressed not only in the epithelium but also in the whole depth of mesenchyme under the third mammary gland-forming area (n = 29/30; 96.6%) (Fig. 2 E, E′, U, and U′).
Bmp4 overexpression led to changes in Tbx3 and Tbx15 expression patterns (Fig. 2 I, I′, K, and K′). Overexpression of Bmp4 completely abolished Tbx15 expression in the flank region (n = 30/30; 100%) (Fig. 2 K and K′), and Tbx3 expression was almost completely inhibited except in the mesenchyme along the DV border between the somite-forming area and dorsal flank (n = 29/30; 96.6%) (Fig. 2 I and I′). These results suggest that Bmp4 signaling regulates the extent of expression of T-box genes along the DV axis of the flank.
There were also changes in Tbx15 and Bmp4 expression after Tbx3 overexpression (n = 30/30; 100%) (Fig. 2 Q, Q′, S, and S′). Tbx15 expression was expanded into both dorsal and ventral mesenchyme at the site of the epithelial thickening that marked the mammary line (compare Fig. 2 D, D′, S, and S′). Bmp4 expression, in contrast, was inhibited where Tbx3 was overexpressed in the flank region (n = 30/30; 100%) (compare Fig. 2 A, A′, Q, and Q′).
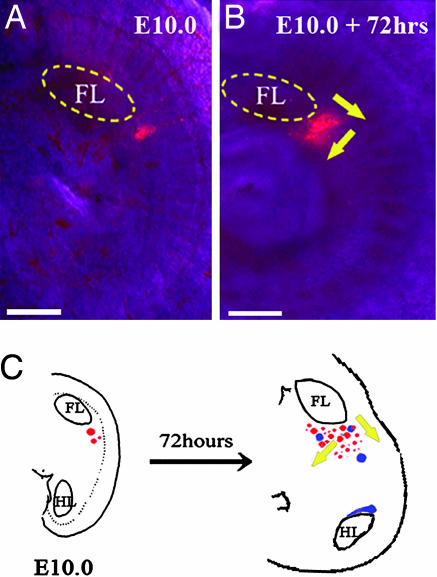
So what is the relationship between the DV boundary at which the limbs develop and that at which mammary glands form? To address this question, we used the lipophilic dye 1,19-dioctadecyl-3,3,39,39-tetramethyl indocarbocyanine perchloride (DiI) to follow cell fate during early mammary gland formation (Fig. 3). We labeled cells with DiI in the flank just posterior to the limb bud at the same DV level as the limb bud. After 72 h of culture the patch of DiI-labeled cells had extended not only posteriorly along the flank but also more ventrally to occupy the area of the forming mammary glands (Fig. 3).
Fig. 3.
Tracing cell movement by DiI microinjection. (A) DiI injected into the ventral–lateral flank posterior to the forelimb bud of E10.0. (B) After culturing the flank for 72 h in in vitro organ culture DiI labeling was monitored to trace the fate of marked cells. (C) Schematic diagrams showing experiments and results together with the location of the mammary glands. FL, forelimb; HL, hindlimb. Red spots, DiI; blue spots, mammary buds; yellow arrows, displacement of DiI-labeled cells. (Scale bars: 125 μm.)
Discussion
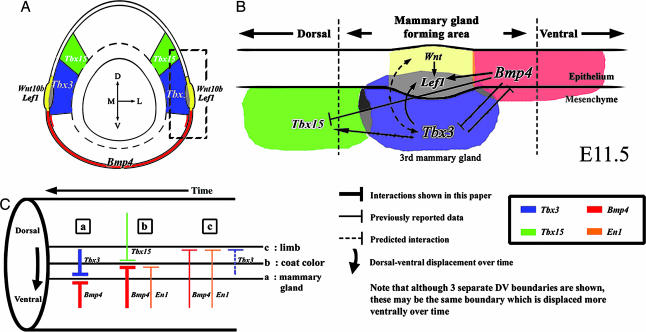
From our results we propose a model for DV patterning of mammary glands. We have shown that expression domains of Bmp4, Lef1, Tbx3, Tbx15, and Wnt10b are specifically localized to different DV levels around the body at the time when mammary gland development is initiated at E11.5 (Fig. 4A). Tbx3 is expressed in the epithelium of the mammary bud and the mesenchyme underlying Lef1 and Wnt10b expression, which marks the DV position at which mammary glands develop, whereas Tbx15 and Bmp4 are expressed dorsally and ventrally, respectively. These striking position-dependent patterns of gene expression along the DV body axis just before and during early mammary gland formation suggest that interactions between these genes, in particular Bmp4 and Tbx3, might control body patterning with respect to mammary gland formation.
Fig. 4.
Schematic diagrams showing position-dependent patterns of gene expression along the DV body axis (A), interactions controlling the position of mammary gland development at E11.5 (B), and a comparison of DV patterning with respect to the mammary glands, coat color, and limbs (C). (A) Transverse section showing patterns of gene expression in relation to the DV axis. (B) Regulatory gene interactions that establish DV patterns of gene expression at E11.5 in the mammary gland-forming region and govern the position of mammary gland initiation. Note that it was not elucidated whether these interactions are direct or indirect. (C) Lateral view of the flank showing interactions. (a) Positioning the mammary glands. (b) Determining coat color boundary. (c) Positioning the limbs. Note that although three separate DV boundaries are shown, these may be the same boundary that is displaced ventrally over time. D, dorsal; V, ventral; M, medial; L, lateral.
We tested this hypothesis by overexpressing Bmp4 and Tbx3 in cultured mouse flanks. Our overexpression experiments showed that there is reciprocal negative regulation between Bmp4 and Tbx3 (Fig. 4B) and that overexpression of Tbx3 could induce Lef1 expression and produce a DV extension of the epithelial thickening of the ectoderm characteristic of the mammary placode. Thus, we propose that inhibitory effects of Bmp4 on Tbx3 might establish a DV boundary, which would then serve to confine Lef1 expression and thickened mammary epithelium to a particular position with respect to the DV body axis. Our experiments with NOGGIN suggest that Bmp signaling also plays a role in maintaining Lef1 expression in the mammary placode.
How does Tbx3 induce Lef1 expression and a thickened mammary placode? As in tooth development (17), Lef1 might direct mesenchymal condensation and be involved in the Wnt pathway that induces an epithelial thickening. Wnt10b and Wnt6 are expressed along the DV boundary of the flank and then become confined to the mammary placodes (16, 18). Consistent with a role for Wnt10b, we found that, when Tbx3 was overexpressed, Wnt10b expression was increased in the mesenchyme of the mammary ridge. Thus, Tbx3 may play a crucial role at the DV boundary by controlling Wnt10b and Lef1 expression and mammary gland initiation (Fig. 4B). These data are consistent with observations on Tbx3−/− mouse embryos, in which neither Lef1 nor Wnt10b could be detected in the regions where mammary glands normally form (15, 16). The proposed involvement of Tbx3 in both setting a DV boundary of the body and initiating mammary gland development could explain why mammary glands fail to form in Tbx3−/− embryos and why, in UMS, which is caused by Tbx3 haploinsufficiency, mammary glands are reduced.
There are striking similarities between the mechanisms that we propose for mammary gland positioning and those that control DV coat color and position the limbs (Fig. 4C). We have proposed that antagonistic interactions between Bmp4 and Tbx3 are involved in initiation of mammary gland formation at a particular DV level (Fig. 4C). From this viewpoint, observations on UMS human patients (19) together with absence of mammary glands in Tbx3−/− mouse embryos might be considered in terms of ventralization of the flank. We have also shown that Bmp4 signaling inhibits expression of Tbx15, which has previously been shown to specify dorsal coat color and have a complementary expression pattern to En1 (Fig. 4C) (11). Work by others has shown that, in the absence of Tbx15, the belly coat color extends more dorsally and therefore again could be considered to be due to (partial) ventralization of the flank (11). Finally, previous work has shown that Bmp4 signaling upstream of En1 also specifies the ventral ectodermal compartment and controls ventral limb pattern (13). Limb bud development occurs much earlier than mammary gland development, and, because fingers can be dorsalized in some human patients with UMS (14, 19), we suggest that, at this earlier stage, Tbx3 may act in concert with Bmp4 to specify ventral limb pattern. Indeed, the importance of Bmp signaling in maintaining Tbx3 expression in the developing limb is well documented (20).
It is not clear whether the same DV boundary operates in all three patterning processes. We have shown that cells that participate in mammary gland formation originate more dorsally and then become displaced ventrally. Furthermore, Tbx15 expression has been reported to extend more ventrally as development proceeds. Therefore, it is possible that the same boundary is used but by means of different target genes, including two gene members of the Tbx family, and at successive times in development.
Materials and Methods
All experiments were performed according to the guidelines of the Intramural Animal Use and Care Committee of Yonsei University College of Dentistry.
Animals.
Adult Institute of Cancer Research mice were housed in a temperature-controlled room (22°C) under artificial illumination (lights on from 0500 to 1700 hours) and 55% relative humidity. The mice had access to food and water ad libitum. Embryos were obtained from time-mated pregnant mice. E0 was designated as the day a vaginal plug was confirmed. Embryos at developmental stages E10.0, E10.5, and E11.5 were used in this study.
in Vitro Organ Culture.
Institute of Cancer Research mouse embryos were isolated at E10.0 and placed in culture medium (BGJb; Sigma, St. Louis, MO) augmented with 0.5% penicillin/streptomycin and 0.2% ascorbic acid as previously described (6). Briefly, individual embryos were dissected into left and right halves by using fine tungsten needles to bisect the neural tube. The left flank was the experimental tissue, and the right acted as control. Each flank tissue was placed on filter membranes (Track-etch, 1.0-μm pore; Whatman Nuclepore), which were supported on stainless steel grids in sterile culture dishes, and cultured at the air–medium interface at 37°C and 7.5% CO2 for 48 and 72 h. Culture medium was replaced at 24 h. Tissues were then fixed and processed for in situ hybridization. At least 30 explants were used in each experiment.
DiI Microinjection.
DiI (Molecular Probes, Eugene, OR) is a vital, highly fluorescent, lipophilic dye belonging to the carbocyanine dye family. After DiI microinjection into designated regions of the flank, the migration pattern of DiI-labeled cells was observed at 72 h by using fluorescence microscopy (MZ-FLIII; LEICA, Jena, Germany).
In Situ Hybridization.
In situ hybridization was performed as previously described by Kim et al. (21). Section in situ hybridization was performed as previously described on wax sections by using standard protocols (21). Briefly, embryos were fixed in 4% paraformaldehyde, embedded in paraffin wax, and sectioned at 12 μm. The following DNA plasmids were used as templates for the synthesis of digoxigenin-labeled RNA probes: Bmp4 and Lef1 (from Yi-Ping Chen, Tulane University, New Orleans, LA); Tbx3 and Tbx15 (from Gregory S. Barsh, Stanford University School of Medicine, Stanford, CA); and Wnt10b.
Bead Implantation.
Affigel Blue beads (Bio-Rad), 150 μm in diameter, were washed with PBS and then soaked in 0.5 mg/ml human recombinant NOGGIN (Regeneron). Beads were implanted in the flank region along the mammary line of E10.0 mouse embryos.
Expression Constructs.
Constructs were pEGFP-N1 and pIRES-DsRed, both of which have been optimized for generating proteins with brighter fluorescence (Clontech). Bmp4 was inserted into the blunted EcoRI/HindIII sites of pEGFP-N1, and Tbx3 was inserted into the XhoI/BamHI sites of pIRES-DsRed.
Electroporation.
Plasmid DNA was purified by using a plasmid purification kit (Qiagen, Valencia, CA) and dissolved in T1/4E (10 mM Tris·HCl, pH 8.0/0.25 mM EDTA). Fast Green at 1/10,000 (Sigma) was added to the DNA solution for visualization within the tissue. A microcapillary needle was used to inject ≈1 μg/μl DNA into the flank mesenchyme, after which 20-ms current pulses of 25 V were applied with an electroporator. The experimental group comprised the left flank electroporated with either Bmp4 in pEGFP-N1 or Tbx3 in pIRES-DsRed construct. The right flank was electroporated with constructs containing only fluorescent proteins (EGFP and DsRed) and used as controls.
Acknowledgments
We thank Dr. Christine Campbell (State University of New York, Buffalo, NY) for the Tbx3 full-length cDNA. This research was supported by Grant R13-2003-13 from the Basic Research Program of the Korea Science and Engineering Foundation and a Royal Society Korea–U.K. joint project grant (to H.-S.J. and C.T.). C.T. receives additional support from The Royal Society. M.C.E. was supported by an Anatomical Society Research Scholarship.
Abbreviations
- DV
dorsoventral
- DiI
1,19-dioctadecyl-3,3,39,39-tetramethyl indocarbocyanine perchloride
- UMS
ulnar-mammary syndrome
- En
embryonic day n
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Wolpert L. Development (Cambridge, UK) 2003;130:4497–4500. doi: 10.1242/dev.00728. [DOI] [PubMed] [Google Scholar]
- 2.Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, Rice R, Spencer-Dene B, Mailleux AA, Rice DP, et al. Development (Cambridge, UK) 2006;133:2325–2335. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- 3.Merkel F. Henle's Grundriss der Anatomiedes des Menschen, Atlas. 4th Ed. Braunschweig, Germany: Friendrich Vieweg und Sohn; 1901. p. 247. [Google Scholar]
- 4.Kollmann J Integmentum commune et Organa sensuum. Handatlas der Entwicklungsgeschichte des Menschen, Zweiter Teil. Jena, Germany: Gustav Fischer; 1907. Fig 654. [Google Scholar]
- 5.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 6.Eblaghie MC, Song S-J, Kim J-Y, Akita K, Tickle C, Jung H-S. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Differentiation. 2003;71:1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- 8.Jones CM, Dale L, Hogan BL, Wright CV, Smith JC. Development (Cambridge, UK) 1996;122:1545–1554. doi: 10.1242/dev.122.5.1545. [DOI] [PubMed] [Google Scholar]
- 9.Tonegawa A, Funayama N, Ueno N, Takahashi Y. Development (Cambridge, UK) 1997;124:1975–1984. doi: 10.1242/dev.124.10.1975. [DOI] [PubMed] [Google Scholar]
- 10.Altabef M, Clarke JD, Tickle C. Development (Cambridge, UK) 1997;124:4547–4556. doi: 10.1242/dev.124.22.4547. [DOI] [PubMed] [Google Scholar]
- 11.Candille SI, Van Raamsdonk CD, Chen C, Kuijper S, Chen-Tsai Y, Russ A, Meijlink F, Barsh GS. PLoS Biol. 2004;2:30–42. doi: 10.1371/journal.pbio.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C. Development (Cambridge, UK) 1997;124:2325–2334. doi: 10.1242/dev.124.12.2325. [DOI] [PubMed] [Google Scholar]
- 13.Pizette S, Abate-Shen C, Niswander L. Development (Cambridge, UK) 2001;128:4463–4474. doi: 10.1242/dev.128.22.4463. [DOI] [PubMed] [Google Scholar]
- 14.Bamshad M, Le T, Watkins WS, Dixon ME, Kramer BE, Roeder AD, Carey JC, Root S, Schinzel A, Van Maldergem L, et al. Am J Hum Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Development (Cambridge, UK) 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 16.Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S. Dev Dyn. 2004;229:349–356. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T, Ito Y, Xu X, Han J, Bringas P, Jr, Maeda T, Slavkin HC, Grosschedl R, Chai Y. Dev Biol. 2005;278:130–143. doi: 10.1016/j.ydbio.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Lane TF, Leder P. Oncogene. 1997;15:2133–2144. doi: 10.1038/sj.onc.1201593. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki G, Ogata T, Ishii T, Hasegawa T, Sato S, Matsuo N. Am J Med Genet. 2002;110:365–369. doi: 10.1002/ajmg.10447. [DOI] [PubMed] [Google Scholar]
- 20.Tumpel S, Sanz-Ezquerro JJ, Isaac A, Eblaghie MC, Dobson J, Tickle C. Dev Biol. 2002;250:251–262. [PubMed] [Google Scholar]
- 21.Kim J-Y, Cho SW, Lee MJ, Hwang HJ, Lee JM, Lee SI, Muramatsu T, Shimono M, Jung H-S. Cell Tissue Res. 2005;320:409–415. doi: 10.1007/s00441-005-1091-y. [DOI] [PubMed] [Google Scholar]