Abstract
The establishment of the primitive streak and its derivative germ layers, mesoderm and endoderm, are prerequisite steps in the formation of many tissues. To model these developmental stages in vitro, an ES cell line was established that expresses CD4 from the foxa2 locus in addition to GFP from the brachyury locus. A GFP-Bry+ population expressing variable levels of CD4-Foxa2 developed upon differentiation of this ES cell line. Analysis of gene-expression patterns and developmental potential revealed that the CD4-Foxa2hiGFP-Bry+ population displays characteristics of the anterior primitive streak, whereas the CD4-Foxa2loGFP-Bry+ cells resemble the posterior streak. Using this model, we were able to demonstrate that Wnt and TGF-β/nodal/activin signaling simultaneously were required for the generation of the CD4-Foxa2+GFP-Bry+ population. Wnt or low levels of activin-induced a posterior primitive streak population, whereas high levels of activin resulted in an anterior streak fate. Finally, sustained activin signaling was found to stimulate endoderm commitment from the CD4-Foxa2+GFP-Bry+ ES cell population. These findings demonstrate that the early developmental events involved in germ-layer induction in the embryo are recapitulated in the ES cell model and uncover insights into the signaling pathways involved in the establishment of mesoderm and endoderm.
Keywords: gastrulation, mesoderm, endoderm
To be able to fully exploit the potential of embryonic stem (ES) cells in basic biology and regenerative medicine, it is essential to recapitulate the critical lineage induction events of the early embryo in this model system. One of the earliest commitment steps in embryogenesis is the formation of the primary germ layers, mesoderm, endoderm, and ectoderm, the founder populations of all somatic cell types in the body (1). Mesoderm and endoderm are formed during gastrulation, a process that involves the movement of undifferentiated epiblast cells through a structure called the primitive streak (PS). The PS can be subdivided into distinct regions, posterior, middle, and anterior, based on lineage development and gene-expression patterns. With respect to developmental potential, distinct subpopulations of mesoderm are induced in each of the different regions, whereas definitive endoderm forms from the anterior PS of the early-PS and mid-PS stages (2–5). These observations suggest that the different regions of the PS constitute different signaling environments that are responsible for the induction of specific lineages.
To address questions regarding early commitment by using the ES cell model, it is essential to be able to track and isolate germ-layer populations from the differentiation cultures. By using an ES cell line with the green fluorescence protein (GFP) targeted to the PS and early mesodermal-specific gene brachyury (6) (GFP-Bry ES cells), it has been possible to quantify mesoderm induction and isolate and characterize different mesodermal populations (7, 8). In addition, brachyury-expressing cells were found to give rise to endoderm derivatives (9). Endoderm was induced with a high concentration of activin A, a factor that binds the same receptor as nodal and likely mimics nodal signaling. Recent studies have confirmed the endoderm-inducing properties of activin in mouse ES cell differentiation cultures (10, 11). The ability of activin to induce endoderm is not restricted to mouse cells as it has been shown to induce similar populations in human ES cell differentiation cultures (12). These observations illustrate the utility of using appropriately marked ES cells for monitoring the earliest induction events in differentiation cultures and identifying the molecules that regulate them.
To further investigate the earliest stages of mesoderm and endoderm formation in the ES cell model, we targeted the human CD4 cDNA to the foxa2 locus in the GFP-Bry ES cells. Foxa2 was selected as it is expressed in the anterior PS, containing definitive endoderm precursors, as well as in most mature endoderm-derived tissues (13–15). In this article, we demonstrate that this ES cell line can be used to isolate cell populations with gene-expression profiles and developmental potentials similar to the posterior and anterior PS of the embryo. We also show that activation of both the Wnt and nodal signaling pathways are required together for the formation of PS like cells in the ES cell differentiation cultures. Finally, we have uncovered two distinct roles for activin signaling in early germ-layer formation. The first is the induction of the PS population, and the second is in the specification of endoderm from the PS.
Results
Generation of an ES Cell Line That Expresses Human CD4 from the Foxa2 Locus.
The foxa2 targeting construct (Fig. 6A, which is published as supporting information on the PNAS web site) contains a truncated version of the human CD4 cDNA that lacks most of the intracellular signaling domain (16). GFP-Bry ES cells were electroporated with the foxa2 targeting vector, and appropriately targeted clones were identified by Southern blot analysis (Fig. 1A). Two targeted clones were generated and transiently transfected with a Cre recombinase expression vector to remove the puro gene. Both clones had a normal karyotype. The targeted clones are hereinafter referred to as CD4-Foxa2/GFP-Bry ES cells.
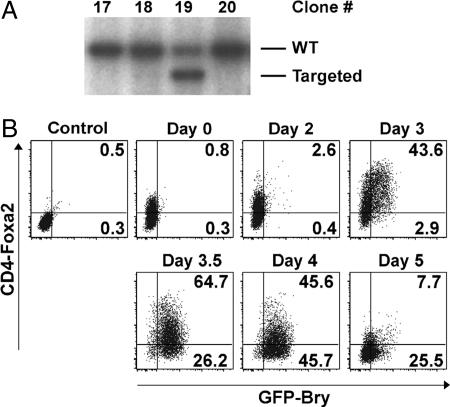
Fig. 1.
Targeting human CD4 to the Foxa2 locus. (A) Southern blot analysis depicting a targeted clone (#19). (B) Flow cytometric analysis of CD4-Foxa2 and GFP-Bry expression in EBs differentiated for varying periods of time. Day 0 is undifferentiated ES cells. Control represents unstained ES cells.
CD4 was expressed at low to undetectable levels on undifferentiated ES cells (Fig. 1B). A kinetic analysis of serum-induced embryoid body (EB) differentiation revealed that the levels of CD4 increased concurrently with the onset of GFP-Bry expression, and by day 3 all GFP+ cells expressed CD4. By day 3.5 of differentiation, CD4 expression decreased substantially and continued to decline to undetectable levels by day 5. GFP expression persisted at high levels to day 4 and then showed a sharp reduction by day 5 [displayed as mean fluorescence intensity (MFI)] (Fig. 6 B). The expression of endogenous foxa2 and brachyury paralleled the patterns of CD4 and GFP, respectively (Fig. 6C). To further characterize the CD4 expression pattern, chimeric mice were generated with CD4-Foxa2/GFP-Bry ES cells, and the resulting gastrulating embryos were costained for CD4 and Foxa2 (Fig. 7A, which is published as supporting information on the PNAS web site). CD4 expression colocalized with Foxa2 in the anterior PS, whereas it was absent from the anterior visceral endoderm. This is the expected pattern because ES cells do not contribute to extra-embryonic endoderm (17). These findings demonstrate that CD4 expression recapitulates expression of foxa2 both in vitro and in vivo and, as such, provides a valid marker for monitoring anterior PS and endoderm development.
Modeling PS Development by Using the CD4-Foxa2/GFP-Bry ES Cell Line.
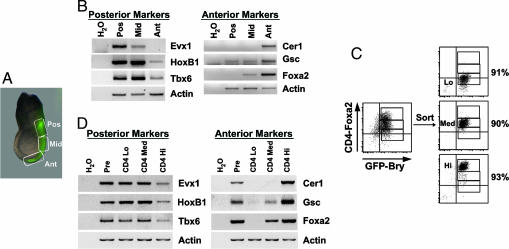
During serum-induced differentiation, GFP expression varied over a relatively narrow range (≈1 log), whereas the expression pattern of CD4-Foxa2 was broad, showing a 2-log difference in levels (Fig. 1B). One interpretation of this pattern is that the brachyury-expressing population is the EB equivalent of the PS with the CD4-Foxa2hi- and CD4-Foxa2lo-expressing cells representing the anterior and posterior regions of the streak, respectively. Consistent with this interpretation is the observation that other genes found in and around the PS, including cer1, gsc, hoxB1, evx1, wnt3, and nodal (5, 18–21), were expressed in the EBs at the same stage that brachyury and foxa2 were expressed (Fig. 6C). In contrast, expression of the ectodermal marker Pax6 (22) was down-regulated with the onset of brachyury and foxa2 expression. To test the interpretation that the CD4-Foxa2 subpopulations represent different regions of the PS, CD4hiGFP+, CD4medGFP+, and CD4loGFP+ fractions were isolated by cell sorting (Fig. 2C) and compared with the posterior, middle, and anterior PS regions from embryonic day 7.25 embryos (Fig. 2A) by gene-expression analyses. The known posterior PS genes evx1, hoxB1, and tbx6 (23) were expressed in the posterior, middle, and anterior regions of the mouse PS in a pattern similar to that found in the CD4loGFP+, CD4medGFP+, and CD4hiGFP+ fractions (Fig. 2 B and D). In addition, the anterior PS markers cer1, gsc, and foxa2 showed highest expression in the anterior PS or CD4hiGFP+ cells. The pattern of foxa2 expression corresponds well to CD4 levels. To determine whether the differences in message levels reflect differences in Foxa2 protein, CD4hi and CD4lo cells were stained with an anti-Foxa2 antibody (Fig. 7B). Foxa2 was detected only in the CD4hi population. The striking similarity in the expression patterns of these genes in the different regions of the PS and the EB-derived populations supports the interpretation that the CD4-Foxa2+GFP-Bry+-expressing EB population represents the in vitro equivalent of the PS.
Fig. 2.
Gene-expression analysis of regions of the PS and CD4-Foxa2+GFP-Bry+ EB subpopulations. (A) An embryonic day 7.25 mouse GFP-Bry embryo expressing GFP in the PS. Boxes indicate the posterior (Pos), middle (Mid), and anterior (Ant) PS regions. (B) RT-PCR gene-expression analysis of the different regions of the PS. (C) The presort (Pre), CD4loGFP+ (CD4 Lo), CD4medGFP+ (CD4 Med), and CD4hiGFP+ (CD4 Hi) populations were isolated by cell sorting from day 3.25 serum-stimulated EBs. (D) RT-PCR gene-expression analysis of the CD4 Lo, CD4 Med, and CD4 Hi populations.
Developmental Potential of Different CD4-Foxa2-Expressing Populations.
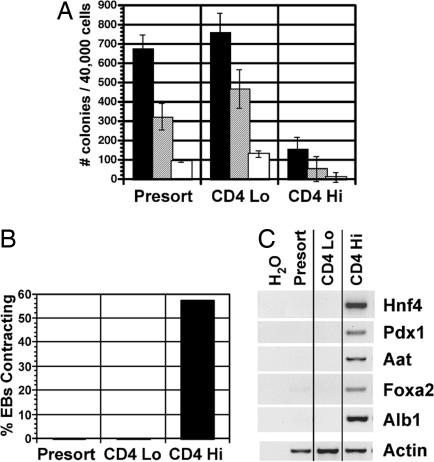
The posterior and anterior regions of the mouse PS display different developmental potentials. At the mid-PS stage, the posterior region gives rise to the yolk sac, whereas the anterior PS forms endoderm and anterior mesoderm, including the cardiac lineage (4, 24). To determine whether the different CD4-expressing populations display developmental potentials similar to the different regions of the PS, CD4lo/medGFP+, and CD4hiGFP+ populations isolated from day 3.25 EBs were assayed for hematopoietic, cardiac, and endoderm potential. As shown in Fig. 3A, the CD4loGFP+ population contained significantly more hematopoietic progenitors than the CD4hiGFP+ population. To assess cardiac potential, the development of contracting cells within aggregates was determined as described (8). Cardiac potential was restricted to the CD4hiGFP+ population (Fig. 3B). The unsorted population also failed to yield contracting aggregates, suggesting that there is a CD4 low or negative population that is inhibitory for cardiac differentiation. To assay endoderm development, the cardiac cultures were maintained under serum-free conditions for an additional 5 days, and endoderm-specific gene expression was analyzed. The expression of hnf4, aat, and alb1, genes associated with hepatocyte specification and pdx1, a marker of early pancreatic development (25, 26), was detected only in the populations generated from the CD4hiGFP+ aggregates, confirming the endoderm potential of these cells (Fig. 3C). Taken together, these findings add support to the interpretation that serum-induced CD4+GFP+ cells represent a population with potential similar to the PS of the embryo. Hereafter, the entire CD4+GFP+ population will be referred to as the ES cell PS population.
Fig. 3.
The functional potential of the CD4loGFP+ and CD4hiGFP+ populations. CD4loGFP+ and CD4hiGFP+ cells were isolated from serum-induced EBs as indicated in Fig. 2 and analyzed as described below. (A) Isolated populations were reaggregated for 24 h in serum-free media and then assayed for hematopoietic potential by colony assay. The number of primitive erythroid (filled bars), macrophage (hatched bars), and macrophage/erythroid (open bars) colonies were counted after 5–7 days of culture. Data are presented as mean number of colonies from three cultures. Bars represent SEM. (B) Isolated populations were reaggregated in serum-free media for 24 h, and the aggregates were allowed to adhere onto matrigel-coated dishes in serum-free media. The proportion of aggregates with contracting cells was scored 1–2 days later. (C) Aggregates were maintained for 7 days on matrigel-coated dishes in serum-free media, and gene expression was analyzed by RT-PCR.
Both Wnt and TGF-β Family Members Are Required for ES Cell PS Induction.
Studies in mice and other organisms have demonstrated the importance of the Wnt and nodal signaling pathways in gastrulation and in endoderm and mesoderm induction (20, 21, 27, 28). Expression of both Wnt3 and nodal was induced in the serum-stimulated EBs (Fig. 6C), suggesting that these factors also may be involved in the development of the ES cell PS population. To investigate this possibility, Wnt signaling was inhibited in the cultures by the addition of DKK1, and TGF-β/nodal/activin signaling was blocked by the addition of the small molecule inhibitor SB-431542 (SB). DKK1 functions by binding the Wnt coreceptors LRP5 and LRP6, which are essential for activation of the β-catenin signaling pathway (29). SB is a specific inhibitor of the receptors ALK4, ALK5, and ALK7 (30) and thus is able to block TGF-β, activin, and nodal but not bone morphogenic protein signaling. Addition of high concentrations of either SB (10 μM) or DKK1 (150 ng/ml) completely blocked the development of the serum-induced ES cell PS population as determined by GFP versus CD4 flow cytometric profile and PS gene expression (Fig. 8, which is published as supporting information on the PNAS web site). Lower concentrations of DKK1 (15 ng/ml) led to a modest increase in the proportion of CD4hiGFP+ cells and a concomitant decrease in the CD4loGFP+ population (Fig. 8A). In contrast, the addition of low concentrations of SB (2 μM) to the cultures blocked the development of the CD4hiGFP+ but not the CD4loGFP+ population (Fig. 8A). These data suggest that both Wnt and TGF-β pathways are required for ES cell PS development.
To determine whether activation of the Wnt and TGF-β/nodal/activin pathways is sufficient to induce the development of the ES cell PS population in the absence of other serum-derived factors, ES cells were differentiated either in the presence of Wnt3a or activin A in serum-free cultures. Activin was used as a surrogate of nodal as it can signal through the same receptor as nodal but is not affected by Lefty1, an inhibitor of nodal (31) that is expressed in the developing EBs (data not shown). We previously have demonstrated that activin can induce both mesoderm and endoderm in serum-free cultures (9). Here, we use an improved serum-free media that supports efficient EB formation and activin-induced CD4-Foxa2 and GFP-Bry expression (data not shown). For all studies detailed below, EB formation was initiated in serum-free media in the absence of factors. At day 2, the various inducers or inhibitors of differentiation were added as described below.
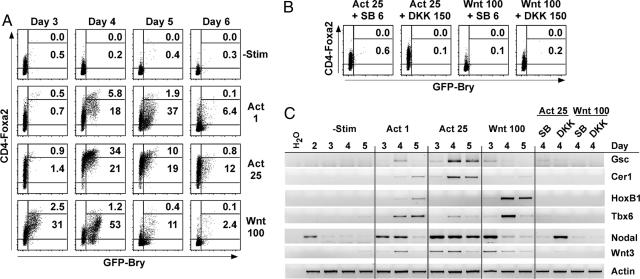
Both activin (25 ng/ml) and Wnt (100 ng/ml) induced the formation of the CD4-Foxa2+GFP-Bry+ population, whereas no significant levels of GFP developed in the absence of factors (Fig. 4A). This concentration of activin stimulation resulted in the generation of a population of cells that expressed GFP and high levels of CD4. In contrast, the GFP+ population induced with Wnt displayed lower levels of CD4. Activin induced the expression of the anterior PS markers gsc and cer1 (Fig. 4C), whereas Wnt was a poor inducer of these markers but did induce expression of the posterior PS markers hoxB1 and tbx6. These findings indicate that activin (25 ng/ml) induced predominantly the ES cell anterior PS population, and Wnt induced the development of the posterior PS.
Fig. 4.
Activin and Wnt can induce the development of PS cells in the absence of serum. ES cells were differentiated in serum-free media alone for 2 days. The EBs were dispersed and reaggregated in serum-free media with activin A (Act; in ng/ml), Wnt3a (Wnt; in ng/ml), SB-431542 (SB; in μM), and DKK1 (DKK; in ng/ml) as indicated. The percentages of CD4hiGFP+ and CD4lo/medGFP+ are indicated on the plots. (A) Kinetics of CD4 and GFP expression in cultures induced with activin or Wnt. (B) Expression of CD4 and GFP in day 4 EB cultures after the indicated treatments. (C) Cells were harvested at the indicated times, and gene-expression analysis was performed by RT-PCR.
To further investigate the role of the strength of TGF-β/nodal/activin signaling, ES cells were stimulated with a lower dose (1 ng/ml) of activin (Fig. 4A). This concentration of activin induced a CD4-Foxa2+GFP-Bry+ population that expressed relatively low levels of CD4. As expected from the CD4 expression pattern, 1 ng/ml of activin induced the posterior markers hoxB1 and tbx6 (Fig. 4C). These data confirm the inhibition studies shown in Fig. 8 and suggest that a strong TGF-β/nodal/activin signal favors an anterior fate, whereas a weaker signal induces the development of a more posterior population.
To confirm the requirement of Wnt and activin/nodal signaling in PS formation from ES cells, downstream effectors of these pathways were inhibited by using RNA interference (32). To examine the requirement for Wnt signaling, a short hairpin RNA (shRNA) against β-catenin was introduced into day 2 EBs cells before addition of low levels of activin and Wnt. Introduction of this hairpin completely blocked GFP-Bry induction (Fig. 9A, which is published as supporting information on the PNAS web site). To block activin/nodal signaling, shRNAs against smad2 and smad3 were used. Introduction of these hairpins individually led to a decrease in GFP-Bry levels, whereas the combination of both hairpins blocked the expression of GFP-Bry. A titration of Smad2 shRNA was used to attenuate activin/nodal signaling in EBs stimulated with higher levels of activin (Fig. 9B). As more shRNA was added to the cultures, there was a progressive loss of CD4-Foxa2hi cells and a general decrease in CD4 levels as indicated by MFI. These data confirm that Wnt and activin/nodal signaling are required for ES cell PS development and that decreasing levels of activin signaling lead to a preferential decrease in anterior PS cells.
Although mutations in either wnt3 or nodal completely block gastrulation and PS induction in vivo, the exact role for each is unclear, considering that wnt3 mutants lack nodal expression (20). In the serum-free ES cell differentiation system, Wnt induced nodal and activin induced wnt3 (Fig. 4C), suggesting that both signaling pathways are involved in the development of the CD4-Foxa2GFP-Bry-expressing population. To further investigate the role of the Wnt and TGF-β/nodal/activin pathways in the induction of the ES cell PS, either activin or Wnt were added to the differentiation cultures with either SB or DKK1 (Fig. 4B). Both inhibitors completely blocked the development of CD4-Foxa2+GFP-Bry+ cells with either inducer. None of the other PS markers (gsc, cer1, hoxB1, and tbx6) were expressed in cells treated with either of the inhibitors (Fig. 4C). To determine whether activin acted earlier than Wnt in the induction of the CD4-Foxa2+GFP-Bry+ population, the day 2 EBs were incubated with activin and DKK1 for 20 h, washed, and then incubated with Wnt and SB. No CD4-Foxa2+GFP-Bry+ cells developed after the sequential activation of these pathways (Fig. 10, which is published as supporting information on the PNAS web site). Addition of the factors/inhibitors in the reverse order, to determine whether Wnt functioned earlier than activin, also failed to induce the CD4-Foxa2+GFP-Bry+ population. These data demonstrate the simultaneous requirement of the Wnt and TGF-β/nodal/activin pathways for PS formation.
To assess the functional potential of the populations induced in serum-free cultures, they were evaluated for hematopoietic progenitor content, mesodermal gene expression, and the ability to generate cells that express endoderm markers. Populations induced with low and high concentrations of activin (1 or 25 ng/ml) or Wnt (10 and 100 ng/ml) as well as those induced with the combination of low concentrations of factors were assayed for hematopoietic potential. For the hematopoietic progenitor assays, 4-day-old EBs induced under the above conditions were dispersed and reaggregated overnight in serum-containing media. Exposure to serum was necessary to obtain hematopoietic colonies, suggesting that factors present in serum were required to specify mesoderm to a hematopoietic fate (9). Populations induced with high concentrations of Wnt or the combination of low levels of both factors displayed the greatest hematopoietic potential (Fig. 11A, which is published as supporting information on the PNAS web site). Wnt or low levels of activin also induced the expression of the somatic and lateral plate mesoderm markers meox1, meox2, foxc1, and foxc2, indicating the development of different types of mesoderm (Fig. 11B) (33–35). To evaluate endoderm potential, day 6 EBs treated as in Fig. 4A were plated onto matrigel for 1 week and then assayed for expression of genes indicative of liver development. In confirmation with our previous findings, endoderm potential was detected only in the population induced with high concentrations of activin (data not shown and ref. 9). Wnt stimulation was unable to induce endoderm gene expression, a finding consistent with the lack of anterior PS markers induced by this factor (data not shown and Fig. 4 A and C). The findings from these functional studies demonstrate that different signaling pathways are required for the induction of different ES cell PS populations.
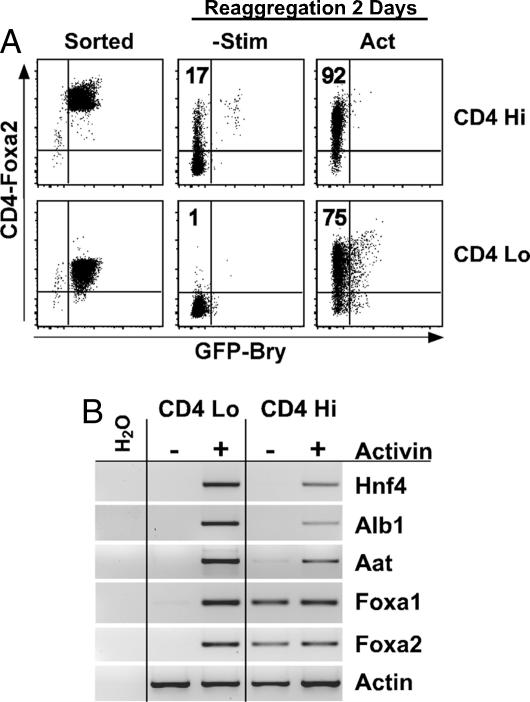
Activin Stimulation Is Required for Endoderm Development from the PS.
The kinetics of CD4-Foxa2 and GFP-Bry expression shown in Fig. 4A suggest a developmental progression from CD4hi GFP-Bry+ anterior PS population to CD4hi GFP-Bryneg definitive endoderm. To formally demonstrate this transition, CD4hiGFP+ cells isolated from day 4 Wnt/activin-induced EBs were reaggregated in the presence or absence of activin (10 ng/ml) for 2 days (Fig. 5A). Aggregates were plated onto matrigel for 5 days and analyzed for endoderm gene expression. The majority of cells aggregated in the presence of activin displayed a CD4-Foxa2+GFP-Bryneg phenotype (Fig. 5A) and expressed genes indicative of endoderm development after culture on matrigel (Fig. 5B). In contrast, most of the population cultured in the absence of activin lost CD4 expression and generated cells that expressed foxa1 and foxa2, but other genes indicative of early hepatocyte differentiation, including alb1, aat, or hnf4, were not found (Fig. 5B). These observations indicate that TGF-β/nodal/activin signaling is important for the specification or maturation of definitive endoderm from the ES cell anterior PS population. To determine whether this activity of activin also could induce endoderm from the posterior PS population, Wnt/activin-induced day 4 CD4loGFP+ cells were reaggregated in the presence or absence of activin (10 ng/ml). Only in the presence of activin did the CD4loGFP+ cells give rise to a CD4hiGFPneg population that matured into cells that expressed genes indicative of endoderm specification (Fig. 5 A and B). These findings demonstrate that the CD4loGFP+ cells induced in serum-free conditions are not yet committed to the mesoderm lineage and can switch developmental fates after high levels of activin stimulation.
Fig. 5.
Activin induces endoderm formation from CD4+GFP+ cells. ES cells were differentiated in serum-free cultures in the presence of activin A (2 ng/ml) and Wnt3a (10 ng/ml). At day 4, the CD4loGFP+ and CD4hiGFP+ cells were isolated and reaggregated in serum-free media alone or with activin (10 ng/ml) as indicated. (A) Expression of GFP and CD4 are shown for sorted cells and 2 days after aggregation. (B) After 2 days of aggregation, aggregates were plated onto dishes. After 5–7 days of culture, gene expression was analyzed by RT-PCR.
Discussion
Through the development and use of the CD4-Foxa2/GFP-Bry ES cell line, we were able to identify a CD4-Foxa2+GFP-Bry+ EB population that shares a number of similarities with the PS of the mouse embryo. Of particular significance was the observation that Wnt and activin/nodal signaling pathways, which are known to be required for PS formation in the early embryo (20, 21), also were required for development of the CD4-Foxa2+GFP-Bry+ population. A complete block in Wnt signaling in the early embryo prevents PS formation, whereas a reduction in signaling in mice expressing decreased levels of the Wnt coreceptors LPR5 and LPR6 results in an expansion of the anterior PS population with a concomitant decrease in the size of the posterior PS (36). This observation suggests that one of the earliest functions of Wnt signaling is to establish posterior PS populations. Our finding that partial Wnt inhibition in the serum-induced cultures resulted in an increase in the size of the CD4hiGFP+ population and a decrease in the size of the CD4loGFP+ population suggests that Wnt functions in a similar fashion in vitro. A specific role for Wnt signaling in the development of this population is provided by our serum-free experiments that demonstrated that Wnt3a preferentially induced a posterior PS population.
Nodal is a well established regulator of gastrulation and germ-layer formation in a number of different species. Mutations of the nodal homologues Squint and Cyclops in zebrafish display a complete loss of endoderm and head and trunk mesoderm (28). In Xenopus, the nodal related Xnr-1 and Xnr-2 genes can induce mesoderm in animal cap explant assays in a dose-dependent manner (37). In addition to the observations that nodal-deficient mice fail to form a PS, more recent studies have shown that mice expressing a hypomorphic allele of nodal with decreased signaling have a truncation of the anterior mesendoderm (38). These findings suggest that different populations of the PS require different levels of nodal signaling, with the anterior region requiring the highest. The inhibition in development of the CD4-Foxa2hiGFP-Bry+ population in the presence of low levels of the TGF-β/nodal/activin inhibitor or by the Smad2 shRNA suggests a similar requirement of high levels of signaling for its formation in vitro. The most direct evidence for differential signaling requirements for the induction of cell types is provided by our serum-free studies, showing that high amounts of activin induce the anterior PS population, whereas low concentrations lead to the development of a population with posterior PS characteristics.
In addition to demonstrating that both Wnt and activin can induce the CD4-Foxa2+GFP-Bry+ population, our serum-free studies have shown that the Wnt and TGF-β/nodal/activin pathways are required simultaneously at the onset of differentiation. Inhibitors of each pathway blocked the induction by factors of the other pathway. Together, these observations indicate that the generation of the PS and its derivatives requires an interplay between these signaling pathways (Fig. 12, which is published as supporting information on the PNAS web site). Although both pathways are required for the initiation of the development of CD4-Foxa2+GFP-Bry+ cells, they ultimately induce different populations. Wnt preferentially induces a posterior PS population, whereas activin induces either anterior or posterior populations, depending on the concentration of factor used. Collectively, these observations highlight the importance of understanding the signaling requirements of different lineages and of using appropriate factors for the generation of specific cell types in ES cell differentiation cultures.
Materials and Methods
Generation of CD4-Foxa2 Targeted ES Cells.
The targeting vector was constructed by modifying a vector previously used to disrupt the foxa2 locus (13) (see Supporting Text, which is published as supporting information on the PNAS web site). The mouse ES cell line GFP-Bry (7) was electroporated with the targeting vector, and clones that had undergone homologous recombination were identified by Southern blot analysis.
ES Cell Maintenance.
ES cells were maintained either in serum containing media with feeder cells (7) or in a modified serum-free/feeder-free culture system (39). The serum-free culture media consisted of 50% Neurobasal media (Invitrogen, Carlsbad, CA) and 50% DMEM/F12 media mixture (Invitrogen) supplemented with 0.5× of both N2 and B27 supplements (Invitrogen), penicillin, streptomycin, 0.05% BSA, leukemia inhibitory factor (1% conditioned medium), human bone morphogenic protein 4 (10 ng/ml; R & D Systems, Minneapolis, MN), and 1.5 × 10−4 M 1-thioglycerol (Sigma, St. Louis, MO).
ES Cell Differentiation.
ES cells were differentiated in either serum-containing media (9) or serum-free media as described below. ES cells were trypsinized and cultured at 1.5 × 105 cells per ml in serum-free differentiation media that consisted of 75% Iscove's modified Dulbecco's medium (Invitrogen) and 25% Ham's F12 media (Invitrogen) supplemented with 0.5× of both N2 and B27 (without retinoic acid) supplements (Invitrogen), penicillin, streptomycin, 0.05% BSA, 2 mM glutamine (Invitrogen), 0.5 mM ascorbic acid (Sigma) and 4.5 × 10−4 M 1-thioglycerol. After 48 h, the EBs were dissociated with trypsin and reaggregated in serum-free differentiation media with the addition of human activin A, mouse Wnt3a, human DKK1 (R & D Systems), or SB (Tocris, Ellisville, MO) as indicated. All data shown are representative examples of at least three independent experiments unless indicated otherwise.
Hematopoietic, Cardiac, and Endoderm Assays.
For the hematopoietic progenitor assay, EBs induced in various conditions were dissociated by trypsin into a single-cell suspension and plated in methylcellulose containing hematopoietic growth factors (9). For the cardiac assay, cells were reaggregated in Iscove's modified Dulbecco's medium with 15% Knockout Serum Replacement (Invitrogen), penicillin, streptomycin, 2 mM glutamine, 0.5 mM ascorbic acid, and 4.5 × 10−4 M 1-thioglycerol at 1–4 × 105 cells per ml in low-cluster tissue-culture dishes (Costar; Corning, Corning, NY). After 48 h, the aggregates were allowed to adhere onto matrigel-coated (Becton Dickinson, Franklin Lakes, NJ) dishes in StemPro 34 (Invitrogen) supplemented with 2 mM glutamine. After an additional 24–48 h, the proportion of EBs containing contracting cells was scored. These cultures also were maintained for 5–7 days, at which time RNA was harvested and analyzed for endoderm-specific gene expression. For the serum-free induced cultures, sorted cells were allowed to reaggregate in serum-free differentiation media as detailed in the results. After 48 h, the EBs were plated onto matrigel-coated tissue-culture dishes in Iscove's modified Dulbecco's medium with 15% FBS supplemented with 2 mM glutamine and dexamethasone 10−7 M. After 5–7 days, the cultures were harvested, and the RNA was isolated.
Embryo Dissections and Explant Cultures.
Male GFP-Bry mice on a mixed genetic background were mated to female Swiss–Webster mice (Taconic, Hudson, NY). Pregnant mice were killed 7.5 days after mating, and the embryos were isolated. Dissections were performed under a fluorescence-equipped dissection microscope (Leica, Bannockburn, IL) to visualize GFP expression. The endoderm was stripped from the embryo by mechanical manipulation with tungsten needles. The PS was isolated and sectioned into posterior, middle, and anterior regions as indicated, and gene expression was analyzed (three to four pieces per group).
Gene-Expression Analysis.
For all ES cell-derived cultures, total RNA was harvested with the RNeasy minikit and treated with RNase-free DNase (Qiagen, Valencia, CA). Then, 0.5–1 μg of RNA was reverse-transcribed into cDNA by using random hexamers and SuperScript II reverse transcriptase kit (Invitrogen). For the dissected PS pieces, total RNA was harvested with the RNA-nano kit with RNase-free DNase (Stratagene, La Jolla, CA), and reverse transcription was performed with random hexamers by using the Sensiscript reverse transcriptase kit (Qiagen). The oligonucleotide sequences for actb (actin), aat, foxa2, hnf4, alb1, brachyury (Bry), pax6, and ipf1 (Pdx1) have been reported previously (9). Other gene-specific oligonucleotides are listed in the Supporting Text.
Flow Cytometry and Cell Sorting.
EBs generated from the CD4-Foxa2/GFP-Bry ES cell line were dissociated by incubation for 1–2 min with trypsin (0.25%)/EDTA (Invitrogen) and stained with anti-human CD4-phycoerythrin or -allophycocyanin (Caltag, Carlsbad, CA). The cells were acquired by using an LSR II or FacsCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) or sorted on a MoFlo cell sorter (Cytomation Systems, Fort Collins, CO), and analysis including MFI calculation was performed with FlowJo software (Tree Star, Ashland, OR).
Supplementary Material
Acknowledgments
We thank Daniel Weinstein (Mount Sinai School of Medicine) for supplying the Foxa2 genomic sequence used to make the targeting vector, and Troy Moore (Open Biosystems, Huntsville, AL) for supplying the shRNAs.
Abbreviations
- PS
primitive streak
- EB
embryoid body
- MFI
mean fluorescence intensity
- SB
SB-431542
- shRNA
short hairpin RNA
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Tam PP, Behringer RR. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- 2.Lawson KA, Meneses JJ, Pedersen RA. Development (Cambridge, UK) 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 3.Lawson KA, Pedersen RA. Ciba Found Symp. 1992;165:3–21. doi: 10.1002/9780470514221.ch2. discussion 21–26. [DOI] [PubMed]
- 4.Tam PP, Beddington RS. Ciba Found Symp. 1992;165:27–41. doi: 10.1002/9780470514221.ch3. discussion 42–49. [DOI] [PubMed]
- 5.Kinder SJ, Tsang TE, Wakamiya M, Sasaki H, Behringer RR, Nagy A, Tam PP. Development (Cambridge, UK) 2001;128:3623–3634. doi: 10.1242/dev.128.18.3623. [DOI] [PubMed] [Google Scholar]
- 6.Kispert A, Herrmann BG. Dev Biol. 1994;161:179–193. doi: 10.1006/dbio.1994.1019. [DOI] [PubMed] [Google Scholar]
- 7.Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Development (Cambridge, UK) 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 8.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Proc Natl Acad Sci USA. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G. Development (Cambridge, UK) 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 10.Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa S. Development (Cambridge, UK) 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 11.Yasunaga M, Tada S, Torikai-Nishikawa S, Nakano Y, Okada M, Jakt LM, Nishikawa S, Chiba T, Era T, Nishikawa S. Nat Biotechnol. 2005;23:1542–1550. doi: 10.1038/nbt1167. [DOI] [PubMed] [Google Scholar]
- 12.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 13.Weinstein DC, Ruiz i, Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 14.Monaghan AP, Kaestner KH, Grau E, Schutz G. Development (Cambridge, UK) 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 15.Kaestner KH, Hiemisch H, Luckow B, Schutz G. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- 16.Bedinger P, Moriarty A, von Borstel RC, II, Donovan NJ, Steimer KS, Littman DR. Nature. 1988;334:162–165. doi: 10.1038/334162a0. [DOI] [PubMed] [Google Scholar]
- 17.Tam PP, Rossant J. Development (Cambridge, UK) 2003;130:6155–6163. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- 18.Forlani S, Lawson KA, Deschamps J. Development (Cambridge, UK) 2003;130:3807–3819. doi: 10.1242/dev.00573. [DOI] [PubMed] [Google Scholar]
- 19.Dush MK, Martin GR. Dev Biol. 1992;151:273–287. doi: 10.1016/0012-1606(92)90232-6. [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Nat Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 21.Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ. Development (Cambridge, UK) 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 22.Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 23.Chapman DL, Agulnik I, Hancock S, Silver LM, Papaioannou VE. Dev Biol. 1996;180:534–542. doi: 10.1006/dbio.1996.0326. [DOI] [PubMed] [Google Scholar]
- 24.Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP. Development (Cambridge, UK) 1999;126:4691–4701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- 25.Zaret KS. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 26.Kim SK, MacDonald RJ. Curr Opin Genet Dev. 2002;12:540–547. doi: 10.1016/s0959-437x(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 27.Slack JM. Curr Biol. 1994;4:116–126. doi: 10.1016/s0960-9822(94)00027-8. [DOI] [PubMed] [Google Scholar]
- 28.Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- 29.Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 30.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Shen MM. Curr Biol. 2004;14:618–624. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- 32.Schaniel C, Li F, Schafer XL, Moore T, Lemischka IR, Paddison PJ. Nat Methods. 2006;3:397–400. doi: 10.1038/nmeth0506-397. [DOI] [PubMed] [Google Scholar]
- 33.Candia AF, Hu J, Crosby J, Lalley PA, Noden D, Nadeau JH, Wright CV. Development (Cambridge, UK) 1992;116:1123–1136. doi: 10.1242/dev.116.4.1123. [DOI] [PubMed] [Google Scholar]
- 34.Kaestner KH, Bleckmann SC, Monaghan AP, Schlondorff J, Mincheva A, Lichter P, Schutz G. Development (Cambridge, UK) 1996;122:1751–1758. doi: 10.1242/dev.122.6.1751. [DOI] [PubMed] [Google Scholar]
- 35.Kume T, Jiang H, Topczewska JM, Hogan BL. Genes Dev. 2001;15:2470–2482. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly OG, Pinson KI, Skarnes WC. Development (Cambridge, UK) 2004;131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 37.Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV. Development (Cambridge, UK) 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- 38.Lowe LA, Yamada S, Kuehn MR. Development (Cambridge, UK) 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- 39.Ying QL, Nichols J, Chambers I, Smith A. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.